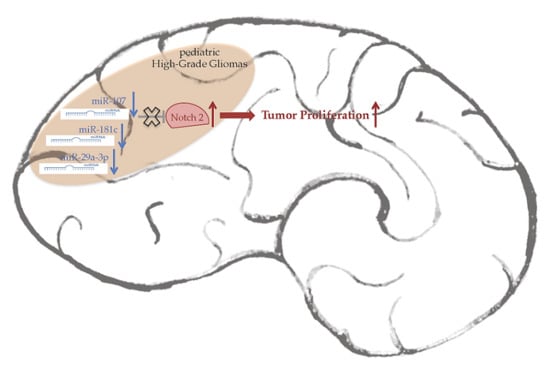
Loss of miR-107, miR-181c and miR-29a-3p Promote Activation of Notch2 Signaling in Pediatric High-Grade Gliomas (pHGGs)
Abstract
1. Introduction
2. Results
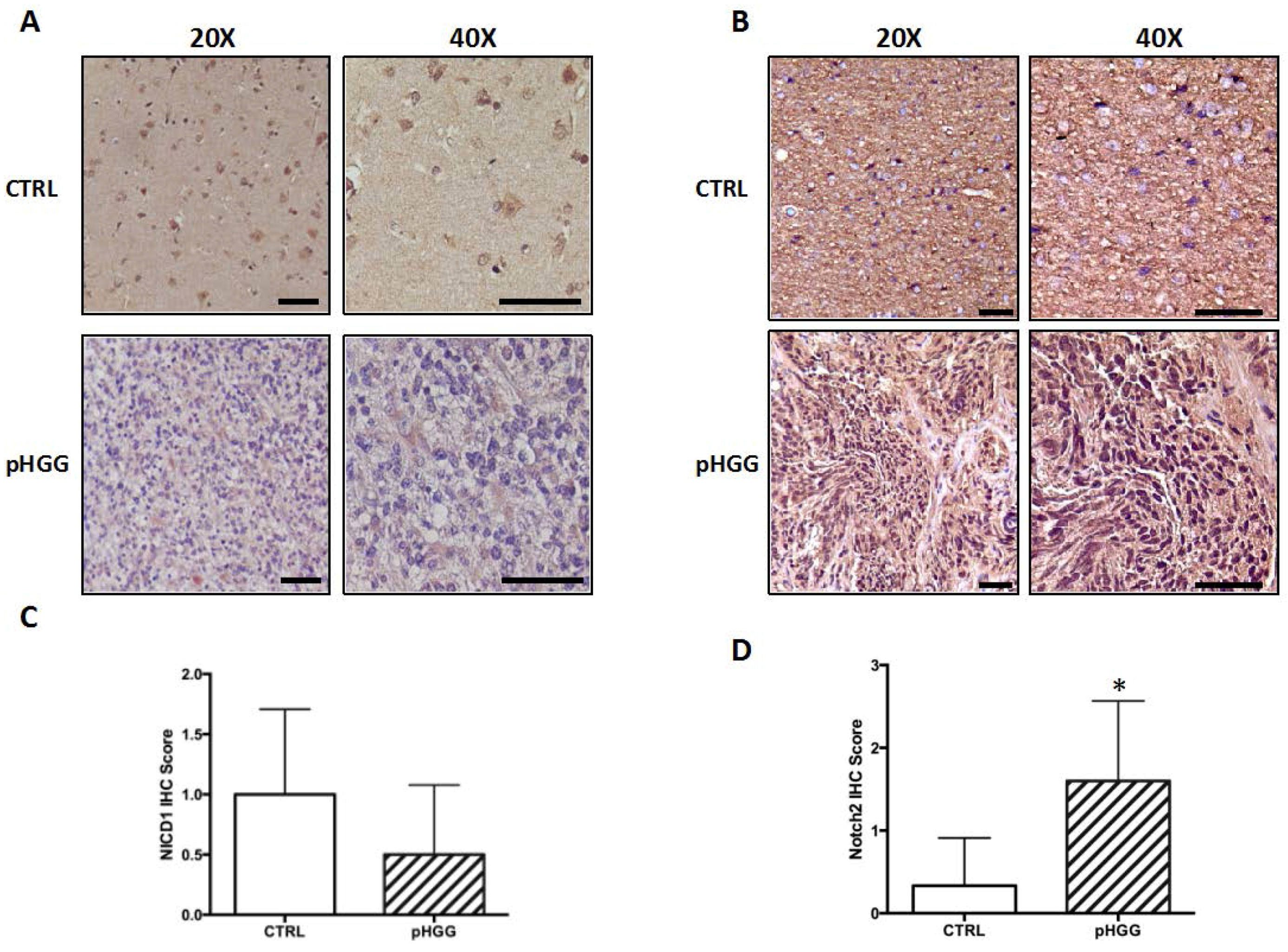
2.1. Notch1 and Notch2 Receptor Expression in pHGG Tumors
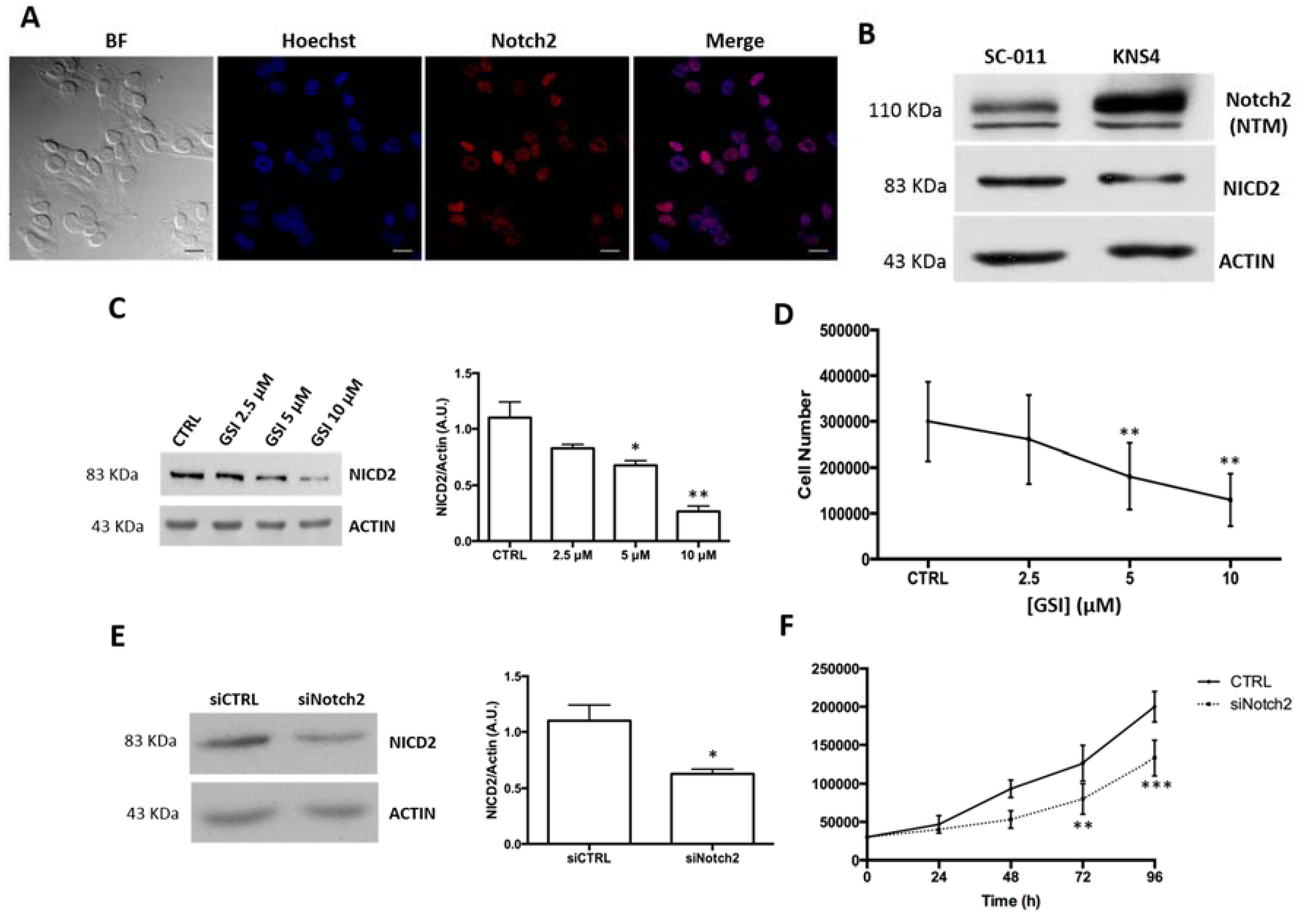
2.2. Notch2 Inhibition Reduces pHGG Cell Proliferation
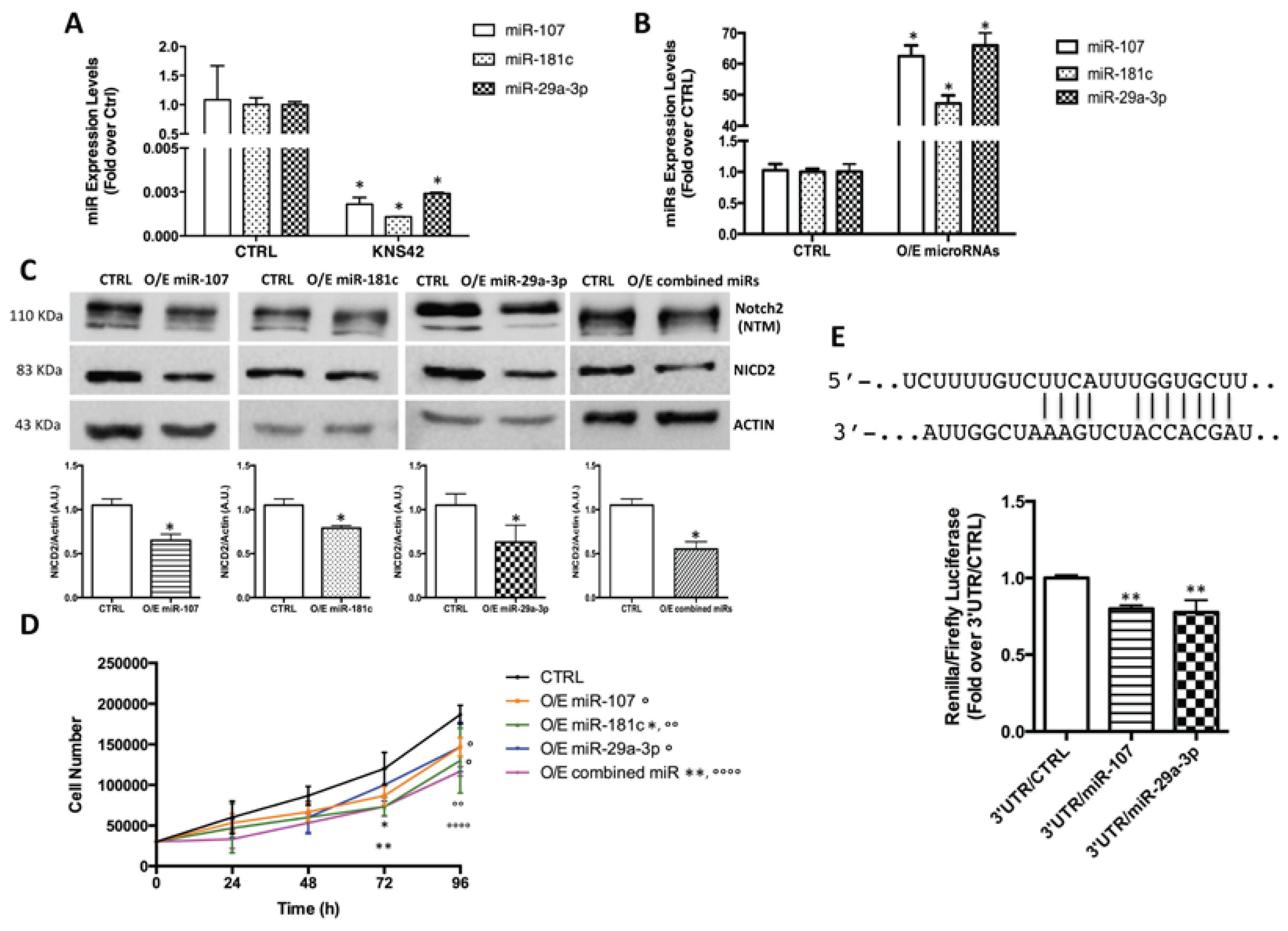
2.3. Notch2 High Levels Are Maintained by Low Levels of miR-107, miR-181c and miR-29a-3p
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Histology
4.3. Notch1 and Notch2 Immunohistochemistry
4.4. Cell Lines and Treatments
4.5. RNA Isolation and qRT-PCR
4.6. Western Blotting
4.7. Immunofluorescence Studies
4.8. Plasmid Construction and Luciferase Reporter Assays
4.9. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sturm, D.; Pfister, S.M.; Jones, D.T. Pediatric gliomas: Current concepts on diagnosis, biology, and clinical management. J. Clin. Oncol. 2017, 35, 2370. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Gittleman, H.; Fulop, J.; Liu, M.; Blanda, R.; Kromer, C.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol. 2015, 17 (Suppl. 4), iv1. [Google Scholar] [CrossRef] [PubMed]
- Juratli, T.A.; Qin, N.; Cahill, D.P.; Filbin, M.G. Molecular pathogenesis and therapeutic implications in pediatric high-grade gliomas. Pharmacol. Ther. 2017. [Google Scholar] [CrossRef] [PubMed]
- Braunstein, S.; Raleigh, D.; Bindra, R.; Mueller, S.; Haas-Kogan, D. Pediatric high-grade glioma: Current molecular landscape and therapeutic approaches. J. Neuro Oncol. 2017, 134, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Miele, E.; Buttarelli, F.; Arcella, A.; Begalli, F.; Garg, N.; Silvano, M.; Po, A.; Baldi, C.; Carissimo, G.; Antonelli, M.; et al. High-throughput microRNA profiling of pediatric high-grade gliomas. Neuro Oncol. 2014, 16, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Louvi, A.; Artavanis-Tsakonas, S. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 2006, 7, 93. [Google Scholar] [CrossRef] [PubMed]
