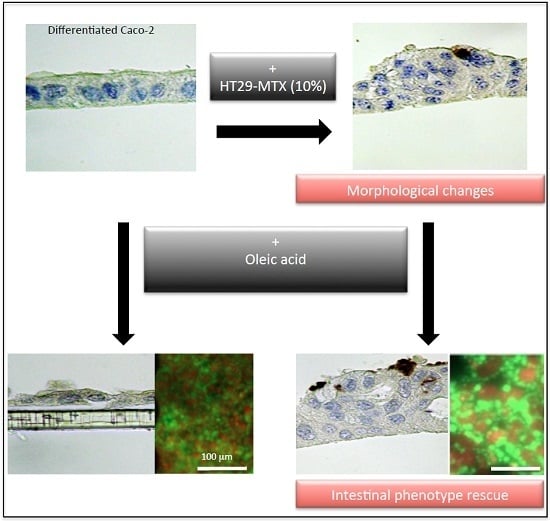
Oleic Acid Uptake Reveals the Rescued Enterocyte Phenotype of Colon Cancer Caco-2 by HT29-MTX Cells in Co-Culture Mode
Abstract
:1. Introduction
2. Results
2.1. Bio-Informatic Dataset Analyses Point out the Role of Fatty Acids in Both Cancer and Intestinal Phenotypes
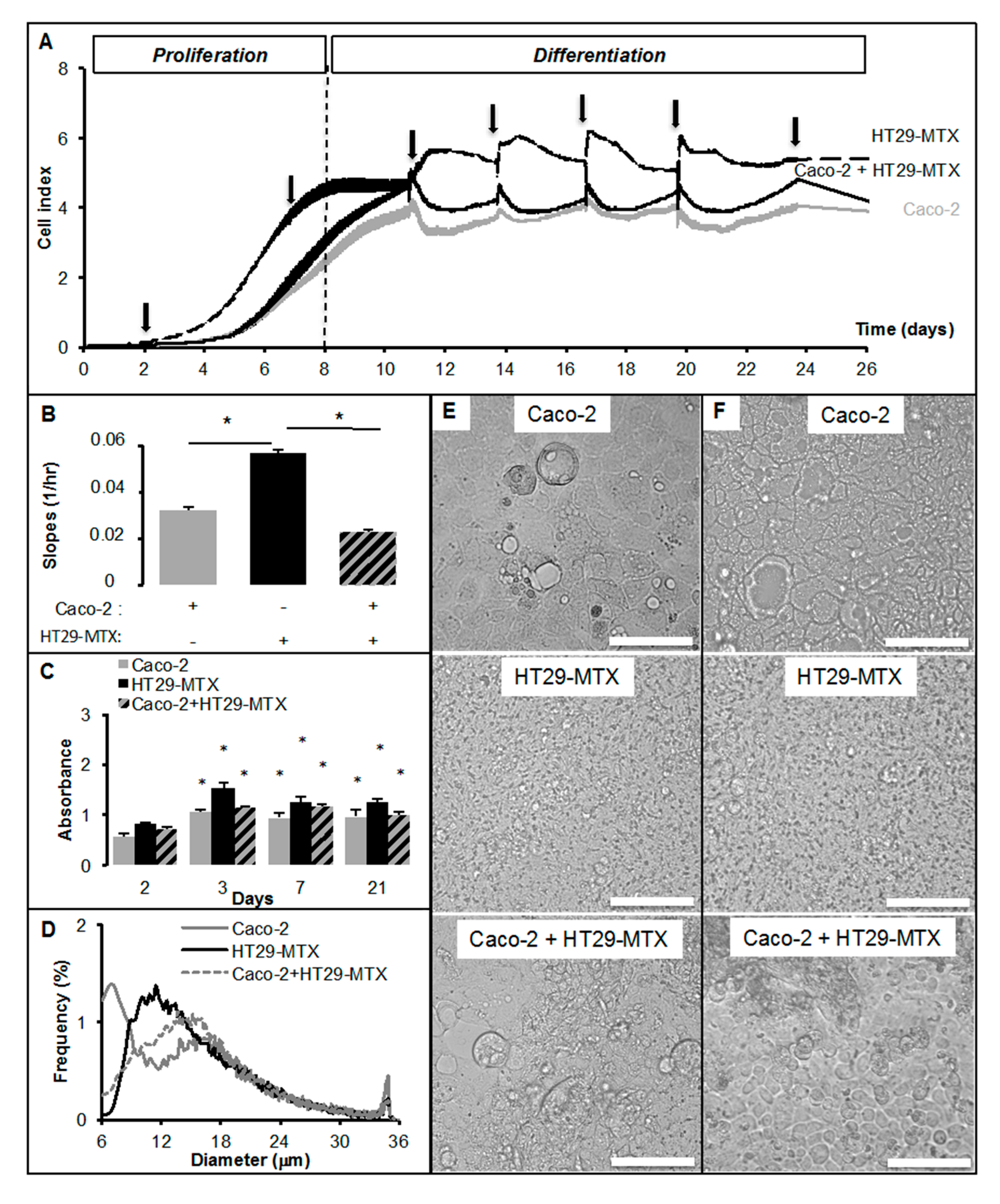
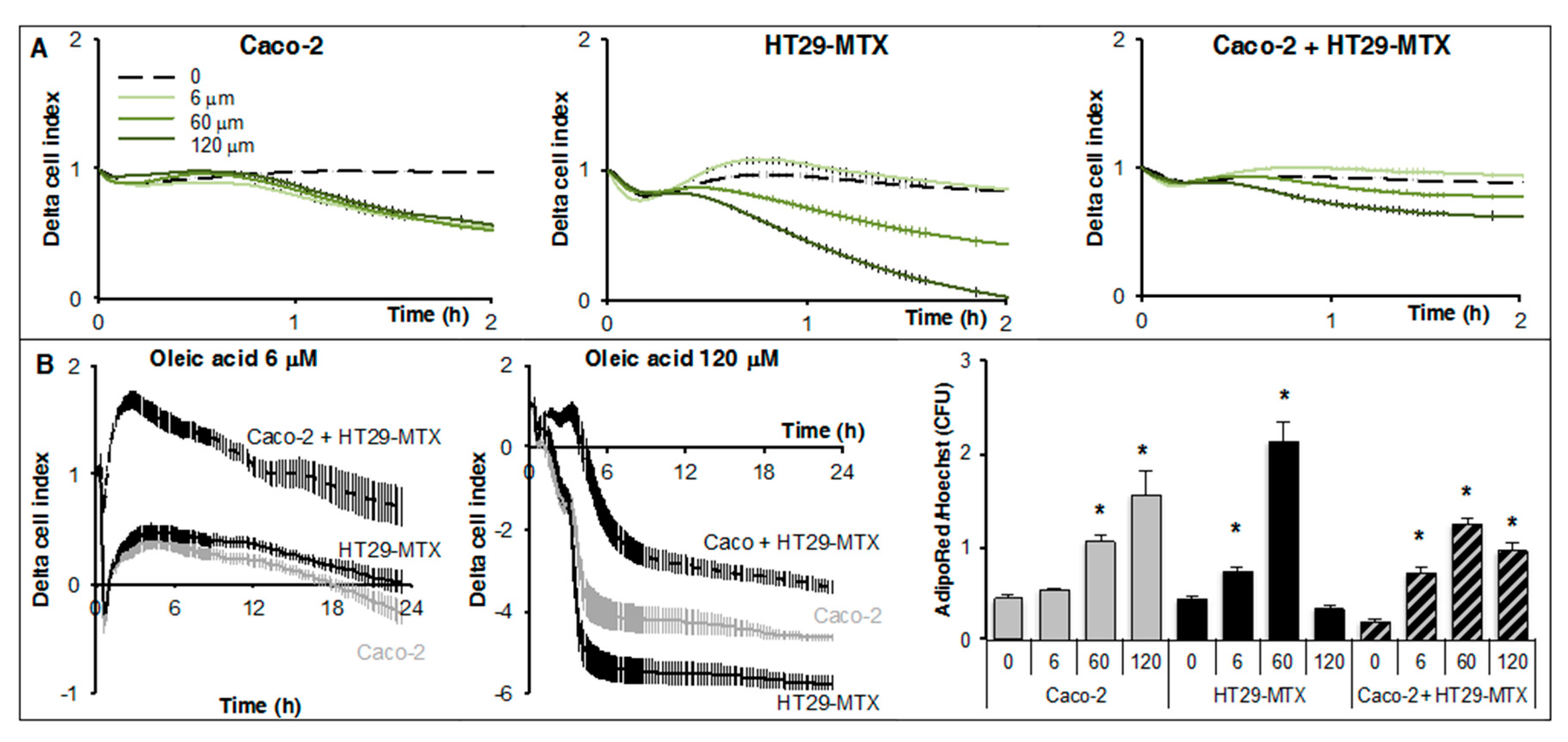
2.2. Real-Time Analysis Reveals the Modulatory Activity of HT29-MTX Cells on Caco-2 Cell Growth and Differentiation
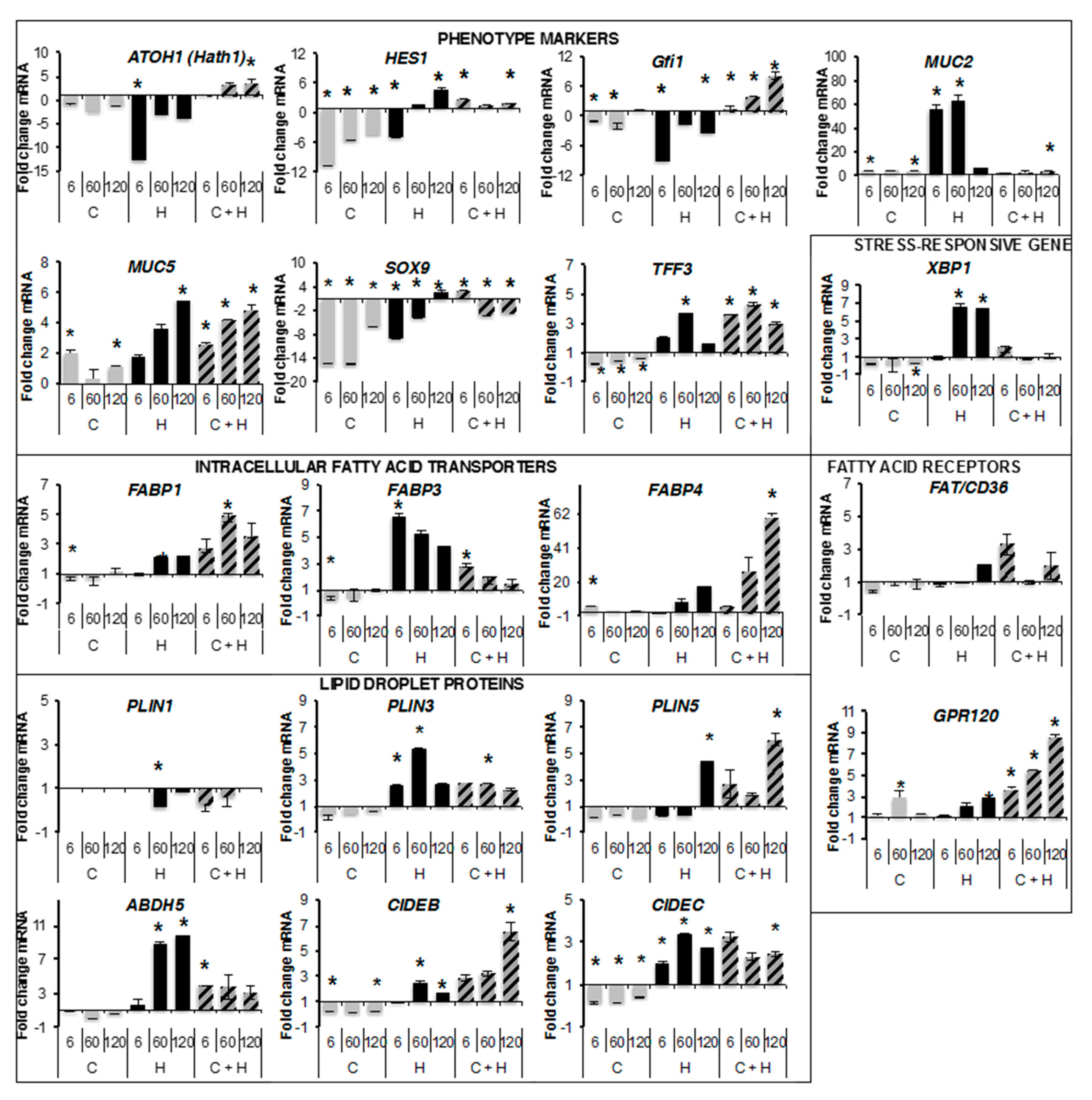
2.3. Quantitative RT-PCR Analyses
2.4. Real-Time Uptake of Oleic Acid by Caco-2, HT29-MTX Cells, and Co-Cultures
2.5. Transcriptional Regulations by Oleic Acid on Caco-2, HT29-MTX Cells and Co-Cultures
3. Discussion
4. Materials and Methods
4.1. Bio-Informatic Dataset Enrichment Analyses
4.2. Cell Culture
4.3. Real-Time Cell Analysis
4.4. Cell Survival and Size Analysis
4.5. Treatments
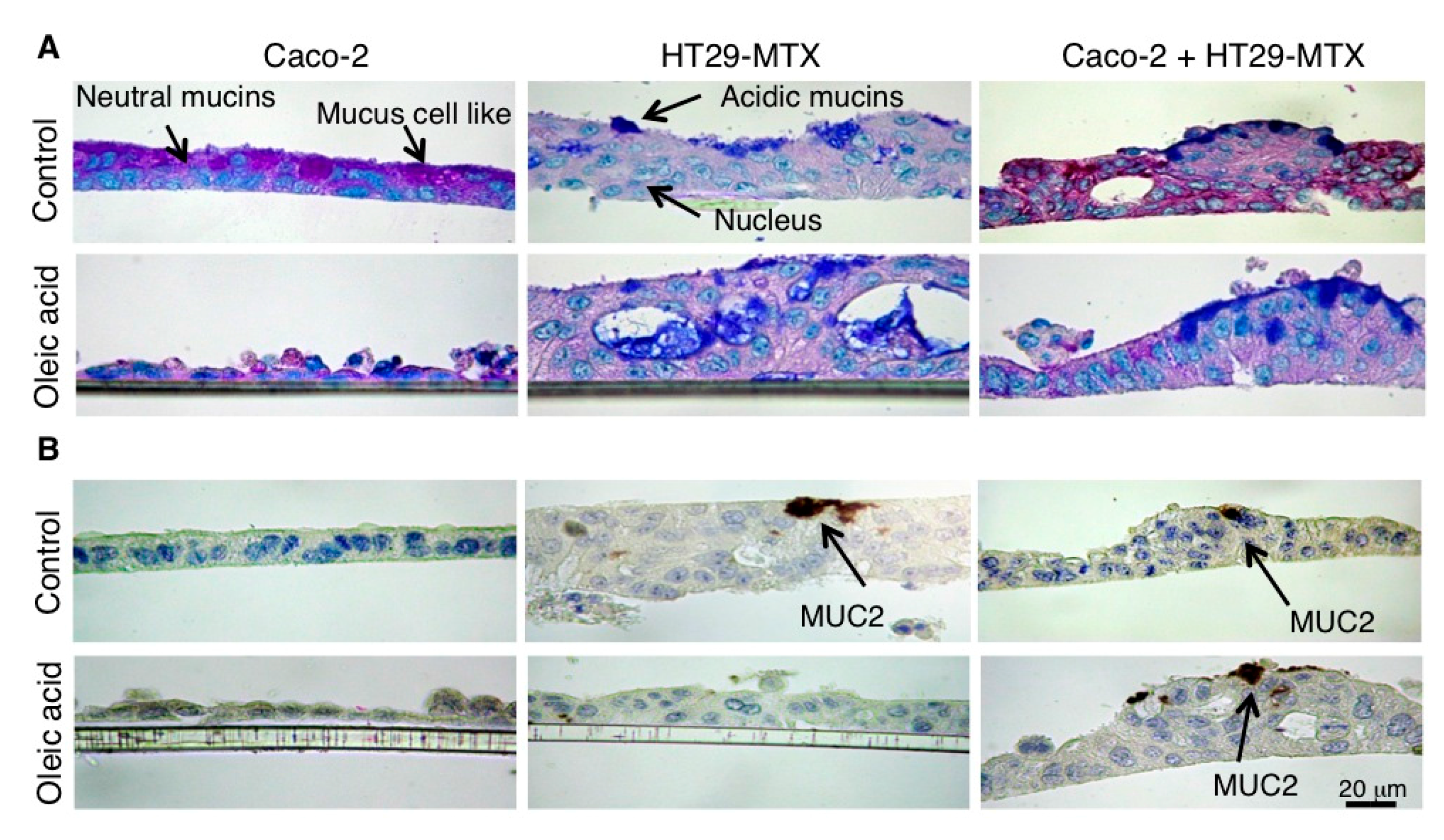
4.6. Histochemistry and Immunochemistry
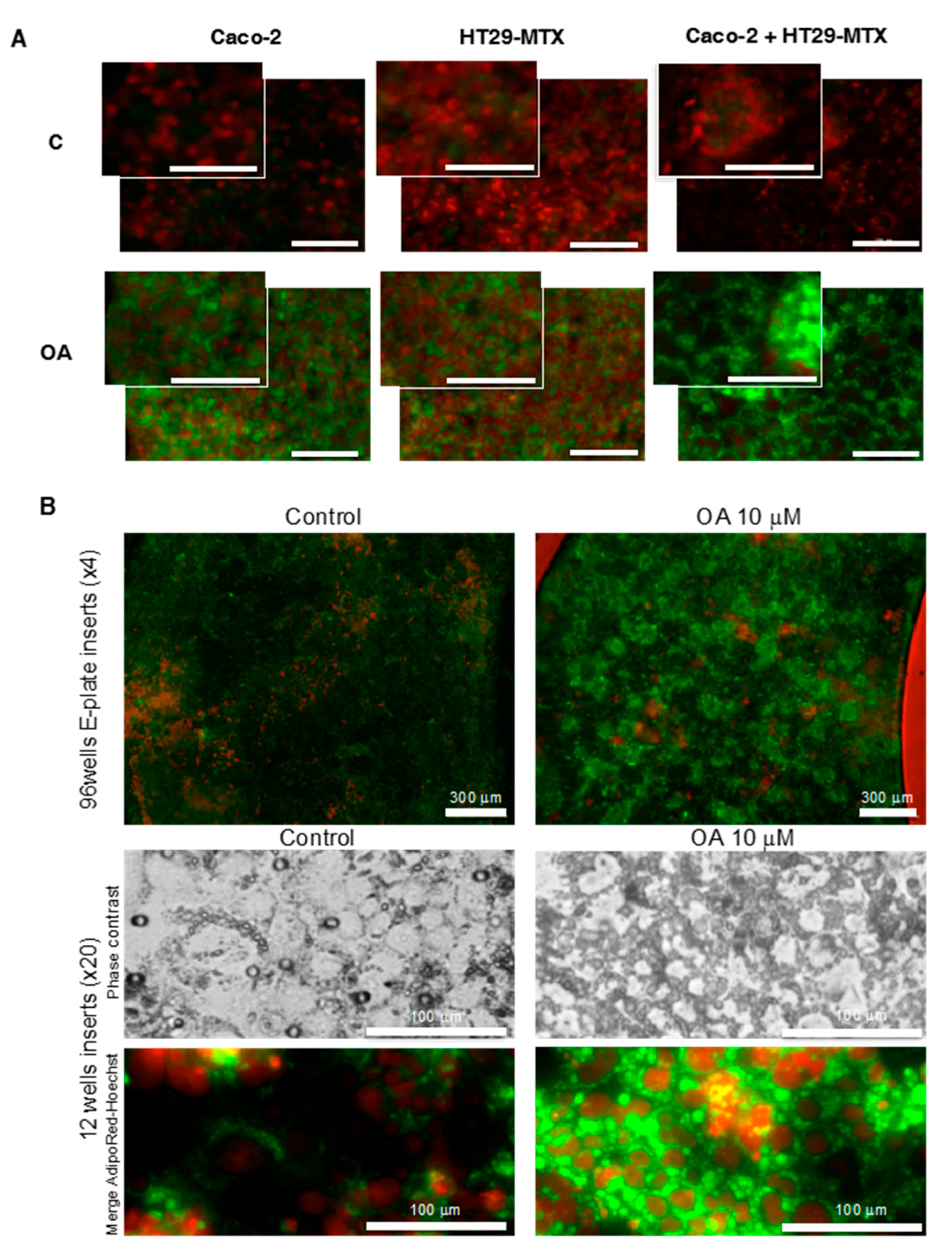
4.7. Fluorescent Cell Lipid Storage Analysis
4.8. qRT-PCR Analysis of Gene Expression
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AB | Alcian Blue |
| ABDH5 | α/β-hydrolase domain-containing protein 5 |
| AMPK | Protein kinase AMP-activated |
| APOA2 | Apoprotein 2 |
| APOA3 | Apoprotein 3 |
| ATOH1 (Hath1) | Helix-loop-helix (HLH) protein |
| BMP4 | Bone Morphogenic Protein 4 |
| BSA | Bovine serum albumin |
| CIDEB and CIDEC | Cell-death activators CIDE-B and C |
| DGAT2 | Diacylglycerol transferase 2 |
| EGF | Epidermal growth Factor |
| FABPs | Fatty acid binding proteins |
| FASN | Fatty acid synthase |
| FAT/CD36 | Fatty acid translocase |
| Gfi1 | Growth factor independence 1 |
| GPR120 | G-protein coupled receptor 120 |
| HES1 | Hairy and enhancer of split-1 |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 |
| Klf4 | Kruppel-like factor 4 |
| LDLR | Low density lipid receptor |
| LIPE | Lipid protein lipase |
| MUC2 | Mucin 2 |
| MUC5AC | Mucin 5AC |
| OA | Oleic acid |
| PAS | Periodic acid-Schiff reaction |
| PLIN1, 3, 5 | Perilipins 1, 3 and 5 |
