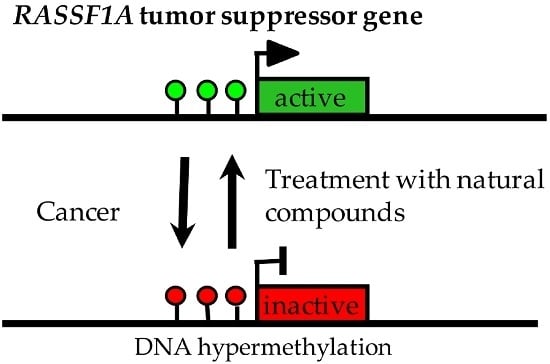
Impact of Natural Compounds on DNA Methylation Levels of the Tumor Suppressor Gene RASSF1A in Cancer
Abstract
1. Introduction
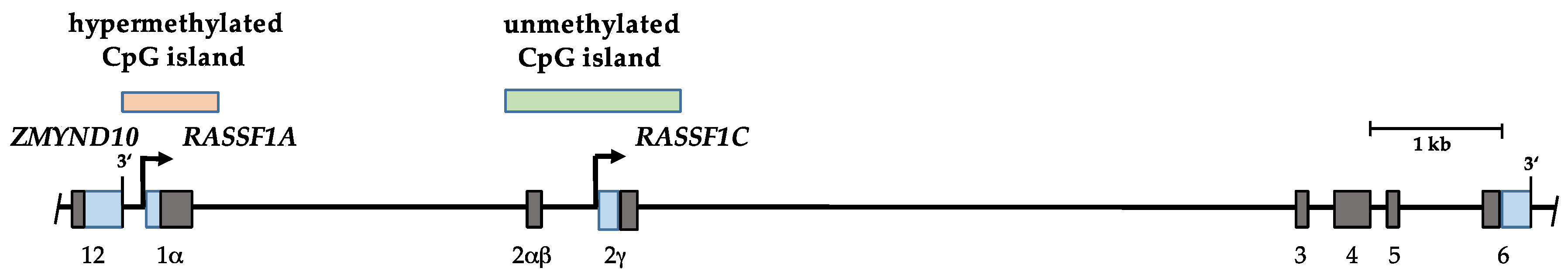
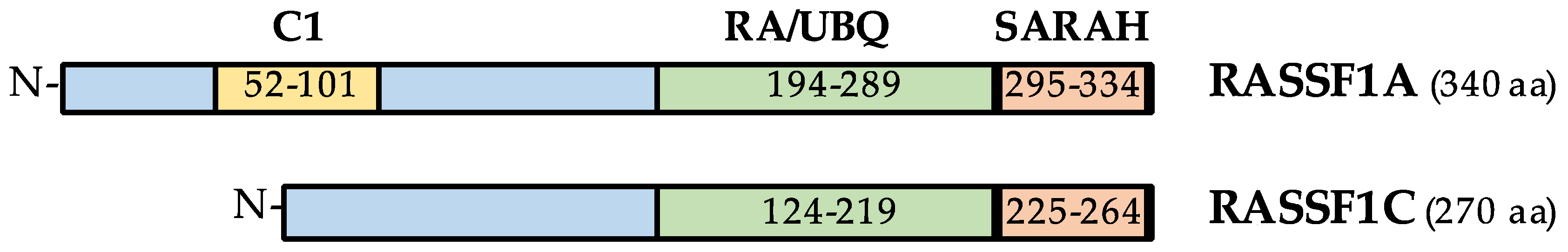
2. The Ras Association Domain Family 1 (RASSF1) Gene
3. Hypermethylation of RASSF1A in Human Cancers
4. Demethylation of RASSF1A by Treatment of Cancer Cells with Cytidine Analogues
5. Effects of Methyl Donors and Vitamins on RASSF1A Methylation
6. Impact of Naturally Occurring Polyphenols on RASSF1A Methylation
7. Effects of Other Natural Compounds on RASSF1A Methylation
8. Conclusions
Acknowledgments
Conflicts of Interest
Abbreviations
| RASSF | Ras Association Domain Family |
| TSG | tumor suppressor gene |
| DNMT | DNA methyltransferase |
| HDAC | histone deacetylase |
| EGCG | epigalloctechin-3-gallate |
| LINE1 | long interspersed nuclear element 1 |
| PEITC | phenethyl isothiocyanate |
| aa | amino acids |
References
- Iida, T.; Suetake, I.; Tajima, S.; Morioka, H.; Ohta, S.; Obuse, C.; Tsurimoto, T. PCNA clamp facilitates action of DNA cytosine methyltransferase 1 on hemimethylated DNA. Genes Cells 2002, 7, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Sharif, J.; Muto, M.; Takebayashi, S.; Suetake, I.; Iwamatsu, A.; Endo, T.A.; Shinga, J.; Mizutani-Koseki, Y.; Toyoda, T.; Okamura, K.; et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting DNMT1 to methylated DNA. Nature 2007, 450, 908–912. [Google Scholar] [CrossRef] [PubMed]
- Richter, A.M.; Pfeifer, G.P.; Dammann, R.H. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim. Biophys. Acta 2009, 1796, 114–128. [Google Scholar] [CrossRef] [PubMed]
- Dammann, R.; Li, C.; Yoon, J.H.; Chin, P.L.; Bates, S.; Pfeifer, G.P. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat. Genet. 2000, 25, 315–319. [Google Scholar] [PubMed]
- Sekido, Y.; Ahmadian, M.; Wistuba, I.I.; Latif, F.; Bader, S.; Wei, M.H.; Duh, F.M.; Gazdar, A.F.; Lerman, M.I.; Minna, J.D. Cloning of a breast cancer homozygous deletion junction narrows the region of search for a 3p21.3 tumor suppressor gene. Oncogene 1998, 16, 3151–3157. [Google Scholar] [CrossRef] [PubMed]
- Kok, K.; Naylor, S.L.; Buys, C.H. Deletions of the short arm of chromosome 3 in solid tumors and the search for suppressor genes. Adv. Cancer Res. 1997, 71, 27–92. [Google Scholar] [PubMed]
- Burbee, D.G.; Forgacs, E.; Zochbauer-Muller, S.; Shivakumar, L.; Fong, K.; Gao, B.; Randle, D.; Kondo, M.; Virmani, A.; Bader, S.; et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J. Natl. Cancer Inst. 2001, 93, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Vavvas, D.; Li, X.; Avruch, J.; Zhang, X.F. Identification of Nore1 as a potential Ras effector. J. Biol. Chem. 1998, 273, 5439–5442. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Vega, S.; Khokhlatchev, A.; Nedwidek, M.; Zhang, X.F.; Dammann, R.; Pfeifer, G.P.; Avruch, J. The putative tumor suppressor RASSF1A homodimerizes and heterodimerizes with the Ras-GTP binding protein Nore1. Oncogene 2002, 21, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Bunney, T.D.; Harris, R.; Gandarillas, N.L.; Josephs, M.B.; Roe, S.M.; Sorli, S.C.; Paterson, H.F.; Rodrigues-Lima, F.; Esposito, D.; Ponting, C.P.; et al. Structural and mechanistic insights into ras association domains of phospholipase C epsilon. Mol. Cell 2006, 21, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Scheel, H.; Hofmann, K. A novel interaction motif, SARAH, connects three classes of tumor suppressor. Curr. Biol. CB 2003, 13, R899–R900. [Google Scholar] [CrossRef] [PubMed]
- Schagdarsurengin, U.; Richter, A.M.; Hornung, J.; Lange, C.; Steinmann, K.; Dammann, R.H. Frequent epigenetic inactivation of RASSF2 in thyroid cancer and functional consequences. Mol. Cancer 2010, 9, 264. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Tommasi, S.; Liu, L.; Yee, J.K.; Dammann, R.; Pfeifer, G.P. RASSF1A is part of a complex similar to the Drosophila Hippo/Salvador/Lats tumor-suppressor network. Curr. Biol. CB 2007, 17, 700–705. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Huang, J.; Dong, J.; Pan, D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 2003, 114, 445–456. [Google Scholar] [CrossRef]
