Systematic Identification, Evolution and Expression Analysis of the Zea mays PHT1 Gene Family Reveals Several New Members Involved in Root Colonization by Arbuscular Mycorrhizal Fungi
Abstract
:1. Introduction
2. Results
2.1. Identification and Properties of Putative Z. mays PHT1 Genes
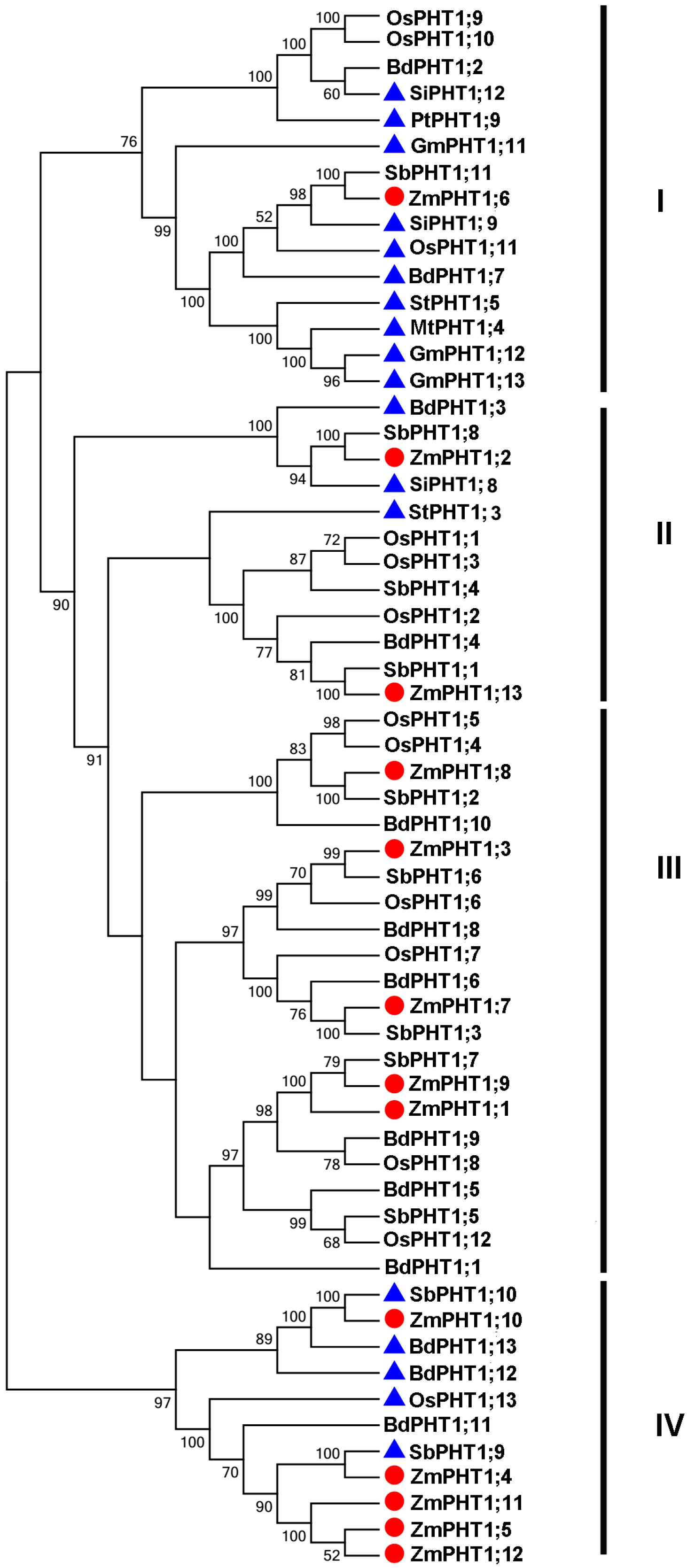
2.2. Phylogenetic Analysis of PHT1 Transporters in Z. mays
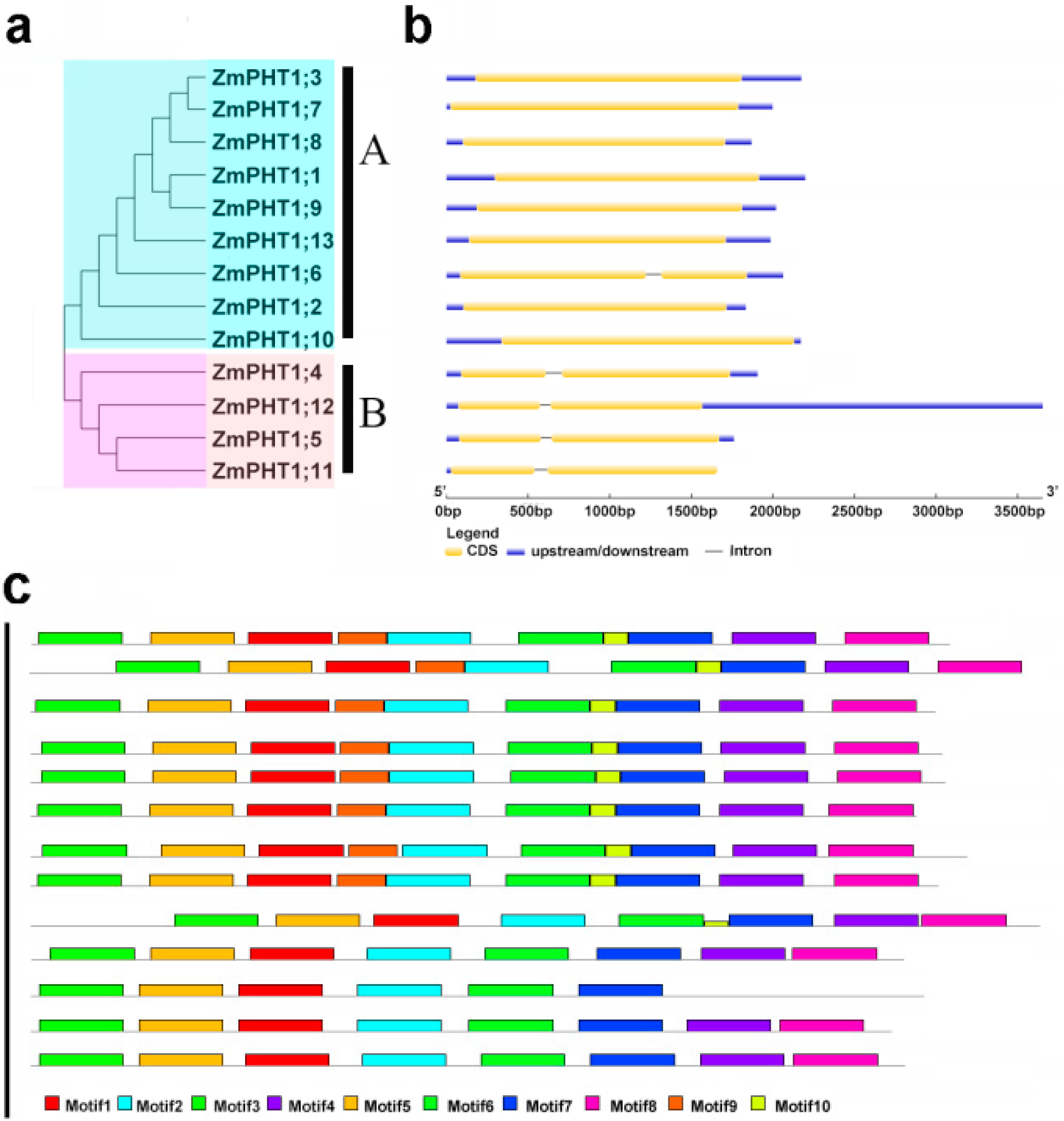
2.3. The Conserved Structure Analysis of Maize PHT1 Proteins
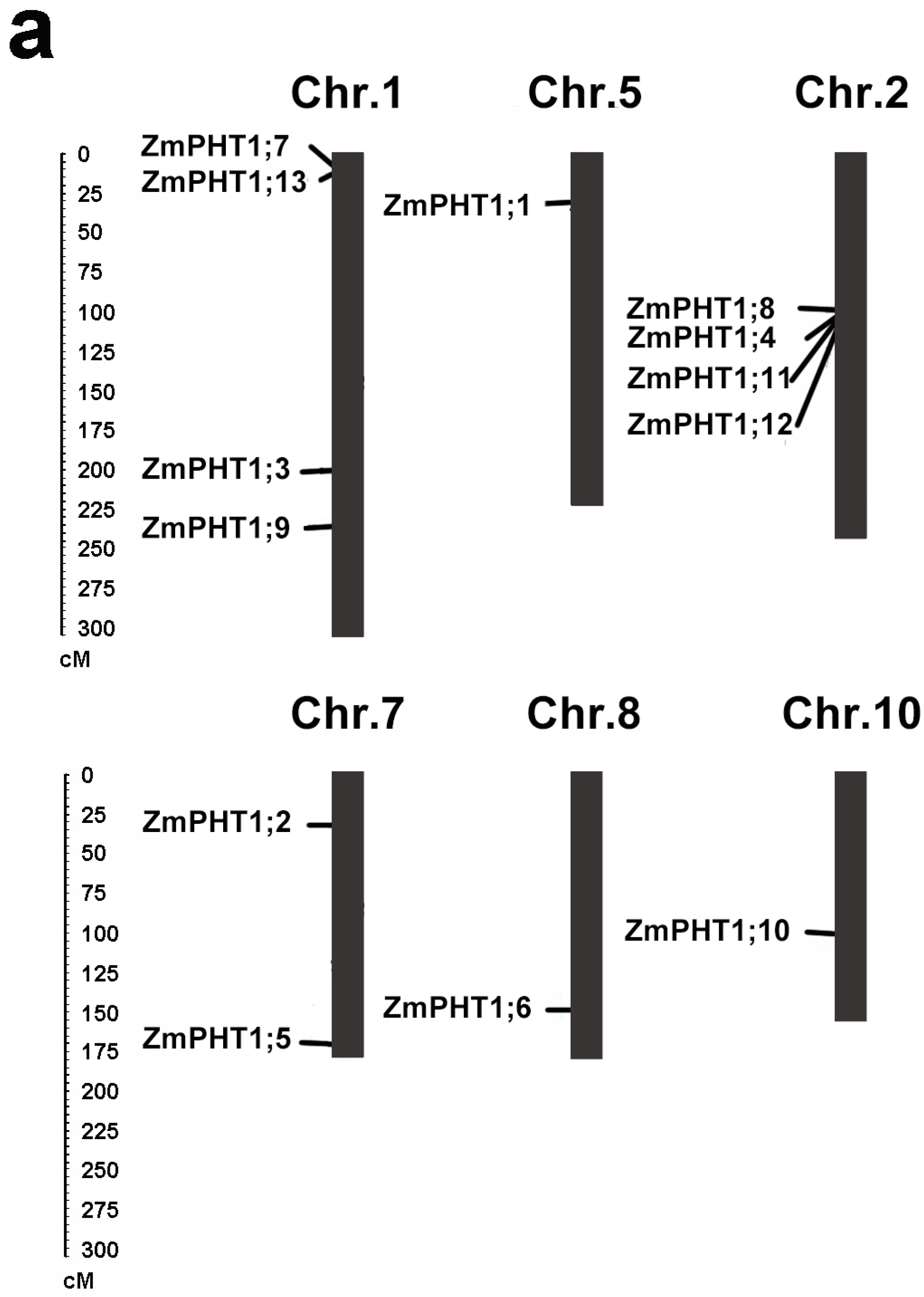
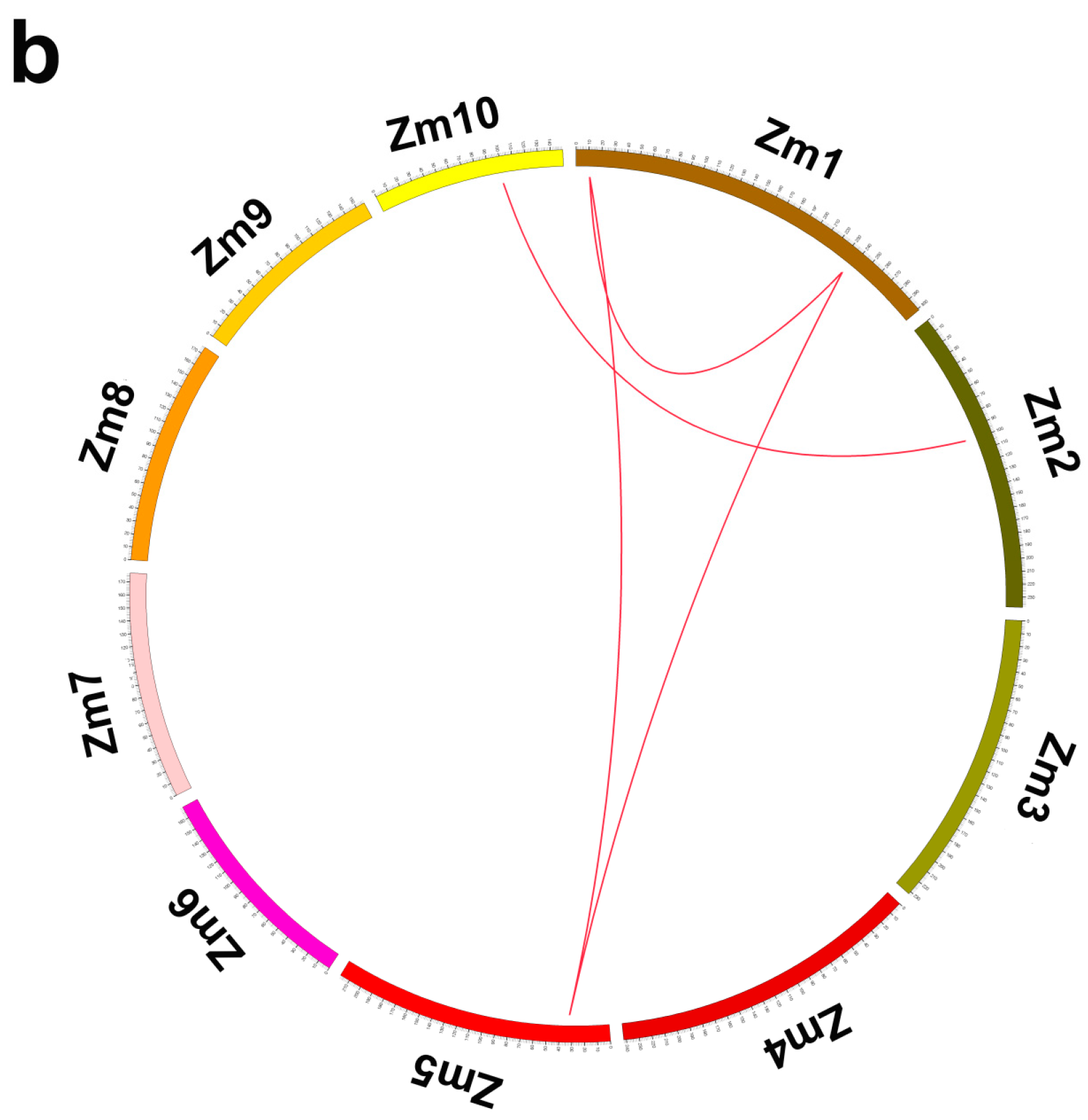
2.4. Distribution and Genomic Duplications of Maize PHT1 Genes
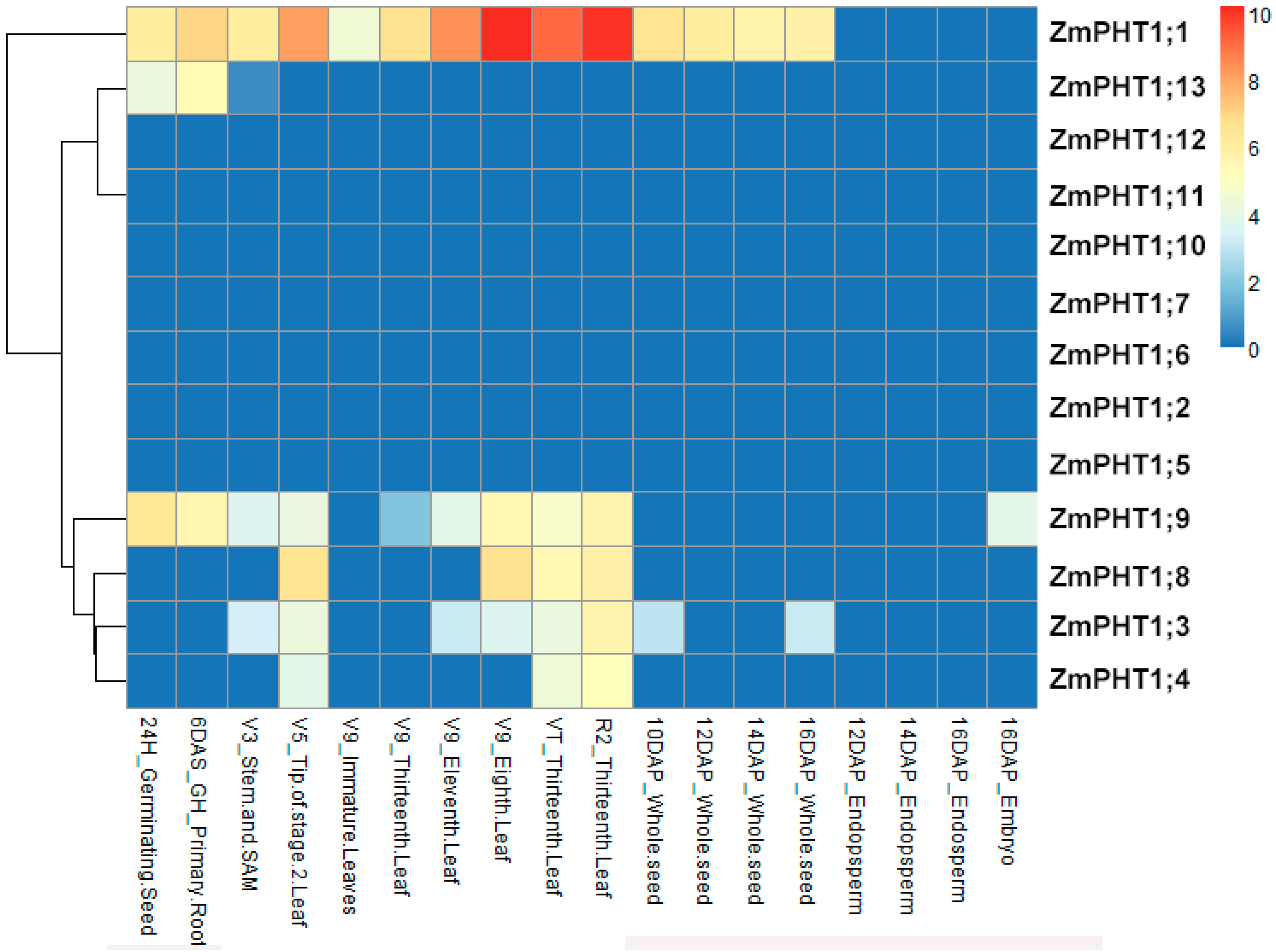
2.5. Tissue Specificity of ZmPHT1 Gene Expressions
2.6. Analysis of the Inorganic Phosphate (Pi) Transport Ability of ZmPHT1 Genes in a Pi-Uptake-Defective Yeast Strain
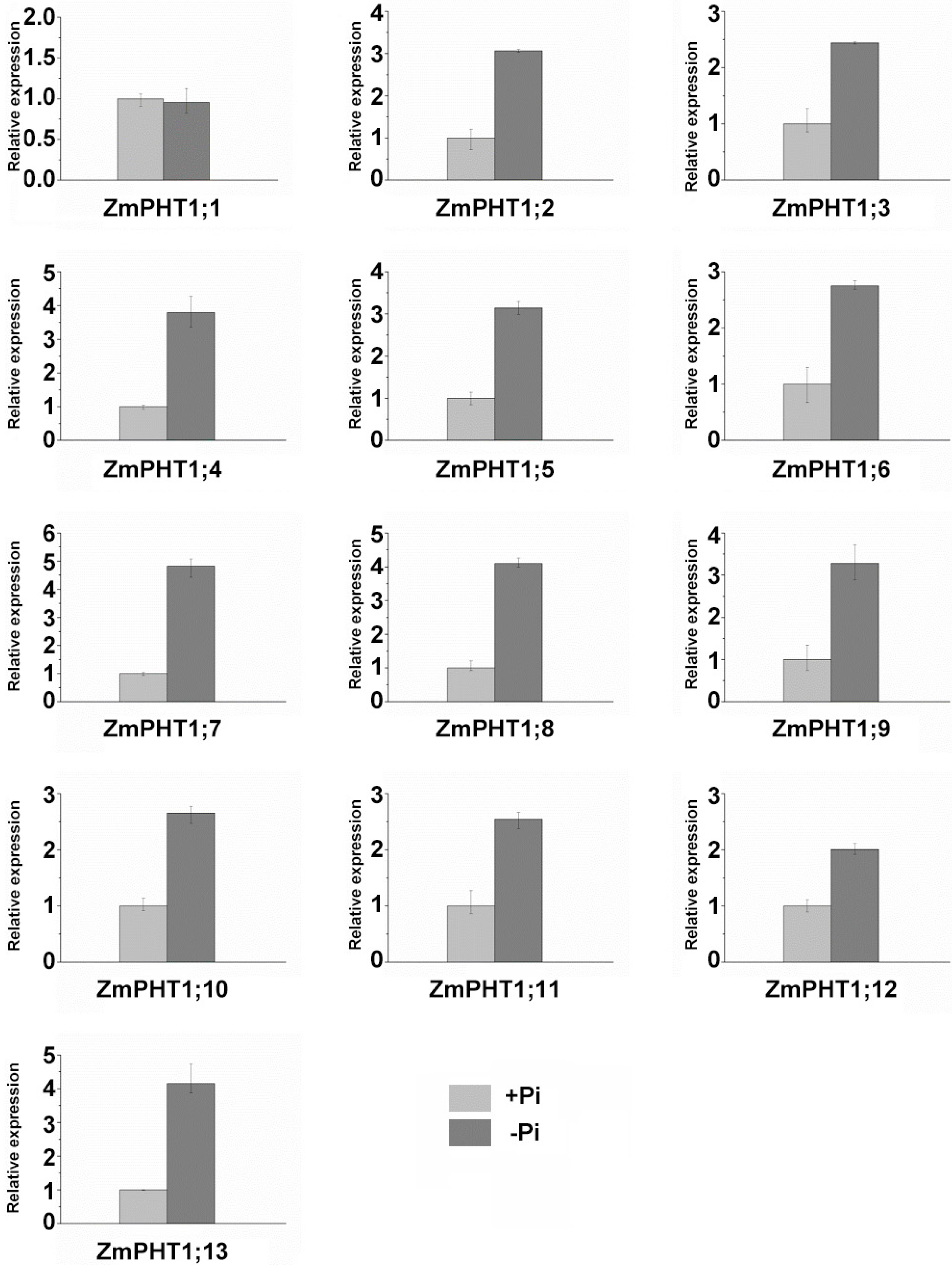
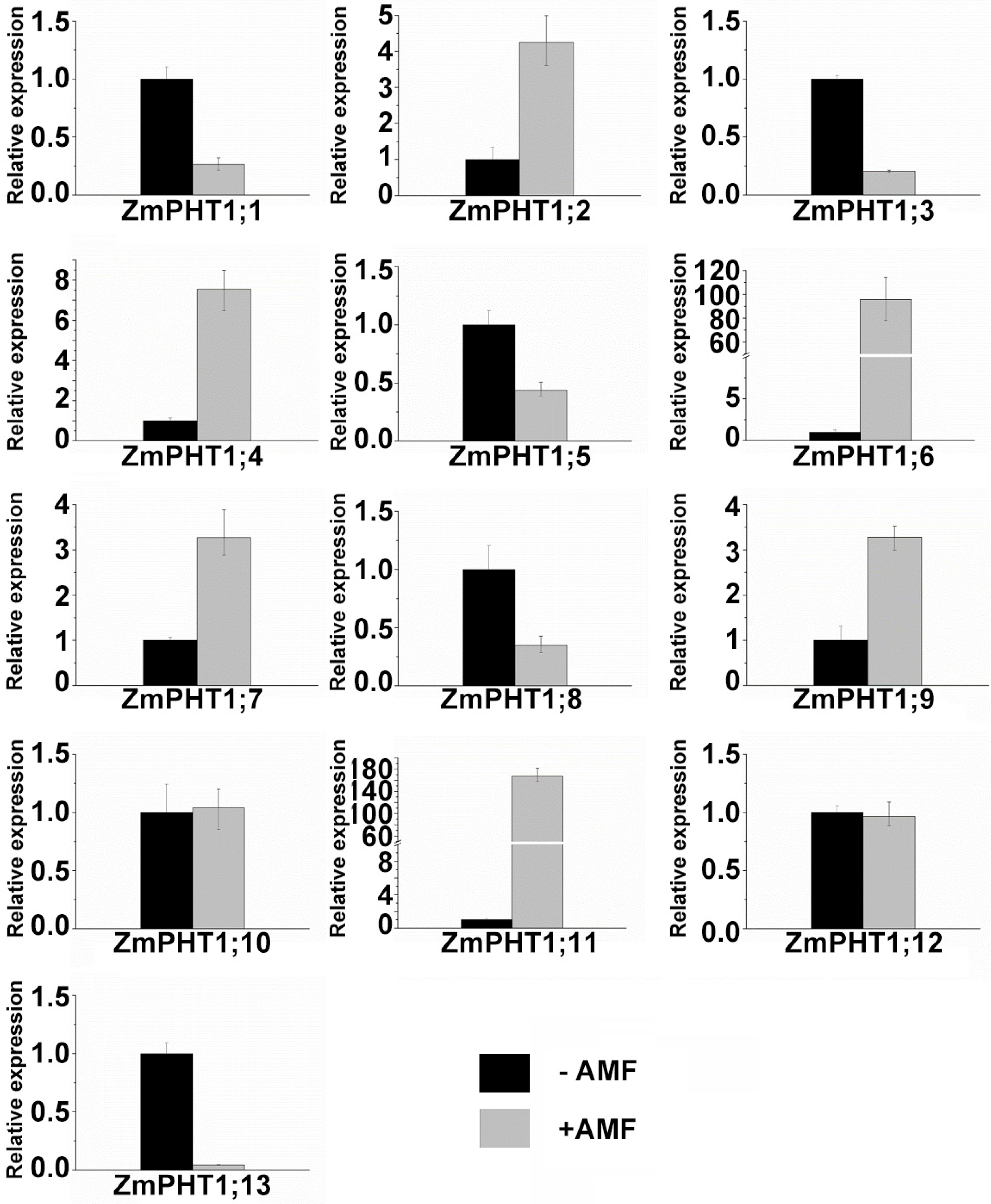
2.7. Influence of Pi Availability and AM Symbiosis on ZmPHT1 Gene Expression
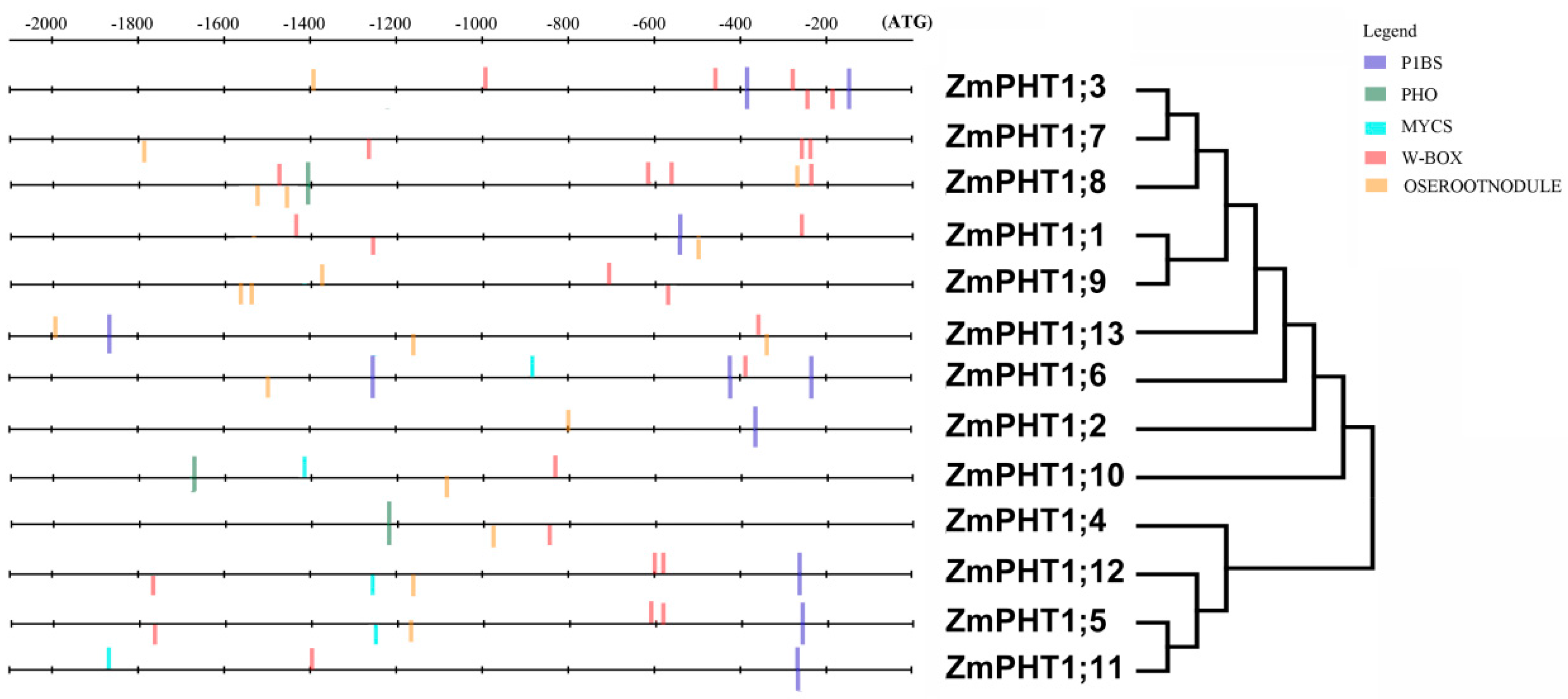
2.8. Identification of the Regulatory Cis-Elements
3. Discussion
3.1. Identification of ZmPHT1 Genes
3.2. Evolutionary Expansion of ZmPHT1 Genes
3.3. ZmPHT1s Response to Pi Availability and AMF Inoculation
3.4. Regulatory Cis-Elements in Promoters of AMF-Inducible ZmPHT1s
4. Materials and Methods
4.1. Bioinformatics Analyses
4.2. Plant Material and Cultivation Conditions
4.3. Detection of Mycorrhizal Colonization
4.4. Total RNA Extraction
4.5. qRT-PCR
4.6. Yeast Manipulations
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AMF | Arbuscular mycorrhizal fungi |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| NJ | Neighbor-joining |
| Ks | The rate of synonymous substitutions |
| Ka | The rate of nonsynonymous substitutions |
| MYCS | Mycorrhizal transcription factor binding sequence |
| P1BS | PHR1 binding site |
References