| PPARs | Peroxisome proliferator activated receptors (alpha, beta/delta and gamma) |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| RTCA | Real-time Cell Analyzer |
| SOX9 | SRY-Box 9 |
| SPDEF | SAM Pointed Domain Containing ETS Transcription Factor |
| TFF3 | Trefoil fator 3 |
| TSPAN8 | Tetraspanin 8 |
| XBP1 | X-box binding protein 1 |
References
- Artursson, P.; Palm, K.; Luthman, K. Caco-2 monolayers in experimental and theoretical predictions of drug transport. Adv. Drug Deliv. Rev. 2001, 46, 27–43. [Google Scholar] [CrossRef]
- Sarmento, B.; Andrade, F.; da Silva, S.B.; Rodrigues, F.; das Neves, J.; Ferreira, D. Cell-based in vitro models for predicting drug permeability. Expert Opin. Drug Metab. Toxicol. 2012, 8, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Scaldaferri, F.; Pizzoferrato, M.; Gerardi, V.; Lopetuso, L.; Gasbarrini, A. The gut barrier: New acquisitions and therapeutic approaches. J. Clin. Gastroenterol. 2012, 46 S12–S17. [Google Scholar] [CrossRef] [PubMed]
- Lenaerts, K.; Bouwman, F.G.; Lamers, W.H.; Renes, J.; Mariman, E.C. Comparative proteomic analysis of cell lines and scrapings of the human intestinal epithelium. BMC Genom. 2007, 8. [Google Scholar] [CrossRef] [PubMed]
- Lesuffleur, T.; Barbat, A.; Dussaulx, E.; Zweibaum, A. Growth adaptation to methotrexate of HT-29 human colon carcinoma cells is associated with their ability to differentiate into columnar absorptive and mucus-secreting cells. Cancer Res. 1990, 50, 6334–6343. [Google Scholar] [PubMed]
- Larhed, A.W.; Artursson, P. Co-cultures of human intestinal goblet (HT29-H) and absorptive (Caco-2) cells for studies of drug and peptide absorption. Eur. J. Pharm. Sci. 1995, 3, 171–183. [Google Scholar] [CrossRef]
- Beduneau, A.; Tempesta, C.; Fimbel, S.; Pellequer, Y.; Jannin, V.; Demarne, F.; Lamprecht, A. A tunable Caco-2/HT29-MTX co-culture model mimicking variable permeabilities of the human intestine obtained by an original seeding procedure. Eur. J. Pharm. Biopharm. 2014, 87, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Hilgendorf, C.; Spahn-Langguth, H.; Regårdh, C.G.; Lipka, E.; Amidon, G.L.; Langguth, P. Caco-2 versus Caco-2/HT29-MTX co-cultured cell lines: Permeabilities via diffusion, inside- and outside-directed carrier-mediated transport. J. Pharm. Sci. 2000, 89, 63–75. [Google Scholar] [CrossRef]
