Comparative Proteomics Analyses of Pollination Response in Endangered Orchid Species Dendrobium Chrysanthum
Abstract
1. Introduction
2. Results and Discussion
2.1. Protein Profiles of D. chrysanthum in Un-Pollination, Self-Pollination and Cross-Pollination
2.2. Identification of the DEP
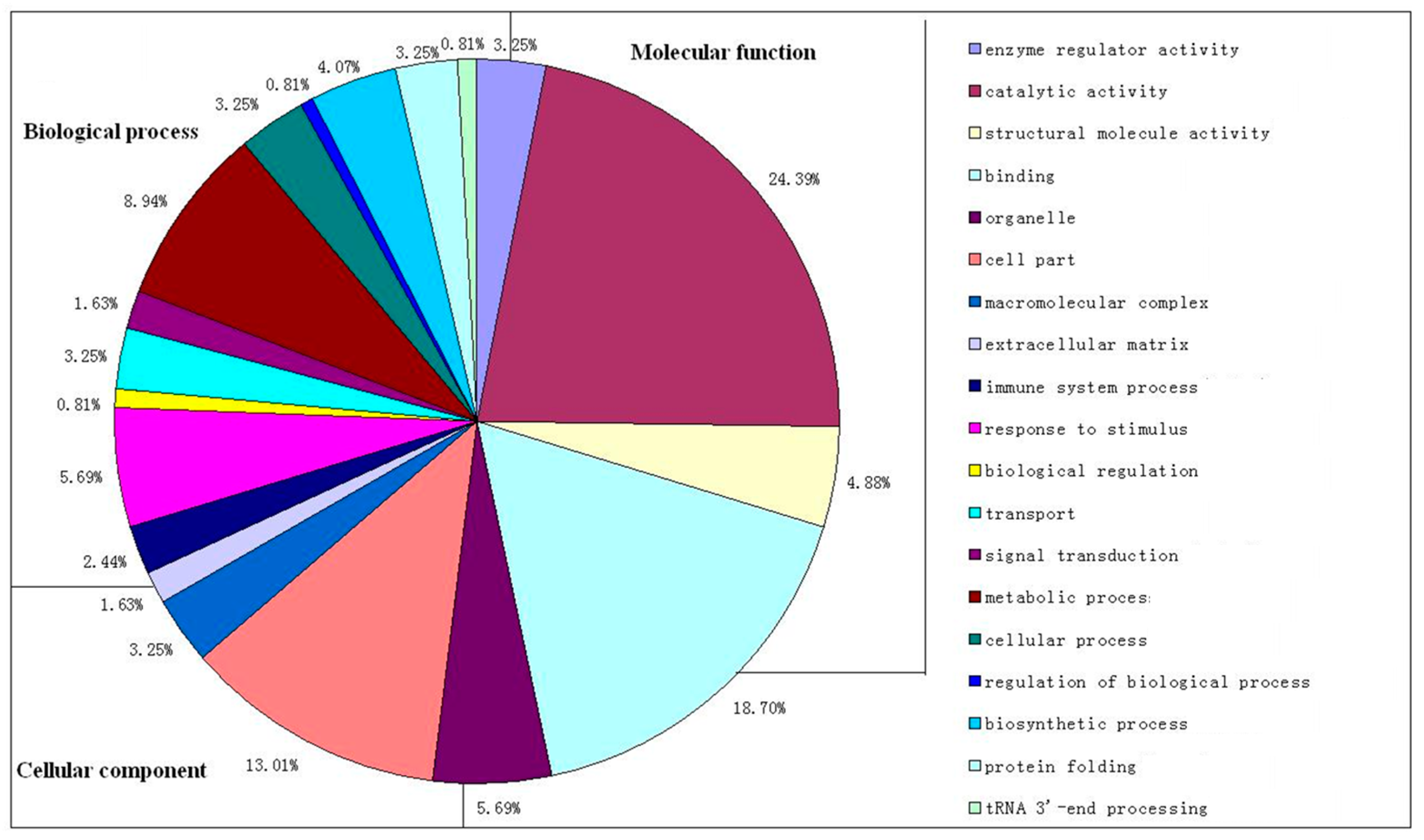
2.3. Gene Ontology Analysis of Differentially Expressed Proteins between Self-Pollination and Cross-Pollination
2.4. Functional Categories of the DEGs
3. Materials and Methods
3.1. Plant Materials
3.2. Protein Extraction and Quantification
3.3. Two-Dimensional Gel Electrophoresis
3.4. Image Acquisition and Data Analysis
3.5. Protein Identification with MALDI-TOF/TOF MS
3.6. Statistical Analysis and Gene Ontology Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lord, E.M.; Russell, S.D. The mechanisms of pollination and fertilization in plants. Annu. Rev. Cell Dev. Biol. 2002, 18, 81–105. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Kubo, K.; Takayama, S. Non-self- and self-recognition models in plant self-incompatibility. Nat. Plants 2016, 2, 16130. [Google Scholar] [CrossRef] [PubMed]
- Ebert, P.R.; Anderson, M.A.; Bernatzky, R.; Altschuler, M.; Clarke, A.E. Genetic polymorphism of self-incompatibility in flowering plants. Cell 1989, 56, 255–262. [Google Scholar] [CrossRef]
- Kalisz, S.; Vogler, D.W.; Hanley, K.M. Context-dependent autonomous self-fertilization yields reproductive assurance and mixed mating. Nature 2004, 430, 884–887. [Google Scholar] [CrossRef] [PubMed]
- Igic, B.; Kohn, J.R. Evolutionary relationships among self-incompatibility RNases. Proc. Nat. Acad. Sci. USA 2001, 98, 13167–13171. [Google Scholar] [CrossRef] [PubMed]
- McClure, B.A.; Gray, J.E.; Anderson, M.A.; Clarke, A.E. Self-incompatibility in Nicotianaalata involves degradation of pollen rRNA. Nature 1990, 347, 757–760. [Google Scholar] [CrossRef]
- Goldraij, A.; Kondo, K.; Lee, C.B.; Hancock, C.N.; Sivaguru, M.; Vazquez-Santana, S.; Kim, S.; Phillips, T.E.; Cruz-Garcia, F.; McClure, B. Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature 2006, 439, 805–810. [Google Scholar] [CrossRef] [PubMed]
- Franklin-Tong, N.V.; Franklin, F.C. Gametophytic self-incompatibility inhibits pollen tube growth using different mechanisms. Trends Plant Sci. 2003, 8, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.G.; Franklin-Tong, V.E. Self-incompatibility triggers programmed cell death in Papaver pollen. Nature 2004, 429, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Takayama, S.; Isogai, A. Self-incompatibility in plants. Annu. Rev. Plant Biol. 2005, 56, 467–489. [Google Scholar] [CrossRef] [PubMed]
- Chapman, L.A.; Goring, D.R. Pollen-pistil interactions regulating successful fertilization in the Brassicaceae. J. Exp. Bot. 2010, 61, 1987–1999. [Google Scholar] [CrossRef] [PubMed]
- Kao, T.H.; Tsukamoto, T. The molecular and genetic bases of S-RNase-based self-incompatibility. Plant Cell 2004, 16, S72–S83. [Google Scholar] [CrossRef] [PubMed]
- McClure, B. New views of S-RNase-based self-incompatibility. Curr. Opin. Plant Biol. 2006, 9, 639–646. [Google Scholar] [CrossRef] [PubMed]
- McClure, B.; Cruz-Garcia, F.; Romero, C. Compatibility and incompatibility in S-RNase-based systems. Ann. Bot. 2011, 108, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Franklin-Tong, V.E.; Holdaway-Clarke, T.L.; Straatman, K.R.; Kunkel, J.G.; Hepler, P.K. Involvement of extracellular calcium influx in the self-incompatibility response of Papaverrhoeas. Plant J. 2002, 29, 333–345. [Google Scholar] [CrossRef] [PubMed]