- Lopez-Arredondo, D.L.; Leyva-González, M.A.; González-Morales, S.I.; López-Bucio, J.; Herrera-Estrella, L. Phosphate Nutrition: Improving Low-Phosphate Tolerance in Crops. Annu. Rev. Plant Biol. 2014, 65, 95–123. [Google Scholar] [CrossRef] [PubMed]
- Javot, H.; Pumplin, N.; Harrison, M.J. Phosphate in the arbuscular mycorrhizal symbiosis: Transport properties and regulatory roles. Plant Cell Environ. 2007, 30, 310–322. [Google Scholar] [CrossRef] [PubMed]
- Tsuzuki, S.; Handa, Y.; Takeda, N.; Kawaguchi, M. Strigolactone-Induced Putative Secreted Protein 1 Is Required for the Establishment of Symbiosis by the Arbuscular Mycorrhizal Fungus Rhizophagus Irregularis. Mol. Plant Microbe Interact. 2016, 29, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Van Der Heijen, M.G.A.; Bardgett, R.; van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 291–310. [Google Scholar] [CrossRef] [PubMed]
- Van Der Heijden, M.G.A.; Streitwolf-Engel, R.; Riedl, R.; Siegrist, S.; Neudecker, A.; Ineichen, K.; Boller, T.; Wiemken, A.; Sanders, I.R. The mycorrhizal contribution to plant productivity, plant nutrition and soil structure in experimental grassland. New Phytol. 2006, 172, 739–752. [Google Scholar] [CrossRef] [PubMed]
- Ayadi, A.; David, P.; Jean-François, A.; Chiarenza, S.; Marie-Christine, T.; Nussaume, L.; Marin, E. Reducing the Genetic Redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 Transporters to Study Phosphate Uptake and Signaling. Plant Physiol. 2015, 167, 1511–1526. [Google Scholar] [CrossRef] [PubMed]
- Walder, F.; Brule, D.; Koegel, S.; Wiemken, A.; Boller, T.; Pierre-Emmanuel, C. Plant phosphorus acquisition in a common mycorrhizal network: Regulation of phosphate transporter genes of the Pht1 family in sorghum and flax. New Phytol. 2015, 205, 1632–1645. [Google Scholar] [CrossRef] [PubMed]