- Pontier, C.; Pachot, J.; Botham, R.; Lenfant, B.; Arnaud, P. HT29-MTX and Caco-2/TC7 monolayers as predictive models for human intestinal absorption: Role of the mucus layer. J. Pharm. Sci. 2001, 90, 1608–1619. [Google Scholar] [CrossRef] [PubMed]
- Mahler, G.J.; Shuler, M.L.; Glahn, R.P. Characterization of Caco-2 and HT29-MTX cocultures in an in vitro digestion/cell culture model used to predict iron bioavailability. J. Nutr. Biochem. 2009, 20, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Carr, D.A.; Peppas, N.A. Assessment of poly (methacrylic acid-co-N-vinyl pyrrolidone) as a carrier for the oral delivery of therapeutic proteins using Caco-2 and HT29-MTX cell lines. J. Biomed. Mater. Res. A 2010, 92, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Guri, A.; Gülseren, I.; Corredig, M. Utilization of solid lipid nanoparticles for enhanced delivery of curcumin in cocultures of HT29-MTX and Caco-2 cells. Food Funct. 2013, 4, 1410–1419. [Google Scholar] [CrossRef] [PubMed]
- Berger, E.; Rome, S.; Vega, N.; Ciancia, C.; Vidal, H. Transcriptome profiling in response to adiponectin in human cancer-derived cells. Physiol. Genom. 2010, 42A, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Berger, E.; Vega, N.; Vidal, H.; Géloën, A. Gene network analysis leads to functional validation of pathways linked to cancer cell growth and survival. Biotechnol. J. 2012, 7, 1395–1404. [Google Scholar] [CrossRef] [PubMed]
- Engle, M.J.; Goetz, G.S.; Alpers, D.H. Caco-2 cells express a combination of colonocyte and enterocyte phenotypes. J. Cell Physiol. 1998, 174, 362–369. [Google Scholar] [CrossRef]
- Berger, E.; Héraud, S.; Mojallal, A.; Lequeux, C.; Weiss-Gayet, M.; Damour, O.; Géloën, A. Pathways commonly dysregulated in mouse and human obese adipose tissue: FAT/CD36 modulates differentiation and lipogenesis. Adipocyte 2015, 4, 161–180. [Google Scholar] [CrossRef] [PubMed]