- McClure, B.A.; Franklin-Tong, V. Gametophytic self-incompatibility: Understanding the cellular mechanisms involved in “self” pollen tube inhibition. Planta 2006, 224, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Doucet, J.; Lee, H.K.; Goring, D.R. Pollen Acceptance or Rejection: A Tale of Two Pathways. Trends Plant Sci. 2016, 21, 1058–1067. [Google Scholar] [CrossRef] [PubMed]
- Stein, J.C.; Howlett, B.; Boyes, D.C.; Nasrallah, M.E.; Nasrallah, J.B. Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc. Nat. Acad. Sci. USA 1991, 88, 8816–8820. [Google Scholar] [CrossRef] [PubMed]
- Nishio, T.; Kusaba, M. Sequence diversity of SLG and SRK in Brassica oleracea L. Ann. Bot. 2000, 85, 141–146. [Google Scholar] [CrossRef]
- Conner, J.; Tantikanjana, T.; Stein, J.C.; Kandasamy, M.K.; Nasrallah, J.B.; Nasrallah, M.E. Transgene-induced silencing of S-locus genes and related genes in Brassica. Plant J. 1997, 11, 809–823. [Google Scholar] [CrossRef]
- Liu, W.; Fan, J.; Li, J.; Song, Y.; Li, Q.; Zhang, Y.; Xue, Y. SCF(SLF)-mediated cytosolic degradation of S-RNase is required for cross-pollen compatibility in S-RNase-based self-incompatibility in Petunia hybrida. Front. Genet. 2014, 5, 228. [Google Scholar] [CrossRef] [PubMed]
- Bosch, M.; Franklin-Tong, V.E. Self-incompatibility in Papaver: Signalling to trigger PCD in incompatible pollen. J. Exp. Bot. 2008, 59, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Johansen, B. Incompatibility in Dendrobium (Orchidaceae). Bot. J. Linn. Soc. 1990, 103, 165–196. [Google Scholar] [CrossRef]
- Pang, S.; Pan, K.; Wang, Y.; Li, W.; Zhang, L.; Chen, Q. Floral morphology and reproductive biology of Dendrobiumjiajiangense (Orchidaceae) in Mt. Fotang, southwestern China. Flora 2012, 207, 469–474. [Google Scholar] [CrossRef]
- Huda, M.K.; Wilcock, C.C. Rapid flower senescence following male function and breeding systems of some tropical orchids. Plant Biol. 2012, 14, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.B. Systematics of Dendrobiinae (Orchidaceae), with special reference to Australian taxa. Bot. J. Linn. Soc. 2011, 166, 105–126. [Google Scholar] [CrossRef]
- Chen, W.; Cheng, X.; Zhou, Z.; Liu, J.; Wang, H. Molecular cloning and characterization of a tropinone reductase from Dendrobiumnobile Lindl. Mol. Biol. Rep. 2013, 40, 1145–1154. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Jiang, Y.; Wang, S.; Jiang, M.; Chen, Z.; Ying, Q.; Wang, H. Molecular Identification of Dendrobium Species (Orchidaceae) Based on the DNA Barcode ITS2 Region and Its Application for Phylogenetic Study. Int. J. Mol. Sci. 2015, 16, 21975–21988. [Google Scholar] [CrossRef] [PubMed]
- Hajong, S.; Kumaria, S.; Tandon, P. Compatible fungi, suitable medium, and appropriate developmental stage essential for stable association of Dendrobium chrysanthum. J. Basic Microbiol. 2013, 53, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, R.; Staden, J.V. Fruit harvesting time and corresponding morphological changes of seed integuments influence in vitro seed germination of Dendrobiumnobile Lindl. Plant Growth Regul. 2010, 60, 237–246. [Google Scholar] [CrossRef]