- Zhao, B.; Tumaneng, K.; Guan, K.L. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat. Cell Biol. 2011, 13, 877–883. [Google Scholar] [CrossRef] [PubMed]
- Sudol, M.; Bork, P.; Einbond, A.; Kastury, K.; Druck, T.; Negrini, M.; Huebner, K.; Lehman, D. Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J. Biol. Chem. 1995, 270, 14733–14741. [Google Scholar] [CrossRef] [PubMed]
- Moroishi, T.; Hansen, C.G.; Guan, K.L. The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer 2015, 15, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Dittfeld, C.; Richter, A.M.; Steinmann, K.; Klagge-Ulonska, A.; Dammann, R.H. The SARAH Domain of RASSF1A and Its Tumor Suppressor Function. Mol. Biol. Int. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, A.P.; Traum, A.; Boettger, T.; Hackstein, H.; Richter, A.M.; Dammann, R.H. The tumor suppressor RASSF1A induces the YAP1 target gene ANKRD1 that is epigenetically inactivated in human cancers and inhibits tumor growth. Oncotarget 2017. [Google Scholar] [CrossRef]
- Steinmann, K.; Sandner, A.; Schagdarsurengin, U.; Dammann, R.H. Frequent promoter hypermethylation of tumor-related genes in head and neck squamous cell carcinoma. Oncol. Rep. 2009, 22, 1519–1526. [Google Scholar] [PubMed]
- Zhang, X.; Guo, C.; Wu, X.; Li, A.X.; Liu, L.; Tsark, W.; Dammann, R.; Shen, H.; Vonderfecht, S.L.; Pfeifer, G.P. Analysis of Liver Tumor-Prone Mouse Models of the Hippo Kinase Scaffold Proteins RASSF1A and SAV1. Cancer Res. 2016, 76, 2824–2835. [Google Scholar] [CrossRef] [PubMed]
- Shivakumar, L.; Minna, J.; Sakamaki, T.; Pestell, R.; White, M.A. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol. Cell. Biol. 2002, 22, 4309–4318. [Google Scholar] [CrossRef] [PubMed]
- Thaler, S.; Hahnel, P.S.; Schad, A.; Dammann, R.; Schuler, M. RASSF1A mediates p21Cip1/Waf1-dependent cell cycle arrest and senescence through modulation of the Raf-MEK-ERK pathway and inhibition of Akt. Cancer Res. 2009, 69, 1748–1757. [Google Scholar] [CrossRef] [PubMed]
- Dammann, R.; Takahashi, T.; Pfeifer, G.P. The CpG island of the novel tumor suppressor gene RASSF1A is intensely methylated in primary small cell lung carcinomas. Oncogene 2001, 20, 3563–3567. [Google Scholar] [CrossRef] [PubMed]
- Helmbold, P.; Lahtz, C.; Herpel, E.; Schnabel, P.A.; Dammann, R.H. Frequent hypermethylation of RASSF1A tumour suppressor gene promoter and presence of Merkel cell polyomavirus in small cell lung cancer. Eur. J. Cancer 2009, 45, 2207–2211. [Google Scholar] [CrossRef] [PubMed]
- Dammann, R.; Strunnikova, M.; Schagdarsurengin, U.; Rastetter, M.; Papritz, M.; Hattenhorst, U.E.; Hofmann, H.S.; Silber, R.E.; Burdach, S.; Hansen, G. CpG island methylation and expression of tumour-associated genes in lung carcinoma. Eur. J. Cancer 2005, 41, 1223–1236. [Google Scholar] [CrossRef] [PubMed]
- Dammann, R.; Yang, G.; Pfeifer, G.P. Hypermethylation of the cpG island of Ras association domain family 1A (RASSF1A), a putative tumor suppressor gene from the 3p21.3 locus, occurs in a large percentage of human breast cancers. Cancer Res. 2001, 61, 3105–3109. [Google Scholar] [PubMed]
- Schagdarsurengin, U.; Wilkens, L.; Steinemann, D.; Flemming, P.; Kreipe, H.H.; Pfeifer, G.P.; Schlegelberger, B.; Dammann, R. Frequent epigenetic inactivation of the RASSF1A gene in hepatocellular carcinoma. Oncogene 2003, 22, 1866–1871. [Google Scholar] [CrossRef] [PubMed]
