Sequencing and Transcriptional Analysis of the Biosynthesis Gene Cluster of Abscisic Acid-Producing Botrytis cinerea
Abstract
:1. Introduction
2. Results
2.1. ABA Production of B. cinerea TB-3-H8
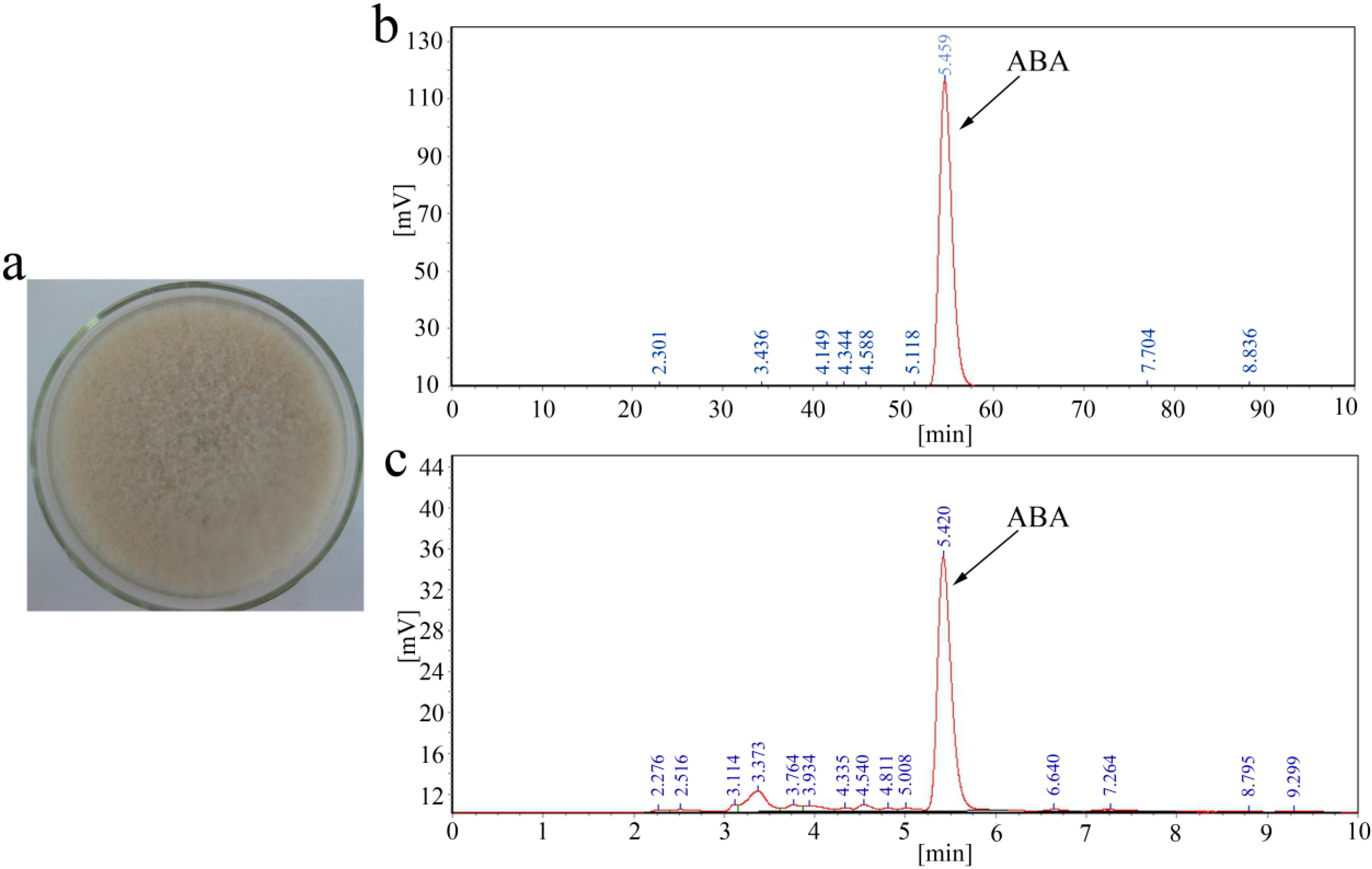
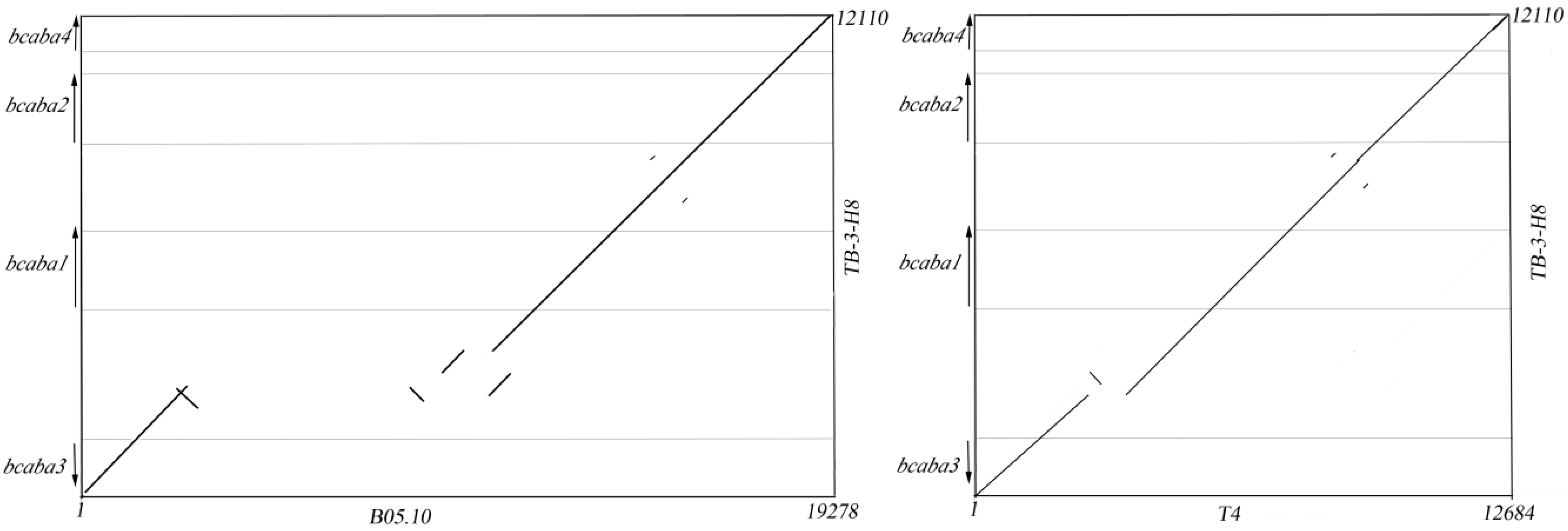
2.2. Sequence Analysis of the ABA Cluster of B. cinerea TB-3-H8
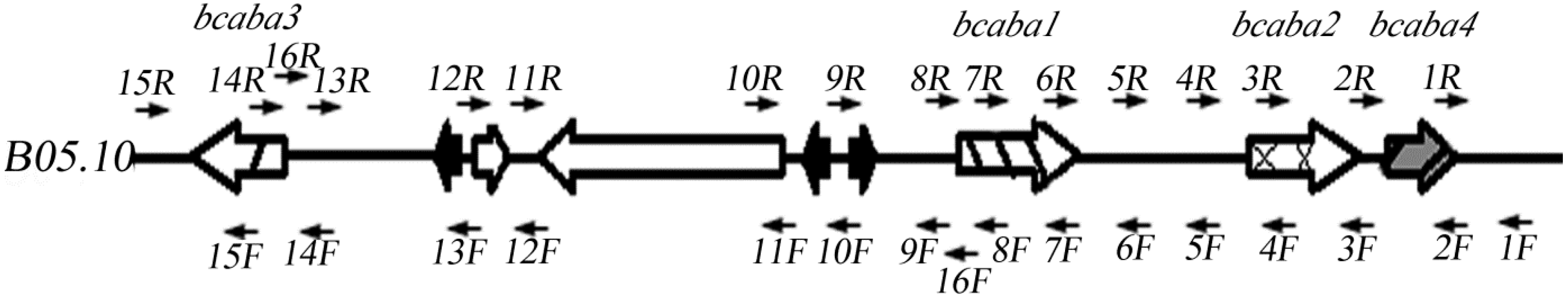
| Primers | Forward (5'-3') | Reverse (5'-3') | Sequence Length (bp) | |
|---|---|---|---|---|
| In B05.10 Supercontig 41 | In TB-3-H8 Genome | |||
| 1 | GAGACGGAGAAGCTAAGGAGG | CATGATGGCAATACACCAGTT | 1046 (71,207–72,252) | 897 |
| 2 | CCATTTGGTTTGAGCTGCTTTG | CCTCCTTAGCTTCTCCGTCTC | 1193 (72,232–73,424) | 1193 |
| 3 | AATTATGCGAAGTCATGTCGA | ACTACCAAAGCAGCTCAAACC | 1240 (73,398–74,637) | 1240 |
| 4 | GGGTTTCACTGCGGAGTCTTG | GACTGATCCCATGCGGCAGAT | 1221 (74,359–75,579) | 1221 |
| 5 | ACTGTAGCCGAGATAGTAGGG | AGGCTTACGATTGGGCTGGAA | 1371 (75,467–76,837) | 1386 |