- Lasky, J.L.; Wu, H. Notch signaling, brain development, and human disease. Pediatr. Res. 2005, 57, 104R. [Google Scholar] [CrossRef] [PubMed]
- Purow, B.W.; Haque, R.M.; Noel, M.W.; Su, Q.; Burdick, M.J.; Lee, J.; Sundaresan, T.; Pastorino, S.; Park, J.K.; Mikolaenko, I. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 2005, 65, 2353–2363. [Google Scholar] [CrossRef] [PubMed]
- Artavanis-Tsakonas, S.; Rand, M.D.; Lake, R.J. Notch signaling: Cell fate control and signal integration in development. Science 1999, 284, 770–776. [Google Scholar] [CrossRef] [PubMed]
- Lubman, O.Y.; Ilagan, M.X.G.; Kopan, R.; Barrick, D. Quantitative dissection of the Notch: CSL interaction: Insights into the Notch-mediated transcriptional switch. J. Mol. Biol. 2007, 365, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Kopan, R.; Ilagan, M.X.G. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Kesari, S.; Rooney, C.; Strack, P.R.; Chen, J.; Shen, H.; Wu, L.; Griffin, J.D. Inhibition of notch signaling blocks growth of glioblastoma cell lines and tumor neurospheres. Genes Cancer 2010, 1, 822–835. [Google Scholar] [CrossRef] [PubMed]
- Bolós, V.; Grego-Bessa, J.; de la Pompa, J.L. Notch signaling in development and cancer. Endocr. Rev. 2007, 28, 339–363. [Google Scholar] [CrossRef] [PubMed]
- Leong, K.G.; Karsan, A. Recent insights into the role of Notch signaling in tumorigenesis. Blood 2006, 107, 2223–2233. [Google Scholar] [CrossRef] [PubMed]