- Karandashov, V.; Bucher, M. Symbiotic phosphate transport in arbuscular mycorrhizas. Trends Plant Sci. 2005, 10, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, B.P.; Kumar, H.; Waight, A.B.; Risenmay, A.J.; Roe-Zurz, Z.; Chau, B.H.; Schlessinger, A.; Bonomi, M.; Harries, W.; Sali, A.; et al. Crystal structure of a eukaryotic phosphate transporter. Nature 2013, 496, 533–536. [Google Scholar] [CrossRef] [PubMed]
- Muchhal, U.S.; Pardo, J.M.; Raghothama, K.G. Phosphate transporters from the higher plant Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1996, 93, 10519–10523. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, F.; Amode, M.R.; Barrell, D.; Beal, K.; Billis, K.; Brent, S.; Carvalho-Silva, D.; Clapham, P.; Coates, G.; Fitzgerald, S.; et al. Ensembl 2015. Nucleic Acids Res. 2015, 43, D662–D669. [Google Scholar] [CrossRef] [PubMed]
- Schnable, P.S. The B73 maize genome: Complexity, diversity, and dynamics (November, pg 1112, 2009). Science 2012, 337, 1040. [Google Scholar] [CrossRef]
- Misson, J.; Thibaud, M.C.; Bechtold, N.; Raghothama, K.; Nussaume, L. Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol. Biol. 2004, 55, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Paszkowski, U.; Kroken, S.; Roux, C.; Briggs, S.P. Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2002, 99, 13324–13329. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Chen, X.; Wang, H.; Liao, D.; Gu, M.; Qu, H.; Sun, S.; Xu, G. Genome-wide investigation and expression analysis suggest diverse roles and genetic redundancy of Pht1 family genes in response to Pi deficiency in tomato. BMC Plant Biol. 2014, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Huang, W.; Liu, F.; Tang, N.; Liu, Y.; Lin, H.; Zhao, B. Functional analysis of the novel mycorrhiza-specific phosphate transporter AsPT1 and PHT1 family from Astragalus sinicus during the arbuscular mycorrhizal symbiosis. New Phytol. 2013, 198, 836–852. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Guo, Y.; Chen, L.; Liang, R.; Gu, M.; Xu, G.; Zhao, J.; Walk, T.; Liao, H. Functional characterization of 14 Pht1 family genes in yeast and their expressions in response to nutrient starvation in soybean. PLoS ONE 2012, 7, e47726. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.J.; Park, Y.S.; Bravo, A.; Bhattarai, K.K.; Daniels, D.A.; Harrison, M.J. Diversity of morphology and function in arbuscular mycorrhizal symbioses in Brachypodium distachyon. Planta 2012, 236, 851–865. [Google Scholar] [CrossRef] [PubMed]
- Ceasar, S.A.; Hodge, A.; Baker, A.; Baldwin, S.A. Phosphate Concentration and Arbuscular Mycorrhizal Colonisation Influence the Growth, Yield and Expression of Twelve PHT1 Family Phosphate Transporters in Foxtail Millet (Setaria italica). PLoS ONE 2014, 9, e108459. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.J.; Dewbre, G.R.; Liu, J. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 2002, 14, 2413–2429. [Google Scholar] [CrossRef] [PubMed]
- Güimil, S.; Chang, H.S.; Zhu, T.; Sesma, A.; Osbourn, A.; Roux, C.; Ioannidis, V.; Oakeley, E.J.; Docquier, M.; Descombes, P.; et al. Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc. Natl. Acad. Sci. USA 2005, 102, 8066–8070. [Google Scholar] [CrossRef] [PubMed]
- Rausch, C.; Daram, P.; Brunner, S.; Jansa, J.; Laloi, M.; Leggewie, G.; Amrhein, N.; Bucher, M. A phosphate transporter expressed in arbuscule-containing cells in potato. Nature 2001, 414, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Nagy, R.; Karandashov, V.; Chague, V.; Kalinkevich, K.; Tamasloukht, M.; Xu, G.; Jakobsen, I.; Levy, A.A.; Amrhein, N.; Bucher, M. The characterization of novel mycorrhiza-specific phosphate transporters from Lycopersicon esculentum and Solanum tuberosum uncovers functional redundancy in symbiotic phosphate transport in solanaceous species. Plant J. 2005, 42, 236–250. [Google Scholar] [CrossRef] [PubMed]