- Dammann, R.; Schagdarsurengin, U.; Liu, L.; Otto, N.; Gimm, O.; Dralle, H.; Boehm, B.O.; Pfeifer, G.P.; Hoang-Vu, C. Frequent RASSF1A promoter hypermethylation and K-ras mutations in pancreatic carcinoma. Oncogene 2003, 22, 3806–3812. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yoon, J.H.; Dammann, R.; Pfeifer, G.P. Frequent hypermethylation of the RASSF1A gene in prostate cancer. Oncogene 2002, 21, 6835–6840. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Dammann, R.; Pfeifer, G.P. Hypermethylation of the CpG island of the RASSF1A gene in ovarian and renal cell carcinomas. Int. J. Cancer 2001, 94, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, C.; Martinez, A.; Zatyka, M.; Agathanggelou, A.; Honorio, S.; Astuti, D.; Morgan, N.V.; Moch, H.; Richards, F.M.; Kishida, T.; et al. Epigenetic inactivation of the RASSF1A 3p21.3 tumor suppressor gene in both clear cell and papillary renal cell carcinoma. Cancer Res. 2001, 61, 7277–7281. [Google Scholar] [PubMed]
- Horiguchi, K.; Tomizawa, Y.; Tosaka, M.; Ishiuchi, S.; Kurihara, H.; Mori, M.; Saito, N. Epigenetic inactivation of RASSF1A candidate tumor suppressor gene at 3p21.3 in brain tumors. Oncogene 2003, 22, 7862–7865. [Google Scholar] [CrossRef] [PubMed]
- Byun, D.S.; Lee, M.G.; Chae, K.S.; Ryu, B.G.; Chi, S.G. Frequent epigenetic inactivation of RASSF1A by aberrant promoter hypermethylation in human gastric adenocarcinoma. Cancer Res. 2001, 61, 7034–7038. [Google Scholar] [PubMed]
- Dammann, R.; Schagdarsurengin, U.; Seidel, C.; Trumpler, C.; Hoang-Vu, C.; Gimm, O.; Dralle, H.; Pfeifer, G.P.; Brauckhoff, M. Frequent promoter methylation of tumor-related genes in sporadic and men2-associated pheochromocytomas. Exp. Clin. Endocrinol. Diabetes 2005, 113, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Schagdarsurengin, U.; Gimm, O.; Hoang-Vu, C.; Dralle, H.; Pfeifer, G.P.; Dammann, R. Frequent epigenetic silencing of the CpG island promoter of RASSF1A in thyroid carcinoma. Cancer Res. 2002, 62, 3698–3701. [Google Scholar] [PubMed]
- Chow, L.S.; Lo, K.W.; Kwong, J.; To, K.F.; Tsang, K.S.; Lam, C.W.; Dammann, R.; Huang, D.P. RASSF1A is a target tumor suppressor from 3p21.3 in nasopharyngeal carcinoma. Int. J. Cancer 2004, 109, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Dammann, R.; Schagdarsurengin, U.; Seidel, C.; Strunnikova, M.; Rastetter, M.; Baier, K.; Pfeifer, G.P. The tumor suppressor RASSF1A in human carcinogenesis: An update. Histol. Histopathol. 2005, 20, 645–663. [Google Scholar] [PubMed]
- Spugnardi, M.; Tommasi, S.; Dammann, R.; Pfeifer, G.P.; Hoon, D.S. Epigenetic inactivation of RAS association domain family protein 1 (RASSF1A) in malignant cutaneous melanoma. Cancer Res. 2003, 63, 1639–1643. [Google Scholar] [PubMed]
- Rastetter, M.; Schagdarsurengin, U.; Lahtz, C.; Fiedler, E.; Marsch, W.; Dammann, R.; Helmbold, P. Frequent intra-tumoural heterogeneity of promoter hypermethylation in malignant melanoma. Histol. Histopathol. 2007, 22, 1005–1015. [Google Scholar] [PubMed]
- Helmbold, P.; Lahtz, C.; Enk, A.; Herrmann-Trost, P.; Marsch, W.; Kutzner, H.; Dammann, R.H. Frequent occurrence of RASSF1A promoter hypermethylation and Merkel cell polyomavirus in Merkel cell carcinoma. Mol. Carcinog. 2009, 48, 903–909. [Google Scholar] [CrossRef] [PubMed]
- Seidel, C.; Bartel, F.; Rastetter, M.; Bluemke, K.; Wurl, P.; Taubert, H.; Dammann, R. Alterations of cancer-related genes in soft tissue sarcomas: Hypermethylation of RASSF1A is frequently detected in leiomyosarcoma and associated with poor prognosis in sarcoma. Int. J. Cancer 2005, 114, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Toyooka, S.; Maitra, A.; Maruyama, R.; Toyooka, K.O.; Timmons, C.F.; Tomlinson, G.E.; Mastrangelo, D.; Hay, R.J.; Minna, J.D.; et al. Aberrant promoter methylation and silencing of the RASSF1A gene in pediatric tumors and cell lines. Oncogene 2002, 21, 4345–4349. [Google Scholar] [CrossRef] [PubMed]
- Kuzmin, I.; Liu, L.; Dammann, R.; Geil, L.; Stanbridge, E.J.; Wilczynski, S.P.; Lerman, M.I.; Pfeifer, G.P. Inactivation of RAS association domain family 1A gene in cervical carcinomas and the role of human papillomavirus infection. Cancer Res. 2003, 63, 1888–1893. [Google Scholar] [PubMed]