- Pinheiro, F.; Cafasso, D.; Cozzolino, S.; Scopece, G. Transitions between self-compatibility and self-incompatibility and the evolution of reproductive isolation in the large and diverse tropical genus Dendrobium (Orchidaceae). Ann. Bot. 2015, 116, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.C.; Huang, J.; Zhang, Y.Q.; Li, P.X.; Zhang, G.Q.; Xu, Q.; Chen, L.J.; Wang, J.Y.; Luo, Y.B.; Liu, Z.J. Lack of S-RNase-Based Gametophytic Self-incompatibility in Orchids Suggests That This System Evolved after the Monocot-Eudicot Split. Front. Plant Sci. 2017, 8, 1106–1119. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.H.; Ji, Z.H.; Wood, J.J.; Wood, H.P. Dendrobium; Flora of China; Scientific Press: Beijing, China, 2009; pp. 367–397. [Google Scholar]
- Li, M.; Sha, A.; Zhou, X.; Yang, P. Comparative proteomic analyses reveal the changes of metabolic features in soybean (Glycine max) pistils upon pollination. Sex. Plant Reprod. 2012, 25, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Samuel, M.A.; Tang, W.Q.; Jamshed, M.; Northey, J.; Patel, D.; Smith, D.; Siu, K.W.M.; Muench, D.G.; Wang, Z.Y.; Goring, D.R. Proteomic analysis of Brassica stigmatic proteins following the self-incompatibility reaction reveals a role for microtubule dynamics during pollen responses. Mol. Cell Proteom. 2011, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.L.; Xu, M.; Ma, F.F.; Chen, H.; Xu, X.H.; Gao, X.Q.; Zhang, X.S. Comparative proteomic analysis reveals similar and distinct features of proteins in dry and wet stigmas. Proteomics 2012, 12, 1983–1998. [Google Scholar] [CrossRef] [PubMed]
- Calder, D.M.; Slater, A.T. The Stigma of Dendrobium speciosum Sm. (Orchidaceae): A New Stigma Type Comprising Detached Cells within a Mucilaginous Matrix. Ann. Bot. 1985, 55, 297–307. [Google Scholar] [CrossRef]
- Slater, A.T.; Calder, D.M. Fine structure of the wet, detached cell stigma of the orchid Dendrobium speciosum Sm. Sex. Plant Reprod. 1990, 3, 61–69. [Google Scholar] [CrossRef]
- Yue, X.; Gao, X.Q.; Zhang, X.S. Circadian rhythms synchronise intracellular calcium dynamics and atp production for facilitating arabidopsis pollen tube growth. Plant Signal. Behav. 2015, 10, e1017699. [Google Scholar] [CrossRef] [PubMed]
- Brugière, N.; Dubois, F.; Limamia, A.M.; Lelandais, M.; Roux, Y.; Sangwan, R.S.; Hirel, B. Glutamine synthetase in the phloem plays a major role in controlling proline production. Plant Cell 1999, 11, 1995–2011. [Google Scholar] [CrossRef] [PubMed]
- Canovas, F.M.; Avila, C.; Canton, F.R.; Canas, R.A.; de la Torre, F. Ammonium assimilation and amino acid metabolism in conifers. J. Exp. Bot. 2007, 58, 2307–2318. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.; Pissarra, J.; Sunkel, C.; Salema, R. Tissue-specific distribution of glutamine synthetase in potato tubers. Ann. Bot. 1996, 77, 429–432. [Google Scholar] [CrossRef]
- Sakurai, N.; Hayakawa, T.; Nakamura, T.; Yamaya, T. Changes in the cellular localization of cytosolic glutamine synthetase protein in vascular bundles of rice leaves at various stages of development. Planta 1996, 200, 306–311. [Google Scholar] [CrossRef]
- Ma, H. Plant reproduction: GABA gradient, guidance and growth. Curr. Biol. 2003, 13, R834–R836. [Google Scholar] [CrossRef] [PubMed]
- Palanivelu, R.; Brass, L.; Edlund, A.F.; Preuss, D. Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 2003, 114, 47–59. [Google Scholar] [CrossRef]
- Yang, Z. GABA, a new player in the plant mating game. Dev. Cell 2003, 5, 185–186. [Google Scholar] [CrossRef]
- Wang, L.; Wang, C.; Ge, T.T.; Wang, J.J.; Liu, T.K.; Hou, X.L.; Li, Y. Expression analysis of self-incompatibility associated genes in non-heading Chinese cabbage. Genet. Mol. Res. 2014, 13, 5025–5035. [Google Scholar] [CrossRef] [PubMed]