- Glassop, D.; Smith, S.E.; Smith, F.W. Cereal phosphate transporters associated with the mycorrhizal pathway of phosphate uptake into roots. Planta 2005, 222, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Nagy, R.; Vasconcelos, M.J.; Zhao, S.; McElver, J.; Bruce, W.; Amrhein, N.; Raghothama, K.G.; Bucher, M. Differential regulation of five Pht1 phosphate transporters from maize (Zea mays L.). Plant Biol. 2006, 8, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Drijber, R.A.; Li, X.; Miller, D.N.; Wienhold, B.J. Arbuscular mycorrhizal fungi differ in their ability to regulate the expression of phosphate transporters in maize (Zea mays L.). Mycorrhiza 2013, 23, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Sekhon, R.S.; Briskine, R.; Hirsch, C.N.; Myers, C.L.; Springer, N.M.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. Maize Gene Atlas Developed by RNA Sequencing and Comparative Evaluation of Transcriptomes Based on RNA Sequencing and Microarrays. PLoS ONE 2013, 8, e61005. [Google Scholar] [CrossRef] [PubMed]
- Rubio, V.; Linhares, F.; Solano, R.; Martín, A.C.; Iglesias, J.; Leyva, A.; Paz-Ares, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Yuan, J.; Chang, X.; Yang, M.; Zhang, L.; Lu, K.; Lian, X. The Phosphate Transporter Gene OsPht1;4 Is Involved in Phosphate Homeostasis in Rice. PLoS ONE 2015, 10, e0126186. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, B.; Li, J.; Zhang, L.; Wang, Y.; Zheng, H.; Lu, M.; Chen, J. Hsf and Hsp gene families in Populus: Genome-wide identification, organization and correlated expression during development and in stress responses. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Coe, E.; Nelson, W.; Bharti, A.L.; Engler, F.; Butler, E.; Kim, H.; Goicoechea, J.L.; Chen, M.; Lee, S.; et al. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 2007, 3, 1254–1263. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Chang, X.J.; Ye, Y.; Xie, W.B.; Wu, P.; Lian, X.M. Comprehensive Sequence and Whole-Life-Cycle Expression Profile Analysis of the Phosphate Transporter Gene Family in Rice. Mol. Plant 2011, 4, 1105–1122. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Jakobsen, I.; Grønlund, M.; Smith, F.A. Roles of Arbuscular Mycorrhizas in Plant Phosphorus Nutrition: Interactions between Pathways of Phosphorus Uptake in Arbuscular Mycorrhizal Roots Have Important Implications for Understanding and Manipulating Plant Phosphorus Acquisition. Plant Physiol. 2011, 156, 1050–1057. [Google Scholar] [CrossRef] [PubMed]
- Lota, F.; Wegmüller, S.; Buer, B.; Sato, S.; Bräutigam, A.; Hanf, B.; Bucher, M. The cis-acting CTTC-P1BS module is indicative for gene function of LjVTI12, a Qb-SNARE protein gene that is required for arbuscule formation in Lotus japonicus. Plant J. 2013, 74, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Feng, L.; Zhu, Y.; Li, Y.; Yan, H.; Xiang, Y. Comparative genomic analysis of the WRKY III gene family in Populus, Grape, Arabidopsis and Rice. Biol. Direct 2015, 10, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, 49. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Cheng, Y.; Jin, J.; Jin, X.; Jiang, H.; Yan, H.; Cheng, B. Genome Duplication and Gene Loss Affect the Evolution of Heat Shock Transcription Factor Genes in Legumes. PLoS ONE 2014, 9, e102825. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Medina-Rivera, A.; Defrance, M.; Sand, O.; Herrmann, C.; Castro-Mondragon, J.A.; Delerce, J.; Jaeger, S.; Blanchet, C.; Vincens, P.; Caron, C.; et al. RSAT 2015: Regulatory Sequence Analysis Tools. Nucleic Acids Res. 2015, 43, W50–W56. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bai, J.; Liu, H.; Sun, Y.; Shi, X.; Ren, Z. Overexpression of a Maize Transcription Factor ZmPHR1 Improves Shoot Inorganic Phosphate Content and Growth of Arabidopsis under Low-Phosphate Conditions. Plant Mol. Biol. Rep. 2013, 31, 665–677. [Google Scholar] [CrossRef]