- Avramouli, A.; Tsochas, S.; Mandala, E.; Katodritou, E.; Ioannou, M.; Ritis, K.; Speletas, M. Methylation status of RASSF1A in patients with chronic myeloid leukemia. Leuk. Res. 2009, 33, 1130–1132. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.G.; Qiu, G.H.; Fu, L.; Waites, E.R.; Srivastava, G.; Heys, D.; Agathanggelou, A.; Latif, F.; Grundy, R.G.; Mann, J.R.; et al. Frequent epigenetic inactivation of the RASSF1A tumor suppressor gene in Hodgkin’s lymphoma. Oncogene 2004, 23, 1326–1331. [Google Scholar] [CrossRef] [PubMed]
- Schagdarsurengin, U.; Gimm, O.; Dralle, H.; Hoang-Vu, C.; Dammann, R. CpG island methylation of tumor-related promoters occurs preferentially in undifferentiated carcinoma. Thyroid 2006, 16, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Grawenda, A.M.; O’Neill, E. Clinical utility of RASSF1A methylation in human malignancies. Br. J. Cancer 2015, 113, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.H.; Li, Y.Y.; Han, J.Z.; Zhou, L.Y.; Lv, Y.Q.; Zhang, H.L.; Zhao, L. Gene methylation as a powerful biomarker for detection and screening of non-small cell lung cancer in blood. Oncotarget 2017, 8, 31692–31704. [Google Scholar] [CrossRef] [PubMed]
- Strunnikova, M.; Schagdarsurengin, U.; Kehlen, A.; Garbe, J.C.; Stampfer, M.R.; Dammann, R. Chromatin inactivation precedes de novo DNA methylation during the progressive epigenetic silencing of the RASSF1A promoter. Mol. Cell. Biol. 2005, 25, 3923–3933. [Google Scholar] [CrossRef] [PubMed]
- Kiehl, S.; Zimmermann, T.; Savai, R.; Pullamsetti, S.S.; Seeger, W.; Bartkuhn, M.; Dammann, R.H. Epigenetic silencing of downstream genes mediated by tandem orientation in lung cancer. Sci. Rep. 2017, 7, 3896. [Google Scholar] [CrossRef] [PubMed]
- Moore, D.J.; Onoufriadis, A.; Shoemark, A.; Simpson, M.A.; Zur Lage, P.I.; De Castro, S.C.; Bartoloni, L.; Gallone, G.; Petridi, S.; Woollard, W.J.; et al. Mutations in ZMYND10, a gene essential for proper axonemal assembly of inner and outer dynein arms in humans and flies, cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2013, 93, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Tian, Y.; Chlenski, A.; Salwen, H.R.; Lu, Z.; Raj, J.U.; Yang, Q. Valproic acid shows a potent antitumor effect with alteration of DNA methylation in neuroblastoma. Anticancer Drugs 2012, 23, 1054–1066. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, L.; Fong, P.; Yang, J.; Zhang, Z.; Yin, S.; Jiang, S.; Liu, X.; Ju, H.; Huang, L.; et al. An association between overexpression of DNA methyltransferase 3B4 and clear cell renal cell carcinoma. Oncotarget 2017, 8, 19712–19722. [Google Scholar] [CrossRef] [PubMed]
- Palakurthy, R.K.; Wajapeyee, N.; Santra, M.K.; Gazin, C.; Lin, L.; Gobeil, S.; Green, M.R. Epigenetic silencing of the RASSF1A tumor suppressor gene through HOXB3-mediated induction of DNMT3B expression. Mol. Cell 2009, 36, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Soejima, K.; Fang, W.; Rollins, B.J. DNA methyltransferase 3b contributes to oncogenic transformation induced by SV40T antigen and activated Ras. Oncogene 2003, 22, 4723–4733. [Google Scholar] [CrossRef] [PubMed]
- Momparler, R.L. Epigenetic therapy of cancer with 5-aza-2′-deoxycytidine (decitabine). Semin. Oncol. 2005, 32, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Singh, B.N.; Huang, Q.; Li, Z.; Gao, Y.; Mishra, P.; Hwa, Y.L.; Li, J.; Dowdy, S.C.; Jiang, S.W. DNA hypermethylation as a chemotherapy target. Cell Signal. 2011, 23, 1082–1093. [Google Scholar] [CrossRef] [PubMed]
- Issa, J.P.; Kantarjian, H.M.; Kirkpatrick, P. Azacitidine. Nat. Rev. Drug Discov. 2005, 4, 275–276. [Google Scholar] [CrossRef] [PubMed]
- Flotho, C.; Claus, R.; Batz, C.; Schneider, M.; Sandrock, I.; Ihde, S.; Plass, C.; Niemeyer, C.M.; Lubbert, M. The DNA methyltransferase inhibitors azacitidine, decitabine and zebularine exert differential effects on cancer gene expression in acute myeloid leukemia cells. Leukemia 2009, 23, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Kaminskas, E.; Farrell, A.T.; Wang, Y.C.; Sridhara, R.; Pazdur, R. FDA drug approval summary: Azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist 2005, 10, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Hackanson, B.; Daskalakis, M. Decitabine. Recent Results Cancer Res. 2014, 201, 269–297. [Google Scholar] [PubMed]