- Aster, J.C.; Pear, W.S. Notch signaling in leukemia. Curr. Opin. Hematol. 2001, 8, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Aster, J.C.; Pear, W.S.; Blacklow, S.C. Notch signaling in leukemia. Annu. Rev. Pathol. Mech. Dis. 2008, 3, 587–613. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.P.; Qi, S.T.; Feng, W.F.; Zhang, G.Z.; Zhang, H.P.; Tian, J.J. Interference of Notch 2 inhibits the progression of gliomas and induces cell apoptosis by induction of the cell cycle at the G0/G1 phase. Mol. Med. Rep. 2015, 11, 734–738. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, X.; Tian, W.; Wang, J. Short hairpin RNA targeting Notch2 inhibits U87 human glioma cell proliferation by inducing cell cycle arrest and apoptosis in vitro and in vivo. Mol. Med. Rep. 2014, 10, 2843–2850. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fouladi, M.; Stewart, C.F.; Olson, J.; Wagner, L.M.; Onar-Thomas, A.; Kocak, M.; Packer, R.J.; Goldman, S.; Gururangan, S.; Gajjar, A. Phase I trial of MK-0752 in children with refractory CNS malignancies: A pediatric brain tumor consortium study. J. Clin. Oncol. 2011, 29, 3529–3534. [Google Scholar] [CrossRef] [PubMed]
- Bracken, C.P.; Scott, H.S.; Goodall, G.J. A network-biology perspective of microRNA function and dysfunction in cancer. Nat. Rev. Genet. 2016, 17, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Bax, D.A.; Little, S.E.; Gaspar, N.; Perryman, L.; Marshall, L.; Viana-Pereira, M.; Jones, T.A.; Williams, R.D.; Grigoriadis, A.; Vassal, G. Molecular and phenotypic characterisation of paediatric glioma cell lines as models for preclinical drug development. PLoS ONE 2009, 4, e5209. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Mao, Z.; Huang, J.; Xie, S.; Liu, T.; Mao, Z. Blocking the NOTCH pathway can inhibit the growth of CD133-positive A549 cells and sensitize to chemotherapy. Biochem. Biophys. Res. Commun. 2014, 444, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, X.-R.; Zhang, R.; Li, P.; Liu, Y.; Yan, K.; Jiang, X.-D. MicroRNA-107 inhibits glioma cell migration and invasion by modulating Notch2 expression. J. Neuro Oncol. 