- Studart-Guimaraes, C.; Gibon, Y.; Frankel, N.; Wood, C.C.; Zanor, M.I.; Fernie, A.R.; Carrari, F. Identification and characterization of the α and β subunits of succinyl CoA ligase of tomato. Plant Mol. Biol. 2005, 59, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.P.; Zhang, Q.Y.; Tang, Z.C.; Su, W.A.; Sun, W.N. Comparative proteomic analysis provides new insights into chilling stress responses in rice. Mol. Cell Proteom. 2006, 5, 484–496. [Google Scholar] [CrossRef] [PubMed]
- Li, M.N.; Xu, W.Y.; Yang, W.Q.; Kong, Z.S.; Xue, Y.B. Genome-wide gene expression profiling reveals conserved and novel molecular functions of the stigma in rice. Plant Physiol. 2007, 144, 1797–1812. [Google Scholar] [CrossRef] [PubMed]
- Holmes-Davis, R.; Tanaka, C.K.; Vensel, W.H.; Hurkman, W.J.; McCormick, S. Proteome mapping of mature pollen of Arabidopsis thaliana. Proteomics 2005, 5, 4864–4884. [Google Scholar] [CrossRef] [PubMed]
- Englbrecht, C.C.; Schoof, H.; Bohm, S. Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genom. 2004, 5, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Jung, B.G.; Lee, K.O.; Lee, S.S.; Chi, Y.H.; Jang, H.H.; Kang, S.S.; Lee, K.; Lim, D.; Yoon, S.C.; Yun, D.J.; et al. A Chinese cabbage cDNA with high sequence identity to phospholipid hydroperoxide glutathione peroxidases encodes a novel isoform of thioredoxin-dependent peroxidase. J. Biol. Chem. 2002, 277, 12572–12578. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Sun, Y.; Wang, Y.; Long, C.; Li, Y.; Li, Y. Proteomic analysis of the low mutation rate of diploid male gametes induced by colchicine in Ginkgo biloba L. PLoS ONE 2013, 8, e76088. [Google Scholar] [CrossRef] [PubMed]
- McInnis, S.M.; Desikan, R.; Hancock, J.T.; Hiscock, S.J. Production of reactive oxygen species and reactive nitrogen species by angiosperm stigmas and pollen: Potential signalling crosstalk? New Phytol. 2006, 172, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Zafra, A.; Rodriguez-Garcia, M.I.; Alche, J.D. Cellular localization of ROS and NO in olive reproductive tissues during flower development. BMC Plant Biol. 2010, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Yamamoto, R.; Shimamura, S.; Hiraga, S.; Nakayama, N.; Nakamura, T.; Yukawa, K.; Hachinohe, M.; Matsumoto, H.; Komatsu, S. Cytosolic ascorbate peroxidase 2 (cAPX 2) is involved in the soybean response to flooding. Phytochemistry 2008, 69, 1295–1303. [Google Scholar] [CrossRef] [PubMed]
- Vierstra, R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Kao, T.H. Identification and characterization of components of a putative petunia S-locus F-box-containing E3 ligase complex involved in S-RNase-based self-incompatibility. Plant Cell 2006, 18, 2531–2553. [Google Scholar] [CrossRef] [PubMed]
- Durso, N.A.; Cyr, R.J. A calmodulin-sensitive interaction between microtubules and a higher plant homolog of elongation factor-1a. Plant Cell 1994, 6, 893–905. [Google Scholar] [CrossRef] [PubMed]
- Olmsted, J.B. Microtubule-associated proteins. Annu. Rev. Cell Dev. B 1986, 2, 421. [Google Scholar] [CrossRef] [PubMed]
- Chapin, S.J.; Bulinski, J.C. Microtubule stabilization by assembly-promoting microtubule-associated proteins: A repeat performance. Cell Motil. Cytoskelet. 1992, 23, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Hirokawa, N. Microtubule organization and dynamics dependent on microtubule-associated proteins. Curr. Opin. Cell Biol. 1994, 6, 74–81. [Google Scholar] [CrossRef]
- Drechsel, D.N.; Hyman, A.A.; Cobb, M.H.; Kirschner, M.W. Modulation of the dynamic instability of tubulin assembly by the microtubule-associated protein tau. Mol. Biol. Cell 1992, 3, 1141–1154. [Google Scholar] [CrossRef] [PubMed]