- Kantarjian, H.M.; Roboz, G.J.; Kropf, P.L.; Yee, K.W.L.; O’Connell, C.L.; Tibes, R.; Walsh, K.J.; Podoltsev, N.A.; Griffiths, E.A.; Jabbour, E.; et al. Guadecitabine (SGI-110) in treatment-naive patients with acute myeloid leukaemia: Phase 2 results from a multicentre, randomised, phase 1/2 trial. Lancet Oncol. 2017, 18, 1317–1326. [Google Scholar] [CrossRef]
- Foulks, J.M.; Parnell, K.M.; Nix, R.N.; Chau, S.; Swierczek, K.; Saunders, M.; Wright, K.; Hendrickson, T.F.; Ho, K.K.; McCullar, M.V.; et al. Epigenetic drug discovery: Targeting DNA methyltransferases. J. Biomol. Screen. 2012, 17, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Munck, J.; Tang, J.; Taverna, P.; Wang, Y.; Miller, D.F.; Pilrose, J.; Choy, G.; Azab, M.; Pawelczak, K.S.; et al. The novel, small-molecule DNA methylation inhibitor SGI-110 as an ovarian cancer chemosensitizer. Clin. Cancer Res. 2014, 20, 6504–6516. [Google Scholar] [CrossRef] [PubMed]
- Balch, C.; Yan, P.; Craft, T.; Young, S.; Skalnik, D.G.; Huang, T.H.; Nephew, K.P. Antimitogenic and chemosensitizing effects of the methylation inhibitor zebularine in ovarian cancer. Mol. Cancer Ther. 2005, 4, 1505–1514. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, M.; Momparler, L.F.; Raynal, N.J.; Bernstein, M.L.; Momparler, R.L. Inhibition of cytidine deaminase by zebularine enhances the antineoplastic action of 5-aza-2′-deoxycytidine. Cancer Chemother. Pharmacol. 2009, 63, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Zwergel, C.; Valente, S.; Mai, A. DNA Methyltransferases Inhibitors from Natural Sources. Curr. Top. Med. Chem. 2016, 16, 680–696. [Google Scholar] [CrossRef] [PubMed]
- Stefanska, B.; Karlic, H.; Varga, F.; Fabianowska-Majewska, K.; Haslberger, A. Epigenetic mechanisms in anti-cancer actions of bioactive food components—The implications in cancer prevention. Br. J. Pharmacol. 2012, 167, 279–297. [Google Scholar] [CrossRef] [PubMed]
- Ottini, L.; Rizzolo, P.; Siniscalchi, E.; Zijno, A.; Silvestri, V.; Crebelli, R.; Marcon, F. Gene promoter methylation and DNA repair capacity in monozygotic twins with discordant smoking habits. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015, 779, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Pirouzpanah, S.; Taleban, F.A.; Mehdipour, P.; Atri, M. Association of folate and other one-carbon related nutrients with hypermethylation status and expression of RARB, BRCA1, and RASSF1A genes in breast cancer patients. J. Mol. Med. 2015, 93, 917–934. [Google Scholar] [CrossRef] [PubMed]
- Vineis, P.; Chuang, S.C.; Vaissiere, T.; Cuenin, C.; Ricceri, F.; Genair, E.C.; Johansson, M.; Ueland, P.; Brennan, P.; Herceg, Z. DNA methylation changes associated with cancer risk factors and blood levels of vitamin metabolites in a prospective study. Epigenetics 2011, 6, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.Z.; Wang, Y.; Ai, N.; Hou, Z.; Sun, Y.; Lu, H.; Welsh, W.; Yang, C.S. Tea polyphenol (–)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003, 63, 7563–7570. [Google Scholar] [PubMed]
- Zhu, W.; Qin, W.; Zhang, K.; Rottinghaus, G.E.; Chen, Y.C.; Kliethermes, B.; Sauter, E.R. Trans-resveratrol alters mammary promoter hypermethylation in women at increased risk for breast cancer. Nutr. Cancer 2012, 64, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Jiang, A.; Wang, X.; Shan, X.; Li, Y.; Wang, P.; Jiang, P.; Feng, Q. Curcumin Reactivates Silenced Tumor Suppressor Gene RARbeta by Reducing DNA Methylation. Phytother. Res. 2015, 29, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Xie, Z.; Wu, L.C.; Chiu, M.; Lin, J.; Chan, K.K.; Liu, S.; Liu, Z. Reactivation of RASSF1A in breast cancer cells by curcumin. Nutr. Cancer 2012, 64, 1228–1235. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Zhu, W.; Shi, H.; Hewett, J.E.; Ruhlen, R.L.; MacDonald, R.S.; Rottinghaus, G.E.; Chen, Y.C.; Sauter, E.R. Soy isoflavones have an antiestrogenic effect and alter mammary promoter hypermethylation in healthy premenopausal women. Nutr. Cancer 2009, 61, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Vardi, A.; Bosviel, R.; Rabiau, N.; Adjakly, M.; Satih, S.; Dechelotte, P.; Boiteux, J.P.; Fontana, L.; Bignon, Y.J.; Guy, L.; et al. Soy phytoestrogens modify DNA methylation of GSTP1, RASSF1A, EPH2 and BRCA1 promoter in prostate cancer cells. In Vivo 2010, 24, 393–400. [Google Scholar] [PubMed]
- Pan, F.P.; Zhou, H.K.; Bu, H.Q.; Chen, Z.Q.; Zhang, H.; Xu, L.P.; Tang, J.; Yu, Q.J.; Chu, Y.Q.; Pan, J.; et al. Emodin enhances the demethylation by 5-Aza-CdR of pancreatic cancer cell tumor-suppressor genes P16, RASSF1A and ppENK. Oncol. Rep. 2016, 35, 1941–1949. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, L.; Bu, H.Q.; Yu, Q.J.; Jiang, D.D.; Pan, F.P.; Wang, Y.; Liu, D.L.; Lin, S.Z. Effects of emodin on the demethylation of tumor-suppressor genes in pancreatic cancer PANC-1 cells. Oncol. Rep. 2015, 33, 3015–3023. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Z.; Cheng, Y.; Wang, K.L.; Liu, R.; Yang, X.L.; Wen, H.M.; Chai, C.; Liang, J.Y.; Wu, H. Peperomin E reactivates silenced tumor suppressor genes in lung cancer cells by inhibition of DNA methyltransferase. Cancer Sci. 2016, 107, 1506–1519. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Song, W.; Xiao, W. Dioscin induces demethylation of DAPK-1 and RASSF-1alpha genes via the antioxidant capacity, resulting in apoptosis of bladder cancer T24 cells. EXCLI J. 2017, 16, 101–112. [Google Scholar] [PubMed]
- Agarwal, S.; Amin, K.S.; Jagadeesh, S.; Baishay, G.; Rao, P.G.; Barua, N.C.; Bhattacharya, S.; Banerjee, P.P. Mahanine restores RASSF1A expression by down-regulating DNMT1 and DNMT3B in prostate cancer cells. Mol. Cancer 2013, 12, 99. [Google Scholar] [CrossRef] [PubMed]
- Jagadeesh, S.; Sinha, S.; Pal, B.C.; Bhattacharya, S.; Banerjee, P.P. Mahanine reverses an epigenetically silenced tumor suppressor gene RASSF1A in human prostate cancer cells. Biochem. Biophys. Res. Commun. 2007, 362, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, K.D.; Banerjee, P.P.; Jagadeesh, S.; Grindrod, S.C.; Zhang, L.; Paige, M.; Brown, M.L. Fluorescent epigenetic small molecule induces expression of the tumor suppressor ras-association domain family 1A and inhibits human prostate xenograft. J. Med. Chem. 2010, 53, 2376–2382. [Google Scholar] [CrossRef] [PubMed]
- Boyanapalli, S.S.; Li, W.; Fuentes, F.; Guo, Y.; Ramirez, C.N.; Gonzalez, X.P.; Pung, D.; Kong, A.N. Epigenetic reactivation of RASSF1A by phenethyl isothiocyanate (PEITC) and promotion of apoptosis in LNCaP cells. Pharmacol. Res. 2016, 114, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Shorter, K.R.; Felder, M.R.; Vrana, P.B. Consequences of dietary methyl donor supplements: Is more always better? Prog. Biophys. Mol. Biol. 2015, 118, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Anderson, O.S.; Sant, K.E.; Dolinoy, D.C. Nutrition and epigenetics: An interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J. Nutr. Biochem. 2012, 23, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Waterland, R.A.; Jirtle, R.L. Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 2003, 23, 5293–5300. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Gammon, M.D.; Jefferson, E.; Zhang, Y.; Cho, Y.H.; Wetmur, J.G.; Teitelbaum, S.L.; Bradshaw, P.T.; Terry, M.B.; Garbowski, G.; et al. The influence of one-carbon metabolism on gene promoter methylation in a population-based breast cancer study. Epigenetics 2011, 6, 1276–1283. [Google Scholar] [CrossRef] [PubMed]
- Van Engeland, M.; Weijenberg, M.P.; Roemen, G.M.; Brink, M.; De Bruine, A.P.; Goldbohm, R.A.; Van den Brandt, P.A.; Baylin, S.B.; De Goeij, A.F.; Herman, J.G. Effects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: The Netherlands cohort study on diet and cancer. Cancer Res. 2003, 63, 3133–3137. [Google Scholar] [PubMed]