2013, 112, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Akiyama, Y.; Otsubo, T.; Shimada, S.; Yuasa, Y. Involvement of epigenetically silenced microRNA-181c in gastric carcinogenesis. Carcinogenesis 2010, 31, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.-H.; Chang, N.-W.; Shrestha, S.; Hsu, S.-D.; Lin, Y.-L.; Lee, W.-H.; Yang, C.-D.; Hong, H.-C.; Wei, T.-Y.; Tu, S.-J. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2015, 44, D239–D247. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: From mechanism to therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef] [PubMed]
- De Smaele, E.; Ferretti, E.; Gulino, A. MicroRNAs as biomarkers for CNS cancer and other disorders. Brain Res. 2010, 1338, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Dantas-Barbosa, C.; Bergthold, G.; Daudigeos-Dubus, E.; Blockus, H.; Boylan, J.F.; Ferreira, C.; Puget, S.; Abely, M.; Vassal, G.; Grill, J. Inhibition of the NOTCH pathway using γ-secretase inhibitor RO4929097 has limited antitumor activity in established glial tumors. Anti-Cancer Drugs 2015, 26, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Miele, L.; Golde, T.; Osborne, B. Notch signaling in cancer. Curr. Mol. Med. 2006, 6, 905–918. [Google Scholar] [CrossRef] [PubMed]
- Aster, J.C.; Pear, W.S.; Blacklow, S.C. The varied roles of notch in cancer. Annu. Rev. Pathol. Mech. Dis. 2017, 12, 245–275. [Google Scholar] [CrossRef] [PubMed]
- Nowell, C.S.; Radtke, F. Notch as a tumour suppressor. Nat. Rev. Cancer 2017, 17, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Zhang, A.; Jiang, R.; Qiu, M.; Kang, C.; Jia, Z.; Wang, G.; Han, L.; Fan, X.; Pu, P. The different role of Notch1 and Notch2 in astrocytic gliomas. PLoS ONE 2013, 8, e53654. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Qiu, M.; Zhang, Z.; Kang, C.; Jiang, R.; Jia, Z.; Wang, G.; Jiang, H.; Pu, P. The oncogenic roles of Notch1 in astrocytic gliomas in vitro and in vivo. J. Neuro Oncol. 