- Pryer, N.K.; Walker, R.A.; Skeen, V.P.; Bourns, B.D.; Soboeiro, M.F. Brain microtubule-associated proteins modulate microtubule dynamic instability in vitro: Realtime observations using video microscopy. J. Cell Sci. 1992, 103, 965–976. [Google Scholar] [PubMed]
- Gamblin, T.C.; Nachmanoff, K.; Halpain, S.; Williams, R.C., Jr. Recombinant microtubule-associated protein 2c reduces the dynamic instability of individual microtubules. Biochemistry 1996, 35, 12576–12586. [Google Scholar] [CrossRef] [PubMed]
- Gard, D.L.; Kirschner, M.W. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plusend. J. Cell Biol. 1987, 105, 2203–2215. [Google Scholar] [CrossRef] [PubMed]
- Vasquez, R.J.; Gard, D.L.; Cassimeris, L. XMAP from Xenopus eggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. J. Cell Biol. 1994, 127, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Dhamodharan, R.; Wadsworth, P. Modulation of microtubule dynamic instability in vivo by brain microtubule associated proteins. J. Cell Sci. 1995, 108, 1679–1689. [Google Scholar] [PubMed]
- Wang, Y.; Ding, Y.; Wang, S.; Chen, H.; Zhang, H.; Chen, W.; Gu, Z.; Chen, Y.Q. Extract of Syzygiumaromaticum suppress eEF1A protein expression and fungal growth. J. Appl. Microbiol. 2017, 123, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.K.; Carp, M.J.; Zuchman, R.; Ziv, T.; Horwitz, B.A.; Gepstein, S. Proteomics of the response of Arabidopsis thaliana to infection with Alternaria brassicicola. J. Proteom. 2010, 73, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Kessler, S.A.; Shimosato-Asano, H.; Keinath, N.F.; Wuest, S.E.; Ingram, G.; Panstruga, R.; Grossniklaus, U. Conserved Molecular Componentsfor Pollen Tube Reception and Fungal Invasion. Science 2010, 330, 968–971. [Google Scholar] [CrossRef] [PubMed]
- Footitt, S.; Dietrich, D.; Fait, A.; Fernie, A.R.; Holdsworth, M.J.; Baker, A.; Theodoulou, F.L. The COMATOSE ATP-Binding Cassette Transporter IsRequired for Full Fertility in Arabidopsis. Plant Physiol. 2007, 144, 1467–1480. [Google Scholar] [CrossRef] [PubMed]
- Popova, O.B.; Baker, M.R.; Tran, T.P.; Le, T.; Serysheva, I.I. Identification of ATP-Binding Regions in the RyR1 Ca2+ Release Channel. PLoS ONE 2012, 7, e48725. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Chen, C.; Yang, A.; Chen, Y.; Liu, Q.; Ni, J. Redox proteomics identification of specifically carbonylated proteins in the hippocampi of triple transgenic Alzheimer's disease mice at its earliest pathological stage. J. Proteom. 2015, 123, 101–113. [Google Scholar] [CrossRef] [PubMed]
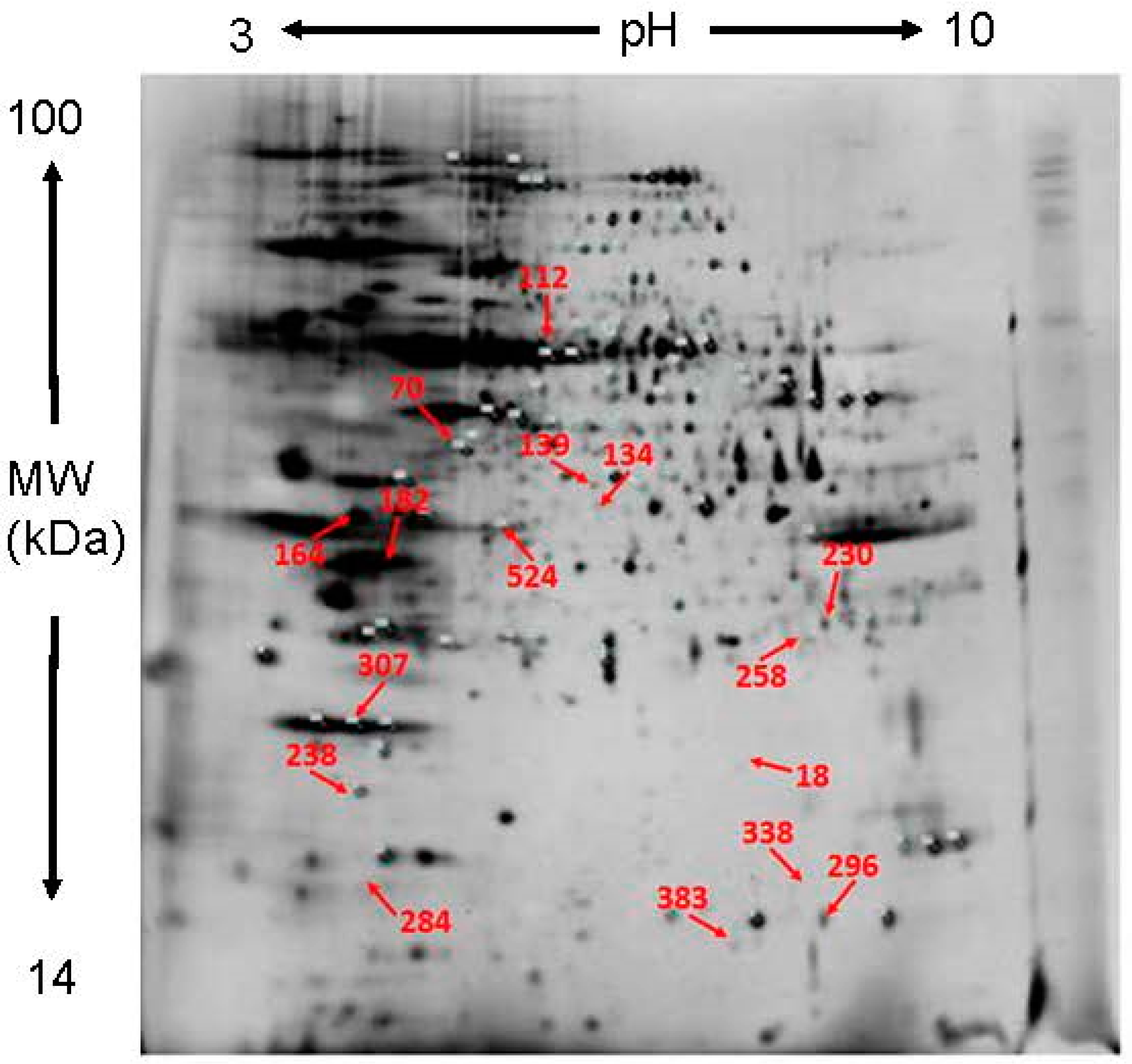
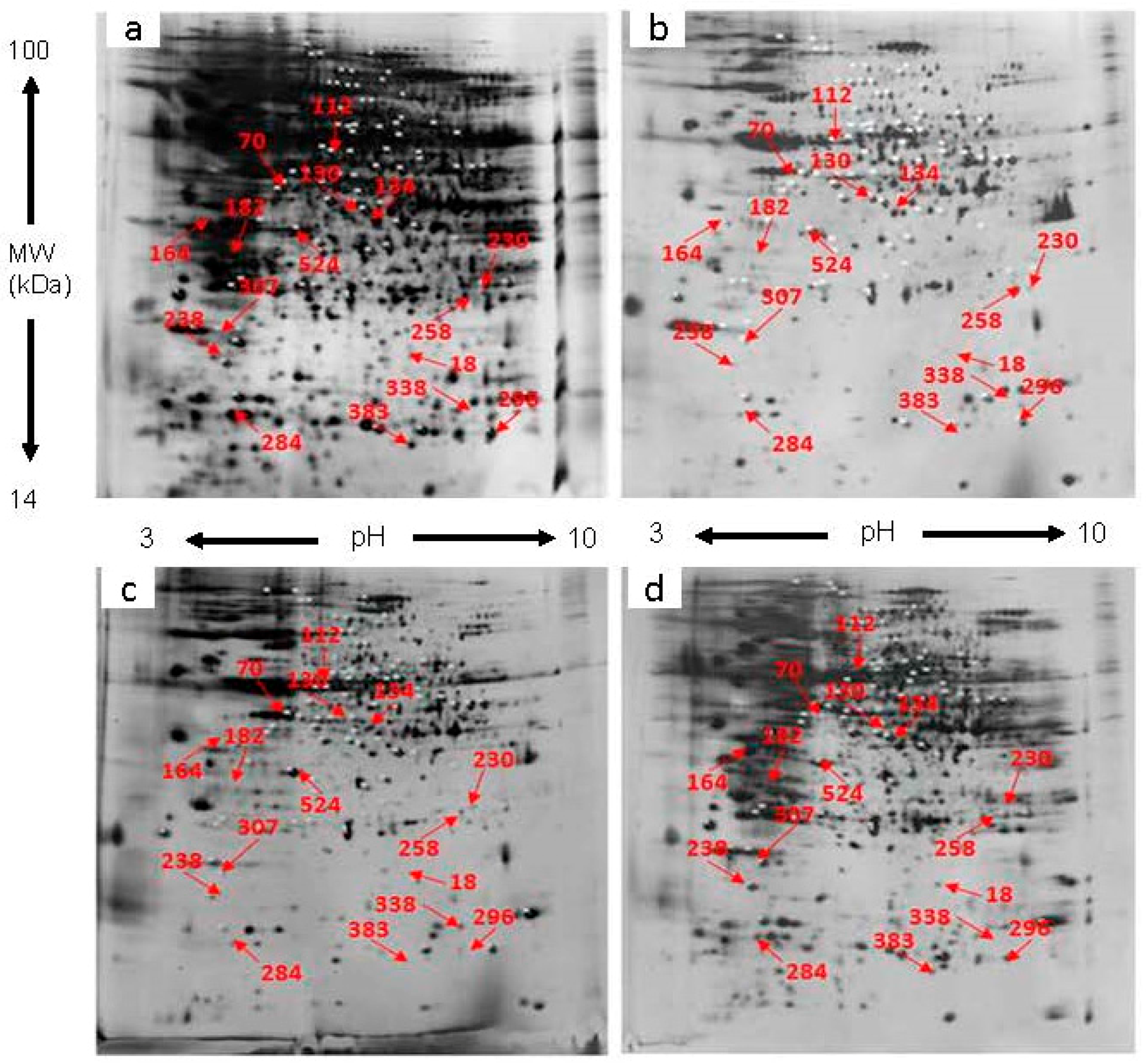
| Spot Number | Protein Name | Accession Number | MW (Da) | Protein PI | Mascot Score | Matched Peptides | Sequence Coverage (%) | Function | Fold Increase (+) or Decrease (−) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SP 2 | SP 4 | CP 2 | CP 4 | |||||||||
| 18 | Dendrobiumnobile cDNA | HO192673 | 27,176 | 8.31 | 408 | 3 | 20 | fatty acid β-oxidation | −31.2 ± 2.23 | −1.8 ± 0.14 | 31.2 ± 2.46 | 20.8 ± 1.22 |
| 31 | Phalaenopsisequestris cDNA clone EFCP035A12 | CB033636 | 24,814 | 9.1 | 235 | 2 | 15 | Phosphopyruvatehydratase activity | −34.4 ± 1.80 | 8.6 ± 0.50 | 15.8 ± 0.87 | 34.4 ± 3.04 |
| 33 | Dendrobiumnobile cDNA | HO193941 | 27,570 | 6.03 | 194 | 2 | 12 | Transferase | −7.6 ± 0.59 | −8.4 ± 0.47 | 4.8 ± 0.43 | 8.4 ± 0.63 |