- Berger, E.; Vega, N.; Weiss-Gayet, M.; Géloën, A. Gene network analysis of glucose linked signaling pathways and their role in human hepatocellular carcinoma cell growth and survival in HuH7 and HepG2 cell lines. BioMed Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Dalerba, P.; Dylla, S.J.; Park, I.K.; Liu, R.; Wang, X.; Cho, R.W.; Hoey, T.; Gurney, A.; Huang, E.H.; Simeone, D.M.; et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 10158–10163. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Ravikumar, T.S.; Yang, W.L. Bone morphogenetic protein-4 inhibits heat-induced apoptosis by modulating MAPK pathways in human colon cancer HCT116 cells. Cancer Lett. 2007, 256, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Gayet, J.; Zhou, X.P.; Duval, A.; Rolland, S.; Hoang, J.M.; Cottu, P.; Hamelin, R. Extensive characterization of genetic alterations in a series of human colorectal cancer cell lines. Oncogene 2001, 20, 5025–5032. [Google Scholar] [CrossRef] [PubMed]
- De Lau, W.; Barker, N.; Clevers, H. Wnt signaling in the normal intestine and colorectal cancer. Front. Biosci. 2007, 12, 471–491. [Google Scholar] [CrossRef] [PubMed]
- Van Beijnum, J.R.; Petersen, K.; Griffioen, A.W. Tumor endothelium is characterized by a matrix remodelling signature. Front. Biosci. 2009, 1, 216–225. [Google Scholar] [CrossRef]
- Bertrand, F.E.; Angus, C.W.; Partis, W.J.; Sigounas, G. Developmental pathways in colon cancer: Crosstalk between WNT, BMP, hedgehog and notch. Cell Cycle 2012, 11, 4344–5431. [Google Scholar] [CrossRef] [PubMed]
- Murgai, M.; Giles, A.; Kaplan, R. Physiological, tumor, and metastatic niches: Opportunities and challenges for targeting the tumor microenvironment. Crit. Rev. Oncog. 2015, 20, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Crotti, S.; Piccoli, M.; Rizzolio, F.; Giordano, A.; Nitti, D.; Agostini, M. Extracellular matrix and colorectal cancer: How surrounding microenvironment affects cancer cell behavior? J. Cell Physiol. 2017, 232, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Crosnier, C.; Stamataki, D.; Lewis, J. Organizing cell renewal in the intestine: Stem cells, signals and combinatorial control. Nat. Rev. Genet. 2006, 7, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Van der Flier, L.G.; Clevers, H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 2009, 71, 241–260. [Google Scholar] [CrossRef] [PubMed]