- Vaissiere, T.; Hung, R.J.; Zaridze, D.; Moukeria, A.; Cuenin, C.; Fasolo, V.; Ferro, G.; Paliwal, A.; Hainaut, P.; Brennan, P.; et al. Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res. 2009, 69, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Nazki, F.H.; Sameer, A.S.; Ganaie, B.A. Folate: Metabolism, genes, polymorphisms and the associated diseases. Gene 2014, 533, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.W.; Liu, X.F.; Bu, R.G.; Chen, X.N.; Ning, L.; Cheng, Y.; Wu, B. Genetic polymorphisms of MTHFR and aberrant promoter hypermethylation of the RASSF1A gene in bladder cancer risk in a Chinese population. J. Int. Med. Res. 2009, 37, 1882–1889. [Google Scholar] [CrossRef] [PubMed]
- Coppede, F.; Migheli, F.; Lopomo, A.; Failli, A.; Legitimo, A.; Consolini, R.; Fontanini, G.; Sensi, E.; Servadio, A.; Seccia, M.; et al. Gene promoter methylation in colorectal cancer and healthy adjacent mucosa specimens: Correlation with physiological and pathological characteristics, and with biomarkers of one-carbon metabolism. Epigenetics 2014, 9, 621–633. [Google Scholar] [CrossRef] [PubMed]
- Supic, G.; Jovic, N.; Kozomara, R.; Zeljic, K.; Magic, Z. Interaction between the MTHFR C677T polymorphism and alcohol--impact on oral cancer risk and multiple DNA methylation of tumor-related genes. J. Dent. Res. 2011, 90, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Premkumar, V.G.; Yuvaraj, S.; Shanthi, P.; Sachdanandam, P. Co-enzyme Q10, riboflavin and niacin supplementation on alteration of DNA repair enzyme and DNA methylation in breast cancer patients undergoing tamoxifen therapy. Br. J. Nutr. 2008, 100, 1179–1182. [Google Scholar] [CrossRef] [PubMed]
- Fazi, F.; Travaglini, L.; Carotti, D.; Palitti, F.; Diverio, D.; Alcalay, M.; McNamara, S.; Miller, W.H., Jr.; Lo Coco, F.; Pelicci, P.G.; et al. Retinoic acid targets DNA-methyltransferases and histone deacetylases during APL blast differentiation in vitro and in vivo. Oncogene 2005, 24, 1820–1830. [Google Scholar] [CrossRef] [PubMed]
- Stefanska, B.; Salame, P.; Bednarek, A.; Fabianowska-Majewska, K. Comparative effects of retinoic acid, vitamin D and resveratrol alone and in combination with adenosine analogues on methylation and expression of phosphatase and tensin homologue tumour suppressor gene in breast cancer cells. Br. J. Nutr. 2012, 107, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Chen, D.; Yang, C.S. Dietary polyphenols may affect DNA methylation. J. Nutr. 2007, 137, 223S–228S. [Google Scholar] [PubMed]
- Mirza, S.; Sharma, G.; Parshad, R.; Gupta, S.D.; Pandya, P.; Ralhan, R. Expression of DNA methyltransferases in breast cancer patients and to analyze the effect of natural compounds on DNA methyltransferases and associated proteins. J. Breast Cancer 2013, 16, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Scoccianti, C.; Ricceri, F.; Ferrari, P.; Cuenin, C.; Sacerdote, C.; Polidoro, S.; Jenab, M.; Hainaut, P.; Vineis, P.; Herceg, Z. Methylation patterns in sentinel genes in peripheral blood cells of heavy smokers: Influence of cruciferous vegetables in an intervention study. Epigenetics 2011, 6, 1114–1119. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Sharma, P.; Capalash, N. DNA methyltransferase-1 inhibitors as epigenetic therapy for cancer. Curr. Cancer Drug Targets 2013, 13, 379–399. [Google Scholar] [CrossRef] [PubMed]