- Winzeler, E.A.; Shoemaker, D.D.; Astromoff, A.; Liang, H.; Anderson, K.; Andre, B.; Bangham, R.; Benito, R.; Boeke, J.D.; Bussey, H.; et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 1999, 285, 901–906. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Transcript ID | Accession Number | Gene Size (bp) | CDS Size (bp) | Protein Length (AA) | Protein Molecular Mass (Da) | pI | Chromosome |
|---|---|---|---|---|---|---|---|---|
| ZmPHT1;1 | GRMZM2G326707_T01 | NM_001279426.1 | 2199 | 1617 | 539 | 58,485.66 | 7.65 | 5 |
| ZmPHT1;2 | GRMZM2G139639_T01 | NM_001156420.1 | 1835 | 1611 | 537 | 58,660.21 | 7.98 | 7 |
| ZmPHT1;3 | GRMZM2G112377_T01 | NM_001112347.2 | 2175 | 1629 | 543 | 57,899.24 | 8.11 | 1 |
| ZmPHT1;4 | GRMZM2G170208_T01 | NM_001279982.1 | 1809 | 1548 | 516 | 57,145.85 | 9.13 | 2 |
| ZmPHT1;5 | GRMZM2G041595_T01 | NM_001112348.1 | 1698 | 1527 | 509 | 56,222.71 | 8.93 | 7 |
| ZmPHT1;6 | GRMZM5G881088_T01 | NM_001112306.1 | 1971 | 1662 | 554 | 60,579.71 | 8.15 | 8 |
| ZmPHT1;7 | GRMZM2G075870_T01 | NM_001157730.1 | 1999 | 1761 | 587 | 63,148.92 | 8.34 | 1 |
| ZmPHT1;8 | GRMZM2G045473_T01 | NM_001139212.1 | 1870 | 1605 | 535 | 58,762.9 | 7.58 | 2 |
| ZmPHT1;9 | GRMZM2G154090_T01 | NM_001112346.1 | 2020 | 1623 | 541 | 58,752.98 | 7.63 | 1 |
| ZmPHT1;10 | GRMZM2G159075_T01 | XM_008664847.1 | 2172 | 1791 | 597 | 65,240.06 | 9.11 | 10 |
| ZmPHT1;11 | GRMZM2G009779_T01 | NM_001112348.1 | 1581 | 1551 | 517 | 57,538.36 | 9.18 | 2 |
| ZmPHT1;12 | GRMZM2G009800_T02 | NM_001112348.1 | 1846 | 1770 | 590 | 66,071.38 | 9.41 | 2 |
| ZmPHT1;13 | GRMZM2G070087_T01 | NM_001196972.1 | 1986 | 1572 | 524 | 57,450.92 | 8.37 | 1 |
| Paralogous Pairs | Ks | Ka | Ka/Ks | Duplication Rate (Mya) | Duplication Type |