- Le Naour, F.; André, M.; Greco, C.; Billard, M.; Sordat, B.; Emile, J.F.; Lanza, F.; Boucheix, C.; Rubinstein, E. Profiling of the tetraspanin web of human colon cancer cells. Mol. Cell. Proteom. 2006, 5, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Bestard-Escalas, J.; Garate, J.; Maimó-Barceló, A.; Fernández, R.; Lopez, D.H.; Lage, S.; Reigada, R.; Khorrami, S.; Ginard, D.; Reyes, J.; et al. Lipid fingerprint image accurately conveys human colon cell pathophysiologic state: A solid candidate as biomarker. Biochim. Biophys. Acta 2016, 1861 Pt A, 1942–1950. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, L.; Ye, X.; Chen, L.; Zhang, L.; Gao, Y.; Kang, J.X.; Cai, C. Characteristics of fatty acid distribution is associated with colorectal cancer prognosis. Prostaglandins Leukot. Essent. Fatty Acids 2013, 88, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Zhu, J.; Wolins, N.E.; Cheng, J.X.; Buhman, K.K. Differential association of adipophilin and TIP47 proteins with cytoplasmic lipid droplets in mouse enterocytes during dietary fat absorption. Biochim. Biophys. Acta 2009, 1791, 1173–1180. [Google Scholar] [CrossRef] [PubMed]
- Kaser, A.; Lee, A.H.; Franke, A.; Glickman, J.N.; Zeissig, S.; Tilg, H.; Nieuwenhuis, E.E.; Higgins, D.E.; Schreiber, S.; Glimcher, L.H.; et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 2008, 134, 743–756. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, C.; Cavia, M.; Alonso-Torre, S.R. Antitumor effect of oleic acid; mechanisms of action: A review. Nutr. Hosp. 2012, 27, 1860–1865. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, C.; Cavia, M.; Alonso-Torre, S.R. Oleic acid inhibits store-operated calcium entry in human colorectal adenocarcinoma cells. Eur. J. Nutr. 2012, 51, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, S.; Miyazaki, Y.; Hiraoka, S.; Nagasawa, Y.; Toyota, M.; Takakura, R.; Kiyohara, T.; Shinomura, Y.; Matsuzawa, Y. PPARgamma agonists inhibit