2010, 97, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Wei, Y.; Wang, J.; Ao, Q.; Gong, K.; Zuo, H. Decreased expression of microRNA-107 predicts poorer prognosis in glioma. Tumor Biol. 2015, 36, 4461–4466. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Lou, S.; Dai, Q.; Mao, D.; Ji, J.; Sun, X. Tumor suppressor miR-181c attenuates proliferation, invasion, and self-renewal abilities in glioblastoma. Neuroreport 2015, 26, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Jiang, X.; Yao, C.; Zhang, L.; Liu, H.; Xia, H.; Wang, Y. Heat shock protein 47 regulated by miR-29a to enhance glioma tumor growth and invasion. J. Neuro Oncol. 2014, 118, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Ren, L.; Sun, C.; Yu, L.; Bian, X.; Zhou, X.; Wen, Y.; Hua, D.; Zhao, S.; Luo, W. miR-29a/b/c function as invasion suppressors for gliomas by targeting CDC42 and predict the prognosis of patients. Br. J. Cancer 2017, 117, 1036–1047. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Shen, K.; Zhao, Y.; Ma, C.; Liu, J.; Ma, J. MiR-92b inhibitor promoted glioma cell apoptosis via targeting DKK3 and blocking the Wnt/beta-catenin signaling pathway. J. Transl. Med. 2013, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Jha, P.; Agrawal, R.; Pathak, P.; Kumar, A.; Purkait, S.; Mallik, S.; Suri, V.; Chand Sharma, M.; Gupta, D.; Suri, A. Genome-wide small noncoding RNA profiling of pediatric high-grade gliomas reveals deregulation of several miRNAs, identifies downregulation of snoRNA cluster HBII-52 and delineates H3F3A and TP53 mutant-specific miRNAs and snoRNAs. Int. J. Cancer 2015, 137, 2343–2353. [Google Scholar] [CrossRef] [PubMed]