| 62 | Dendrobiumnobile cDNA | HO189262 | 29,674 | 5.81 | 368 | 3 | 19 | response to stress | −16.3 ± 1.51 | 16.3 ± 1.15 | 9.5 ± 0.77 | −1.9 ± 0.17 |
| 70 | flavoprotein subunit 2 [Arabidopsis thaliana] | NP_179435 | 70,015 | 5.85 | 189 | 4 | 9 | mitochondrial electron transport, succinate to ubiquinone | −16.1 ± 1.40 | 10.4 ± 0.90 | 16.1 ± 1.39 | 8.7 ± 0.61 |
| 71 | Texas blueweed Helianthus ciliaris CHCL8946 | EL420682 | 31,916 | 5.93 | 80 | 1 | 5 | Zein-binding | −19.4 ± 1.03 | 19.4 ± 1.02 | 9.2 ± 0.60 | 5.1 ± 0.35 |
| 81 | PRUPE_ppa003377mg [Prunuspersica] | EMJ16224 | 62,014 | 5.8 | 179 | 2 | 5 | metabolic process magnesium ion binding | −6.5 ± 0.59 | 6.5 ± 0.33 | 3.0 ± 0.28 | −1.8 ± 0.14 |
| 91 | Phalaenopsisviolacea cDNA | CK857713 | 29,392 | 6.99 | 114 | 1 | 6 | ATP-binding, protein folding | −20.4 ± 1.24 | 18.3 ± 1.53 | 14.3 ± 1.33 | 20.4 ± 1.17 |
| 100 | Triticum aestivum cDNA clone wl1n.pk0005.g10 | CA616775 | 17,053 | 9.3 | 82 | 1 | 9 | transmembrane transport | −29.7 ± 2.22 | 13.3 ± 1.15 | 13.1 ± 0.74 | 29.7 ± 2.44 |
| 107 | Festuca pratensis cDNA clone 29N21 | GO893814 | 23,943 | 10.05 | 76 | 1 | 6 | uncharacterized protein | −46.4 ± 3.34 | 13.6 ± 0.76 | 37.0 ± 2.83 | 46.4 ± 2.99 |
| 112 | 5-methyltetrahydropteroyl triglutamate-homocysteine [Populustrichocarpa] | XP_002319710 | 85,370 | 6.1 | 261 | 4 | 6 | zinc ion binding, methionine biosynthetic process | −91.9 ± 6.03 | 11.3 ± 0.96 | 91.9 ± 6.36 | 48.6 ± 3.53 |
| 127 | monodehydroascorbate reductase [Oncidium hybrid cultivar] | ACJ38541 | 46,809 | 5.26 | 205 | 4 | 17 | oxidoreductase activity | −80.5 ± 6.31 | 19.3 ± 1.48 | 6.7 ± 0.58 | −3.5 ± 0.22 |
| 128 | Malus x domestica cDNA | DT002244 | 24,691 | 8.31 | 73 | 1 | 6 | Acyltransferase | −215.2 ± 11.83 | 71.8 ± 3.97 | 215.2 ± 16.77 | 5.9 ± 0.41 |
| 134 | VITISV_034728 [Vitis vinifera] | CAN70186 | 53,150 | 6.76 | 280 | 5 | 12 | generating NADPH | −4.5 ± 0.29 | 2.0 ± 0.13 | 4.5 ± 0.32 | 2.7 ± 0.16 |
| 139 | PRUPE_ppa003869mg [Prunuspersica] | EMJ11768 | 59,411 | 6.69 | 116 | 2 | 4 | oxidoreductase activity NAD or NADP as acceptor | −12.7 ± 0.75 | 4.2 ± 0.28 | 10.0 ± 0.91 | 12.7 ± 1.31 |
| 151 | Oncidium Gower Ramsey cDNA | HS521850 | 30,518 | 6 | 166 | 2 | 11 | ATP-binding, Metal-binding, succinate-CoA ligase activity | −1.1 ± 0.10 | 2.9 ± 0.27 | 2.0 ± 0.17 | 2.8 ± 0.24 |
| 154 | Soybean Seeds Containing Globular-Stage Embryos Glycine max cDNA | GD856994 | 3073 | 5.69 | 73 | 1 | 55 | protein methyltransferase activity | −15.8 ± 1.42 | 12.8 ± 0.94 | 15.8 ± 1.12 | 14.8 ± 1.34 |
| 161 | Phalaenopsis equestris cDNA clone EFCP035E06 | CB033673 | 20,026 | 7.83 | 390 | 3 | 32 | magnesium ion binding Methionine biosynthesis | −10.4 ± 0.62 | 3.0 ± 0.18 | 10.4 ± 0.78 | 2.0 ± 0.16 |
| 162 | DAFB seeds Malus x domestica cDNA clone AAWA002059 | CN887431 | 21,889 | 11.14 | 70 | 1 | 5 | May play a role in plant defense | −15.2 ± 0.88 | 5.6 ± 0.30 | 10.3 ± 0.81 | 15.2 ± 0.88 |
| 164 | monodehydroascorbate reductase [Oncidium hybrid cultivar] | ACJ38541 | 46,809 | 5.26 | 206 | 4 | 17 | oxidoreductase activity | −343.3 ± 20.59 | 47.3 ± 3.42 | 136.3 ± 7.43 | 343.3 ± 25.51 |
| 165 | Dendrobiumnobile cDNA | HO189275 | 26,298 | 7.88 | 419 | 4 | 22 | phosphoglycerate kinase activity | −13.3 ± 0.72 | 13.3 ± 1.01 | 5.2 ± 0.39 | 2.3 ± 0.17 |
| 178 | Triphysariaversicolor cDNA | EY010367 | 23,141 | 10 | 77 | 1 | 6 | hydrolase activity | −20.5 ± 1.55 | 2.8 ± 0.17 | 11.7 ± 0.94 | 20.5 ± 1.38 |
| 182 | Os02g0735200 [Oryza sativa Japonica Group] | NP_001048045 | 39,405 | 5.51 | 238 | 2 | 14 | High-affinity glutamine synthetase | −35.8 ± 3.07 | 35.8 ± 2.55 | 21.0 ± 2.09 | 20.6 ± 1.62 |