cell growth and suppress the expression of cyclin D1 and EGF-like growth factors in ras-transformed rat intestinal epithelial cells. Int. J. Cancer 2001, 94, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Marin, H.E.; Peraza, M.A.; Billin, A.N.; Willson, T.M.; Ward, J.M.; Kennett, M.J.; Gonzalez, F.J.; Peters, J.M. Ligand activation of peroxisome proliferator-activated receptor beta inhibits colon carcinogenesis. Cancer Res. 2006, 66, 4394–4401. [Google Scholar] [CrossRef] [PubMed]
- De Vogel-van den Bosch, H.M.; Bünger, M.; de Groot, P.J.; Bosch-Vermeulen, H.; Hooiveld, G.J.; Müller, M. PPARalpha-mediated effects of dietary lipids on intestinal barrier gene expression. BMC Genom. 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Sato, O.; Kuriki, C.; Fukui, Y.; Motojima, K. Dual promoter structure of mouse and human fatty acid translocase/CD36 genes and unique transcriptional activation by peroxisome proliferator-activated receptor α and γ ligands. J. Biol. Chem. 2002, 277, 15703–15711. [Google Scholar] [CrossRef] [PubMed]
- Bumpus, N.N.; Johnson, E.F. 5-Aminoimidazole-4-carboxyamide-ribonucleoside (AICAR)-stimulated hepatic expression of Cyp4a10, Cyp4a14, Cyp4a31, and other peroxisome proliferator-activated receptor α-responsive mouse genes is AICAR 5′-monophosphate-dependent and AMP-activated protein kinase-independent. J. Pharmacol. Exp. Ther. 2011, 339, 886–895. [Google Scholar] [CrossRef] [PubMed]
- Lemieux, E.; Boucher, M.J.; Mongrain, S.; Boudreau, F.; Asselin, C.; Rivard, N. Constitutive activation of the MEK/ERK pathway inhibits intestinal epithelial cell differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 301, G719–G730. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.T.; Poirier, H.; Clément, L.; Nassir, F.; Pelsers, M.M.; Petit, V.; Degrace, P.; Monnot, M.C.; Glatz, J.F.; Abumrad, N.A.; et al. Luminal lipid regulates CD36 levels and downstream signaling to stimulate chylomicron synthesis. J. Biol. Chem. 2011, 286, 25201–25210. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, S.; Shahid, R.; Riehl, T.E.; Chandra, R.; Nassir, F.; Stenson, W.F.; Liddle, R.A.; Abumrad, N.A. CD36-dependent signaling mediates fatty acid-induced gut release of secretin and cholecystokinin. FASEB J. 2013, 27, 1191–1202. [Google Scholar] [CrossRef] [PubMed]