- Campbell, F.C.; Collett, G.P. Chemopreventive properties of curcumin. Future Oncol. 2005, 1, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Shu, L.; Khor, T.O.; Lee, J.H.; Boyanapalli, S.S.; Huang, Y.; Wu, T.Y.; Saw, C.L.; Cheung, K.L.; Kong, A.N. Epigenetic CpG demethylation of the promoter and reactivation of the expression of Neurog1 by curcumin in prostate LNCaP cells. AAPS J. 2011, 13, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xu, W.; Huang, Y.; Huang, X.; Xu, L.; Lv, Z. Genistein demethylates the promoter of CHD5 and inhibits neuroblastoma growth in vivo. Int. J. Mol. Med. 2012, 30, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Lu, G.; Shen, H.M.; Chung, M.C.; Ong, C.N. Anti-cancer properties of anthraquinones from rhubarb. Med. Res. Rev. 2007, 27, 609–630. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.L.; Li, N.; Hasegawa, T.; Sakai, J.; Mitsui, T.; Ogura, H.; Kataoka, T.; Oka, S.; Kiuchi, M.; Tomida, A.; et al. Bioactive secolignans from Peperomia dindygulensis. J. Nat. Prod. 2006, 69, 790–794. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cheng, Y.; Wang, N.; Wang, D.M.; Li, Y.W.; Han, F.; Shen, J.G.; Yang, D.P.; Guan, X.Y.; Chen, J.P. Dioscin induces cancer cell apoptosis through elevated oxidative stress mediated by downregulation of peroxiredoxins. Cancer Biol. Ther. 2012, 13, 138–147. [Google Scholar] [PubMed]
- Tachibana, Y.; Kikuzaki, H.; Lajis, N.H.; Nakatani, N. Antioxidative activity of carbazoles from Murraya koenigii leaves. J. Agric. Food Chem. 2001, 49, 5589–5594. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Wright, S.E.; Kim, S.H.; Srivastava, S.K. Phenethyl isothiocyanate: A comprehensive review of anti-cancer mechanisms. Biochim. Biophys. Acta 2014, 1846, 405–424. [Google Scholar] [CrossRef] [PubMed]
| Compound | Effect on RASSF1A | Effect on Other Genes | Mechanism | References |
|---|---|---|---|---|
| folate | no effect [70,71], increased methylation [72] | decreased methylation of RARB [70,71], BRCA1 [71] and CDH1 [70], increased MTHFR methylation [72] | methyl donor | [70,71,72] |
| methionine | decreased methylation | decreased methylation of p16 and MTHFR | methyl donor | [72] |
| vitamin B12 | decreased methylation | increased MTHFR methylation [72] | methyl donor | [71,72] |
| EGCG a | not analyzed | decreased methylation of p16, RARB, MGMT and MLH1 | inhibits DNMT b activity | [73] |
| reseveratol | decreased methylation | no effect on p16, APC and CCND2 methylation | downregulation of DNMT | [74] |
| curcumin | decreased methylation | decreased methylation of RARB [75] | downregulation of DNMT | [76] |
| genistein | no effect | increased methylation of RARB and CCND2 [77], decreased methylation of GSTP1 and EPHB2 [78] | inhibits DNMT | [77,78] |
| emodin | decreased methylation | decreased methylation of p16 and ppENK | downregulation of DNMT | [79,80] |
| peperomin E | decreased methylation | decreased methylation of p16, APC and RUNX3 | inhibits DNMT activity | [81] |
| dioscin | decreased methylation | decreased methylation of DAPK1 | antioxidant | [82] |
| mahanine | decreased methylation | not reported | inhibits DNMT activity | [83,84,85] |
| PEITC c | decreased methylation | not reported | downregulation of DNMT | [86] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dammann, R.H.; Richter, A.M.; Jiménez, A.P.; Woods, M.; Küster, M.; Witharana, C. Impact of Natural Compounds on DNA Methylation Levels of the Tumor Suppressor Gene RASSF1A in Cancer. Int. J. Mol. Sci. 2017, 18, 2160. https://doi.org/10.3390/ijms18102160
Dammann RH, Richter AM, Jiménez AP, Woods M, Küster M, Witharana C. Impact of Natural Compounds on DNA Methylation Levels of the Tumor Suppressor Gene RASSF1A in Cancer. International Journal of Molecular Sciences. 2017; 18(10):2160. https://doi.org/10.3390/ijms18102160
Chicago/Turabian StyleDammann, Reinhard H., Antje M. Richter, Adriana P. Jiménez, Michelle Woods, Miriam Küster, and Chamindri Witharana. 2017. "Impact of Natural Compounds on DNA Methylation Levels of the Tumor Suppressor Gene RASSF1A in Cancer" International Journal of Molecular Sciences 18, no. 10: 2160. https://doi.org/10.3390/ijms18102160
APA StyleDammann, R. H., Richter, A. M., Jiménez, A. P., Woods, M., Küster, M., & Witharana, C. (2017). Impact of Natural Compounds on DNA Methylation Levels of the Tumor Suppressor Gene RASSF1A in Cancer. International Journal of Molecular Sciences, 18(10), 2160. https://doi.org/10.3390/ijms18102160