| 184 | Dendrobiumnobile cDNA | HO197113 | 25,515 | 5.7 | 176 | 1 | 12 | ATP binding, MAP kinase activity | −25.1 ± 1.53 | 9.9 ± 0.84 | 7.2 ± 0.40 | 25.1 ± 2.49 |
| 187 | Solanumhabrochaites cDNA | GT169059 | 40,752 | 8.53 | 246 | 3 | 11 | GTP-binding, protein transport | −10.1 ± 0.82 | 1.2 ± 0.09 | 5.9 ± 0.31 | 10.1 ± 0.89 |
| 204 | Dendrobiumnobile cDNA | HO203854 | 30,487 | 8.35 | 221 | 2 | 13 | tricarboxylic acid cycle malate metabolic process | −1.1 ± 0.10 | 1.8 ± 0.14 | 2.2 ± 0.16 | 3.1 ± 0.23 |
| Dendrobiumnobile cDNA | HO196589 | 23,411 | 7.82 | 161 | 2 | 12 | ||||||
| 217 | Dendrobiumnobile cDNA | HO203393 | 29,572 | 9 | 234 | 3 | 16 | malate metabolic process Oxidoreductase | −14.5 ± 0.81 | 14.5 ± 0.85 | 10.2 ± 0.75 | 14.0 ± 0.83 |
| 221 | peptide ABC transporter substrate -binding protein [Bacilluscereus] | YP_002368400 | 63,740 | 8.69 | 662 | 5 | 13 | Signal, ATP-driven transport Metal-binding | −2.2 ± 0.21 | 2.2 ± 0.15 | 3.9 ± 0.29 | 3.9 ± 0.23 |
| 230 | sunflower Helianthus annuus cDNA clone CCFS4413 | GE489969 | 30,766 | 9.36 | 86 | 1 | 5 | microtubule motor activity | 2.5 ± 0.21 | 10.8 ± 0.90 | 9.0 ± 0.51 | 3.8 ± 0.24 |
| 232 | Vanda hybrid cultivar cDNA | GW392872 | 19,682 | 9.47 | 119 | 1 | 9 | oxidoreductaseactivity, zinc ion binding | −4.6 ± 0.26 | 4.8 ± 0.31 | 3.2 ± 0.27 | 4.8 ± 0.28 |
| 233 | Dendrobiumnobile cDNA | HO195954 | 27,748 | 5.56 | 112 | 1 | 7 | carboxylesterase activity | −13.6 ± 1.19 | 10.0 ± 0.93 | 13.1 ± 0.90 | 2.9 ± 0.21 |
| 238 | Dendrobiumnobile cDNA | HO189451 | 25,061 | 5.47 | 217 | 2 | 13 | Unknown protein | −14.7 ± 1.46 | 8.4 ± 0.51 | 14.7 ± 0.10 | 13.2 ± 0.94 |
| 240 | Mimulusguttatus cDNA clone CCIG14980 | GR000041 | 27,311 | 9.2 | 109 | 1 | 6 | regulation of translational initiation, translation initiation factor activity | 2.4 ± 0.19 | 11.0 ± 0.84 | 12.2 ± 0.77 | 6.7 ± 0.40 |
| 247 | putative enoyl-ACP-reductase protein [Elaeisguineensis] | AEZ00840 | 38,749 | 9.27 | 229 | 3 | 18 | Oxidoreductase, enoyl-[acyl-carrier-protein] reductase (NADH) activity | 2.7 ± 1.67 | 5.0 ± 0.41 | 4.5 ± 0.35 | 5.0 ± 0.33 |
| 249 | Dendrobiumnobile cDNA | HO192097 | 24,344 | 5.22 | 152 | 2 | 11 | response to stress, oxidoreductase activity | 4.4 ± 0.36 | 6.2 ± 0.45 | 8.2 ± 0.64 | 13.7 ± 1.21 |
| Dendrobiumnobile cDNA | HO190191 | 26,789 | 9.13 | 156 | 2 | 11 | ||||||
| 258 | Oncidium Gower Ramsey cDNA | HS521951 | 30,535 | 8.96 | 194 | 2 | 11 | Uncharacterized protein | 1.4 ± 0.09 | 4.0 ± 0.20 | 2.9 ± 0.27 | 6.1 ± 0.41 |
| 261 | Dendrobiumnobile cDNA | HO196032 | 28,575 | 5.99 | 252 | 2 | 11 | Uncharacterized protein | 3.1 ± 0.25 | 2.2 ± 0.20 | 4.5 ± 0.23 | 4.5 ± 0.27 |
| Dendrobiumnobile cDNA | HO201509 | 27,739 | 8.26 | 318 | 2 | 14 | ||||||
| 284 | Dendrobiumnobile cDNA | HO192248 | 23,859 | 5.75 | 99 | 1 | 6 | Uncharacterized protein | 2.3 ± 0.21 | −3.1 ± 0.30 | 3.1 ± 0.22 | 5.2 ± 0.30 |
| 294 | Panicumvirgatum cDNA | JG964858 | 30,004 | 9.4 | 84 | 1 | 5 | transporter activity | 2.0 ± 0.17 | 2.7 ± 0.22 | 4.6 ± 0.38 | 4.6 ± 0.44 |
| 296 | Dendrobiumnobile cDNA | HO189346 | 23,335 | 9.36 | 271 | 2 | 18 | ubiquitin-dependent protein | 3.6 ± 0.32 | −9.2 ± 0.85 | 12.3 ± 1.16 | 12.3 ± 0.91 |
| 303 | fibrillin-like protein [Oncidium hybrid cultivar] | AAY24688 | 34,734 | 5.48 | 85 | 2 | 10 | structural molecule activity | 1.4 ± 0.10 | 2.2 ± 0.14 | 4.2 ± 0.40 | 4.2 ± 0.32 |
| Oncidium hybrid cultivar cDNA | HS524185 | 24,403 | 8.12 | 249 | 2 | 14 | ||||||