- Benoit, B.; Bruno, J.; Kayal, F.; Estienne, M.; Debard, C.; Ducroc, R.; Plaisancié, P. Saturated and unsaturated fatty acids differently modulate colonic goblet cells in vitro and in rat pups. J. Nutr. 2015, 145, 1754–1762. [Google Scholar] [CrossRef] [PubMed]
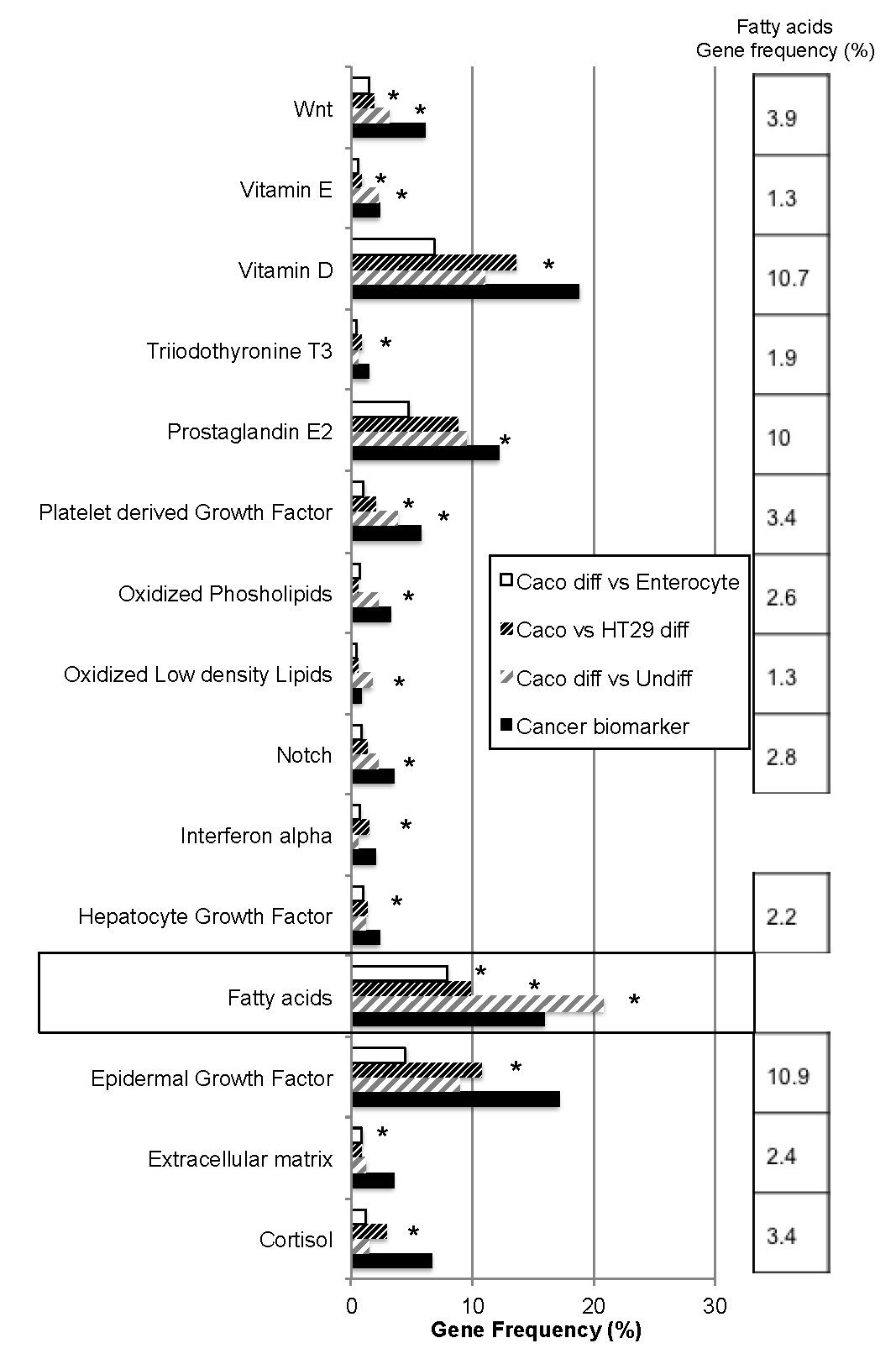
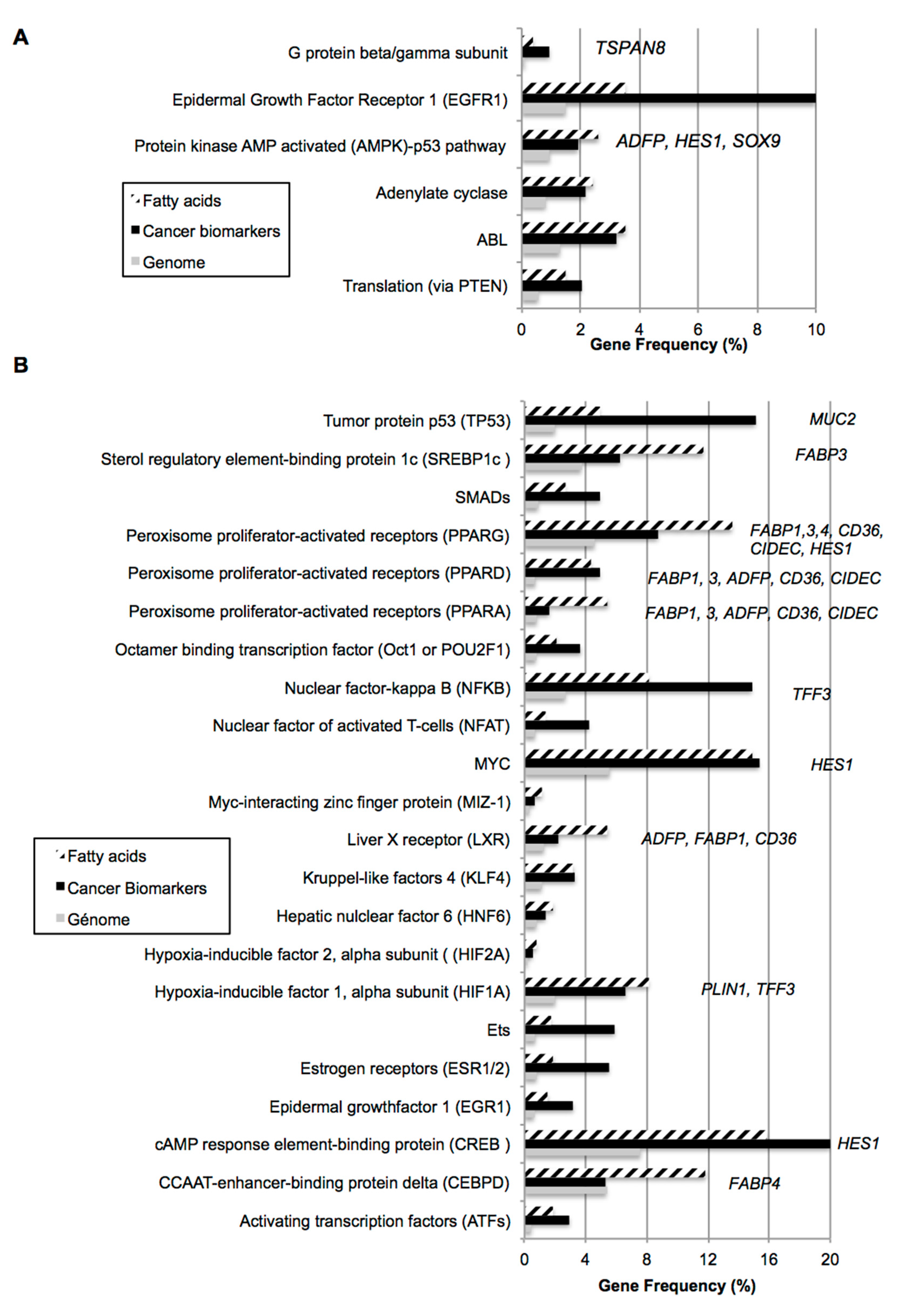
| Gene Dataset Name | Gene Number | Dataset Analysis | GEO | Caco-2 Diff vs. Undiff | Caco-2 Diff vs. HT29-MTX | Caco Diff vs. Entero (ileal + Juj) | Colonocyte | Enterocyte |
|---|---|---|---|---|---|---|---|---|
| Common Gene Frequency % (Number) | ||||||||
| Caco-2 differentiated vs. undifferentiated | 579 | Pellis et al., 2005 | GSE2047 | 13.6 (79) | 51.1 (296) | 1.9 (11) | 2.1 (12) | |
| Caco-2 differentiated vs. HT29-MTX differentiated (3 weeks) | 1371 | −2 < FcLOG < 2 | GSE30292 | 5.8 (79) | 61.7 (846) | 0.9 (12) | 0.9 (13) | |
| Caco-2 differentiated vs. (enterocyte ileal + jejunal) | 6038 | −2 < FcLOG < 2 | GSE30292 | 4.9 (296) | 14.0 (846) | 0.4 (22) | 0.4 (28) | |
| Colonocyte | 103 | COPE (PubMed list) | 10.7 (11) | 11.7 (12) | 21.4 (22) | 48.5 (50) | ||
| Enterocyte | 142 | COPE (PubMed list) | 8.5 (12) | 9.2 (13) | 19.7 (28) | 35.2 (50) | ||
| Gene Symbol | Caco-2 | HT29-MTX | Caco-2/HT29-MTX | Fc vs. Theoric | Fc OA Caco-2/HT29-MTX |
|---|---|---|---|---|---|
| Intestinal phenotype matkers | |||||
| ATOH1 (Hath1) | 1366 ± 30 | 17,481 ± 1590 | 212 ± 29 | −14 | 3.4 ± 0.2 |
| GFi1 | 4.20 ± 0.10 | 56.0 ± 9.4 | 0.7 ± 0.1 | −14 | 3.9 ± 0.1 |
| HES1 | 1260 ± 338 | 1697 ± 36 | 527 ± 29 | −2 | 1.6 ± 0.1 |
| MUC2 | 17.4 ± 1.6 | 1.4 ± 0.2 | 2.0 ± 0.5 | −8 | 2.4 ± 1.7 |
| MUC5 | 1540 ± 10 | 30,661 ± 1314 | 619 ± 27 | −7 | 4.2 ± 0.0 |
| SOX9 | 228 ± 10 | 489 ± 83 | 158 ± 20 | −2 | −3.2 ± 0.0 |
| TFF3 | 580774 ± 2 | 485,507 ± 15,791 | 101,000 ± 882 | −6 | 4.3 ± 0.2 |
| Fatty acid uptake & transport | |||||
| FAT/CD36 | 0.05 ± 0.03 | 1.32 ± 0.35 | 0.05 ± 0.00 | −3 | 1.0 ± 0.1 |
| FABP1 | 263,971 ± 115,167 | 27 ± 2 | 50,176 ± 4078 | −5 | 4.9 ± 0.4 |