|---|---|---|---|---|---|
| ZmPHT1;1/ZmPHT1;9 | 0.16 | 0.03 | 0.19 | 12.36 | Segmental |
| ZmPHT1;1/ZmPHT1;13 | 0.43 | 0.18 | 0.41 | 32.72 | Segmental |
| ZmPHT1;8/ZmPHT1;10 | 0.55 | 0.36 | 0.66 | 42.05 | Segmental |
| ZmPHT1;9/ZmPHT1;13 | 0.10 | 0.28 | 2.76 | 7.91 | Segmental |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, F.; Xu, Y.; Jiang, H.; Jiang, C.; Du, Y.; Gong, C.; Wang, W.; Zhu, S.; Han, G.; Cheng, B. Systematic Identification, Evolution and Expression Analysis of the Zea mays PHT1 Gene Family Reveals Several New Members Involved in Root Colonization by Arbuscular Mycorrhizal Fungi. Int. J. Mol. Sci. 2016, 17, 930. https://doi.org/10.3390/ijms17060930
Liu F, Xu Y, Jiang H, Jiang C, Du Y, Gong C, Wang W, Zhu S, Han G, Cheng B. Systematic Identification, Evolution and Expression Analysis of the Zea mays PHT1 Gene Family Reveals Several New Members Involved in Root Colonization by Arbuscular Mycorrhizal Fungi. International Journal of Molecular Sciences. 2016; 17(6):930. https://doi.org/10.3390/ijms17060930
Chicago/Turabian StyleLiu, Fang, Yunjian Xu, Huanhuan Jiang, Chaosheng Jiang, Yibin Du, Cheng Gong, Wei Wang, Suwen Zhu, Guomin Han, and Beijiu Cheng. 2016. "Systematic Identification, Evolution and Expression Analysis of the Zea mays PHT1 Gene Family Reveals Several New Members Involved in Root Colonization by Arbuscular Mycorrhizal Fungi" International Journal of Molecular Sciences 17, no. 6: 930. https://doi.org/10.3390/ijms17060930
APA StyleLiu, F., Xu, Y., Jiang, H., Jiang, C., Du, Y., Gong, C., Wang, W., Zhu, S., Han, G., & Cheng, B. (2016). Systematic Identification, Evolution and Expression Analysis of the Zea mays PHT1 Gene Family Reveals Several New Members Involved in Root Colonization by Arbuscular Mycorrhizal Fungi. International Journal of Molecular Sciences, 17(6), 930. https://doi.org/10.3390/ijms17060930