- Eguía-Aguilar, P.; Pérezpeña-Díazconti, M.; Benadón-Darszon, E.; de León, F.C.-P.; Gordillo-Domínguez, L.; Torres-García, S.; Sadowinski-Pine, S.; Arenas-Huertero, F. Reductions in the expression of miR-124-3p, miR-128-1, and miR-221-3p in pediatric astrocytomas are related to high-grade supratentorial, and recurrent tumors in Mexican children. Childs Nerv. Syst. 2014, 30, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.-L.; Hsieh, T.-H.; Ng, K.-H.; Tsai, Y.-N.; Tsai, C.-F.; Chao, M.-E.; Liu, D.-J.; Chu, S.-S.; Chen, W.; Liu, Y.-R. Downregulation of miR-137 and miR-6500-3p promotes cell proliferation in pediatric high-grade gliomas. Oncotarget 2016, 7, 19723–19737. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef] [PubMed]
- De Paula Alves Coelho, K.M.; Stall, J.; Júnior, H.F.; Blasius, R.; de França, P.H.C. Evaluation of expression of genes CADM1, TWIST1 and CDH1 by immunohistochemestry in melanocytic lesions. Pathol. Res. Pract. 2017, 213, 1067–1071. [Google Scholar] [CrossRef] [PubMed]
- Catanzaro, G.; Besharat, Z.; Garg, N.; Ronci, M.; Pieroni, L.; Miele, E.; Mastronuzzi, A.; Carai, A.; Alfano, V.; Po, A.; et al. microRNAs-proteomic networks characterizing human medulloblastoma-SLCs. Stem Cells Int. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Po, A.; Silvano, M.; Miele, E.; Capalbo, C.; Eramo, A.; Salvati, V.; Todaro, M.; Besharat, Z.; Catanzaro, G.; Cucchi, D.; et al. Noncanonical GLI1 signalling promotes stemness features and in-vivo growth in lung adenocarcinoma. Oncogene 2017, 36, 4641–4652. [Google Scholar] [CrossRef] [PubMed]
- Po, A.; Begalli, F.; Abballe, L.; Alfano, V.; Besharat, Z.M.; Catanzaro, G.; Vacca, A.; Napolitano, M.; Tafani, M.; Giangaspero, F.; et al. β-Arrestin1/miR-326 Transcription Unit Is Epigenetically Regulated in Neural Stem Cells Where It Controls Stemness and Growth Arrest. Stem Cells Int. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Ronci, M.; Catanzaro, G.; Pieroni, L.; Po, A.; Besharat, Z.M.; Greco, V.; Levi Mortera, S.; Screpanti, I.; Ferretti, E.; Urbani, A. Proteomic analysis of human sonic hedgehog (SHH) medulloblastoma stem-like cells. Mol. Biosyst. 2015, 11, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010, 11, R90. [Google Scholar] [CrossRef] [PubMed]