| FABP3 | 191 ± 18 | 3.2 ± 0.5 | 98 ± 8 | −2 | 2.0 ± 0.5 |
| FABP4 | 4.05 ± 0.62 | 0.69 ± 0.14 | 0.15 ± 0.03 | −25 | 26.7 ± 8.3 |
| GRP120 | 2844 ± 91 | 9007 ± 473 | 312 ± 8 | −11 | 5.4 ± 0.2 |
| Lipid droplet structure | |||||
| ABDH5 (CGI-58) | 507 ± 158 | 165 ± 14 | 184 ± 28 | −3 | 3.7 ± 1.5 |
| PLIN1 | nd | 0.03 ± 0.004 | 0.002 ± 0.001 | −1.6 ± 0.1 | |
| PLIN2 (ADFP) | nd | nd | nd | ||
| PLIN3 (TIP47) | 3199 ± 870 | 1670 ± 63 | 1200 ± 271 | −3 | 2.7 ± 0.1 |
| PLIN5 (oxPAT) | 0.005 ± 0.001 | 0.031 ± 0.008 | 0.001 ± 0.000 | −10 | 1.9 ± 0.1 |
| CIDEA | nd | nd | nd | ||
| CIDEB | 16.1 ± 2.8 | 18.1 ± 2.0 | 3.1 ± 0.1 | −5 | 3.2 ± 0.2 |
| CIDEC | 16,762 ± 2219 | 4837 ± 64 | 2217 ± 49 | −7 | 2.3 ± 0.2 |
| UPR stress response | |||||
| XBP1 | 19,033 ± 634 | 61,810 ± 2713 | 13,454 ± 6654 | −2 | −1.4 ± 0.0 |
| Cancer marker | |||||
| TSPAN8 | 124,836 ± 3825 | 706,666 ± 5070 | 19,088 ± 3500 | −10 | 4.7 ± 0.0 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berger, E.; Nassra, M.; Atgié, C.; Plaisancié, P.; Géloën, A. Oleic Acid Uptake Reveals the Rescued Enterocyte Phenotype of Colon Cancer Caco-2 by HT29-MTX Cells in Co-Culture Mode. Int. J. Mol. Sci. 2017, 18, 1573. https://doi.org/10.3390/ijms18071573
Berger E, Nassra M, Atgié C, Plaisancié P, Géloën A. Oleic Acid Uptake Reveals the Rescued Enterocyte Phenotype of Colon Cancer Caco-2 by HT29-MTX Cells in Co-Culture Mode. International Journal of Molecular Sciences. 2017; 18(7):1573. https://doi.org/10.3390/ijms18071573
Chicago/Turabian StyleBerger, Emmanuelle, Merian Nassra, Claude Atgié, Pascale Plaisancié, and Alain Géloën. 2017. "Oleic Acid Uptake Reveals the Rescued Enterocyte Phenotype of Colon Cancer Caco-2 by HT29-MTX Cells in Co-Culture Mode" International Journal of Molecular Sciences 18, no. 7: 1573. https://doi.org/10.3390/ijms18071573
APA StyleBerger, E., Nassra, M., Atgié, C., Plaisancié, P., & Géloën, A. (2017). Oleic Acid Uptake Reveals the Rescued Enterocyte Phenotype of Colon Cancer Caco-2 by HT29-MTX Cells in Co-Culture Mode. International Journal of Molecular Sciences, 18(7), 1573. https://doi.org/10.3390/ijms18071573