| 307 | ascorbate peroxidase [Oncidium hybrid cultivar] | ACJ38537 | 27,441 | 5.34 | 241 | 2 | 20 | response to oxidative stress | 1.2 ± 0.11 | 2.8 ± 0.22 | 4.1 ± 0.28 | 4.1 ± 0.32 |
| 311 | Dendrobiumnobile cDNA | HO202862 | 29,065 | 5.26 | 655 | 4 | 26 | triose-phosphate isomerase activity | 1.3 ± 0.12 | 3.1 ± 0.21 | 3.7 ± 0.29 | 3.7 ± 0.22 |
| 320 | Oncidium Gower Ramsey cDNA | HS522419 | 32,789 | 5.48 | 79 | 1 | 4 | Zinc phosphodiesterase, Endonuclease | −1.5 ± 0.12 | 2.3 ± 0.19 | 2.2 ± 0.20 | 1.9 ± 0.14 |
| 329 | Coffeaarabica cDNA clone CAET42MIX-CFEZE47TVC | GT010034 | 32,267 | 8.06 | 139 | 1 | 5 | Thaumatin-like protein | 2.1 ± 0.18 | 8.7 ± 0.85 | 3.7 ± 0.25 | 8.1 ± 0.70 |
| 357 | Dendrobiumnobile cDNA | HO198288 | 24,617 | 7.72 | 217 | 3 | 15 | PPIases accelerate the folding of proteins | −15.3 ± 0.91 | 10.3 ± 0.84 | 15.3 ± 1.08 | 12.4 ± 1.09 |
| 360 | unknown [Piceasitchensis] | gi|116779193 | 18,169 | 8.34 | 102 | 2 | 14 | peptidyl-prolylcis-trans isomerase activity | −11.0 ± 0.77 | 11.0 ± 0.83 | 6.0 ± 0.51 | −4.4 ± 0.24 |
| 363 | peroxiredoxin 5 cell rescue protein [Loliumperenne] | AFA36612 | 11,445 | 5.13 | 160 | 2 | 31 | oxidation-reduction process | 2.0 ± 1.48 | 2.5 ± 0.19 | 6.6 ± 0.34 | 6.6 ± 0.62 |
| 383 | Ophrysfusca cDNA clone Ofup2722 | HO849917 | 19,693 | 8.11 | 98 | 1 | 7 | defense response | −9.6 ± 0.91 | 5.6 ± 0.31 | −14.7 ± 1.06 | 14.7 ± 0.79 |
| 388 | Dendrobiumnobile cDNA | HO198066 | 24,490 | 9.24 | 235 | 3 | 17 | defense response | −17.9 ± 1.28 | 11.4 ± 0.88 | 5.6 ± 0.54 | 18.0 ± 1.47 |
| 464 | lettuce serriola Lactuca serriola cDNA clone QGH6B22 | BU007993 | 23,103 | 4.84 | 112 | 1 | 9 | ATP-binding, Formation of phosphoenolpyruvate | −61.9 ± 3.71 | 61.9 ± 4.49 | 50.6 ± 4.57 | −1.1 ± 0.10 |
| 524 | translational elongation factor EF-TuM [Zea mays] | AAG32661 | 48,746 | 5.99 | 390 | 4 | 22 | translation elongation factor activity, GTP catabolic process | −3.1 ± 0.21 | 3.1 ± 0.25 | 2.9 ± 1.74 | 1.5 ± 0.15 |
| 530 | M569_05826, partial [Genliseaaurea] | EPS68937 | 48,363 | 8.25 | 291 | 3 | 13 | transferase activity | −15.0 ± 0.94 | 15.0 ± 1.24 | 15.0 ± 1.18 | 1.6 ± 0.16 |
| 531 | Dendrobiumnobile cDNA | HO193012 | 26,029 | 6.5 | 299 | 2 | 16 | zinc-containing alcohol dehydrogenase family | −4.7 ± 0.35 | 12.2 ± 0.70 | 9.8 ± 0.60 | 1.1 ± 0.08 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Yu, H.; Li, T.; Li, L.; Zhang, G.; Liu, Z.; Huang, T.; Zhang, Y. Comparative Proteomics Analyses of Pollination Response in Endangered Orchid Species Dendrobium Chrysanthum. Int. J. Mol. Sci. 2017, 18, 2496. https://doi.org/10.3390/ijms18122496
Wang W, Yu H, Li T, Li L, Zhang G, Liu Z, Huang T, Zhang Y. Comparative Proteomics Analyses of Pollination Response in Endangered Orchid Species Dendrobium Chrysanthum. International Journal of Molecular Sciences. 2017; 18(12):2496. https://doi.org/10.3390/ijms18122496
Chicago/Turabian StyleWang, Wei, Hongyang Yu, Tinghai Li, Lexing Li, Guoqiang Zhang, Zhongjian Liu, Tengbo Huang, and Yongxia Zhang. 2017. "Comparative Proteomics Analyses of Pollination Response in Endangered Orchid Species Dendrobium Chrysanthum" International Journal of Molecular Sciences 18, no. 12: 2496. https://doi.org/10.3390/ijms18122496
APA StyleWang, W., Yu, H., Li, T., Li, L., Zhang, G., Liu, Z., Huang, T., & Zhang, Y. (2017). Comparative Proteomics Analyses of Pollination Response in Endangered Orchid Species Dendrobium Chrysanthum. International Journal of Molecular Sciences, 18(12), 2496. https://doi.org/10.3390/ijms18122496