- Betel, D.; Wilson, M.; Gabow, A.; Marks, D.S.; Sander, C. The microRNA. org resource: Targets and expression. Nucleic Acids Res. 2008, 36 (Suppl. 1), D149–D153. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Catanzaro, G.; Sabato, C.; Russo, M.; Rosa, A.; Abballe, L.; Besharat, Z.M.; Po, A.; Miele, E.; Bellavia, D.; Chiacchiarini, M.; et al. Loss of miR-107, miR-181c and miR-29a-3p Promote Activation of Notch2 Signaling in Pediatric High-Grade Gliomas (pHGGs). Int. J. Mol. Sci. 2017, 18, 2742. https://doi.org/10.3390/ijms18122742
Catanzaro G, Sabato C, Russo M, Rosa A, Abballe L, Besharat ZM, Po A, Miele E, Bellavia D, Chiacchiarini M, et al. Loss of miR-107, miR-181c and miR-29a-3p Promote Activation of Notch2 Signaling in Pediatric High-Grade Gliomas (pHGGs). International Journal of Molecular Sciences. 2017; 18(12):2742. https://doi.org/10.3390/ijms18122742
Chicago/Turabian StyleCatanzaro, Giuseppina, Claudia Sabato, Michele Russo, Alessandro Rosa, Luana Abballe, Zein Mersini Besharat, Agnese Po, Evelina Miele, Diana Bellavia, Martina Chiacchiarini, and et al. 2017. "Loss of miR-107, miR-181c and miR-29a-3p Promote Activation of Notch2 Signaling in Pediatric High-Grade Gliomas (pHGGs)" International Journal of Molecular Sciences 18, no. 12: 2742. https://doi.org/10.3390/ijms18122742
APA StyleCatanzaro, G., Sabato, C., Russo, M., Rosa, A., Abballe, L., Besharat, Z. M., Po, A., Miele, E., Bellavia, D., Chiacchiarini, M., Gessi, M., Peruzzi, G., Napolitano, M., Antonelli, M., Mastronuzzi, A., Giangaspero, F., Locatelli, F., Screpanti, I., Vacca, A., & Ferretti, E. (2017). Loss of miR-107, miR-181c and miR-29a-3p Promote Activation of Notch2 Signaling in Pediatric High-Grade Gliomas (pHGGs). International Journal of Molecular Sciences, 18(12), 2742. https://doi.org/10.3390/ijms18122742