| 6 | AATGTGGAACTTGCCTTTGGT | CGTTACAACCCTACTATCTCG | 933 (76,809–77,741) | 933 |
| 7 | TCGATTCAATCCATATTATGACAAC | AAGTTCCACATTGCGCTCCATCTC | 1177 (77,730–78,906) | 1177 |
| 8 | CATAATGTGGGCTAACTACTCTGA | ACCGACCTCCAAGTCAGGCATAT | 1034 (78,786–79,819) | 1034 |
| 9 | ATTCGTCTGTGCGTAACCGTGCA | TCAGGTGGTGACGAATACAAGAT | 1554 (79,689–81,242) | No data |
| 10 | GCAGAAAGGGTGCTCAAAGTGTT | TGCACGGTTACGCACAGACGAAT | 986 (81,220–82,205) | No data |
| 11 | TTCCTGTGGTCGAGTTTATTG | AACACTTTGAGCACCCTTTCT | 4500 (82,183–866,682) | No data |
| 12 | TCAGTATGCCTCCTACACGAACA | GCAGCGTACTACAATACGAAACGT | 1005 (86,564–87,568) | No data |
| 13 | AGGCGGTTCCCTCTTAC | ACGGATACACTCGGCACAACG | 2299 (87,402–89,700) | No data |
| 14 | ACGATGTTTGTAGAAAGCCACTG | ATCCTCAAGAGTCGCAGTTCATA | 1226 (89,625–90,850) | 1226 |
| 15 | CGTCGCTGGGACAACATGCTCAA | AAGGCGAGTGAAAGACGAGATGG | 1303 (90,617–91,919) | 1303 |
| 16 | CGCAATGTCAGGAGGGTAGTA | TTATCGCAGAGAATCAGGCAG | 10,943 (79,374–90,316) | 3762 |
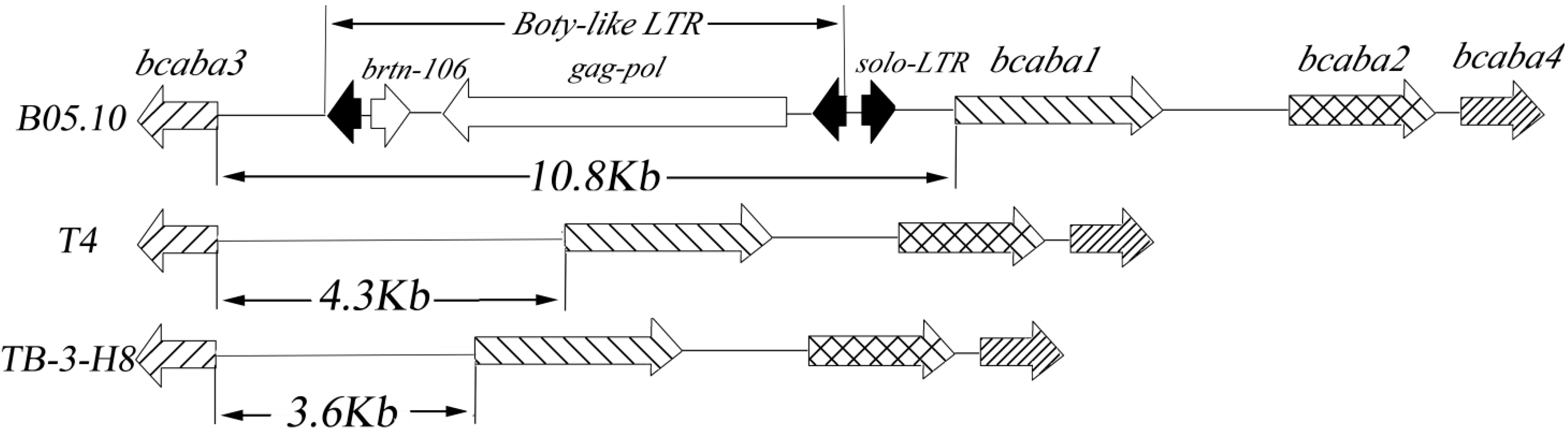
 comparison of SNPs among three strains. Sites in TB-3-H8 were used for standard positions of SNPs. Structure of genes in B05.10;
comparison of SNPs among three strains. Sites in TB-3-H8 were used for standard positions of SNPs. Structure of genes in B05.10;  Structure of gene inTB-3-H8 and T4;
Structure of gene inTB-3-H8 and T4;  Structure of genes in B05.10, TB-3-H8 and T4,
Structure of genes in B05.10, TB-3-H8 and T4,  introns,
introns,  intergenic sequences.
intergenic sequences.
 comparison of SNPs among three strains. Sites in TB-3-H8 were used for standard positions of SNPs. Structure of genes in B05.10;
comparison of SNPs among three strains. Sites in TB-3-H8 were used for standard positions of SNPs. Structure of genes in B05.10;  Structure of gene inTB-3-H8 and T4;
Structure of gene inTB-3-H8 and T4;  Structure of genes in B05.10, TB-3-H8 and T4,
Structure of genes in B05.10, TB-3-H8 and T4,  introns,
introns,  intergenic sequences.
intergenic sequences.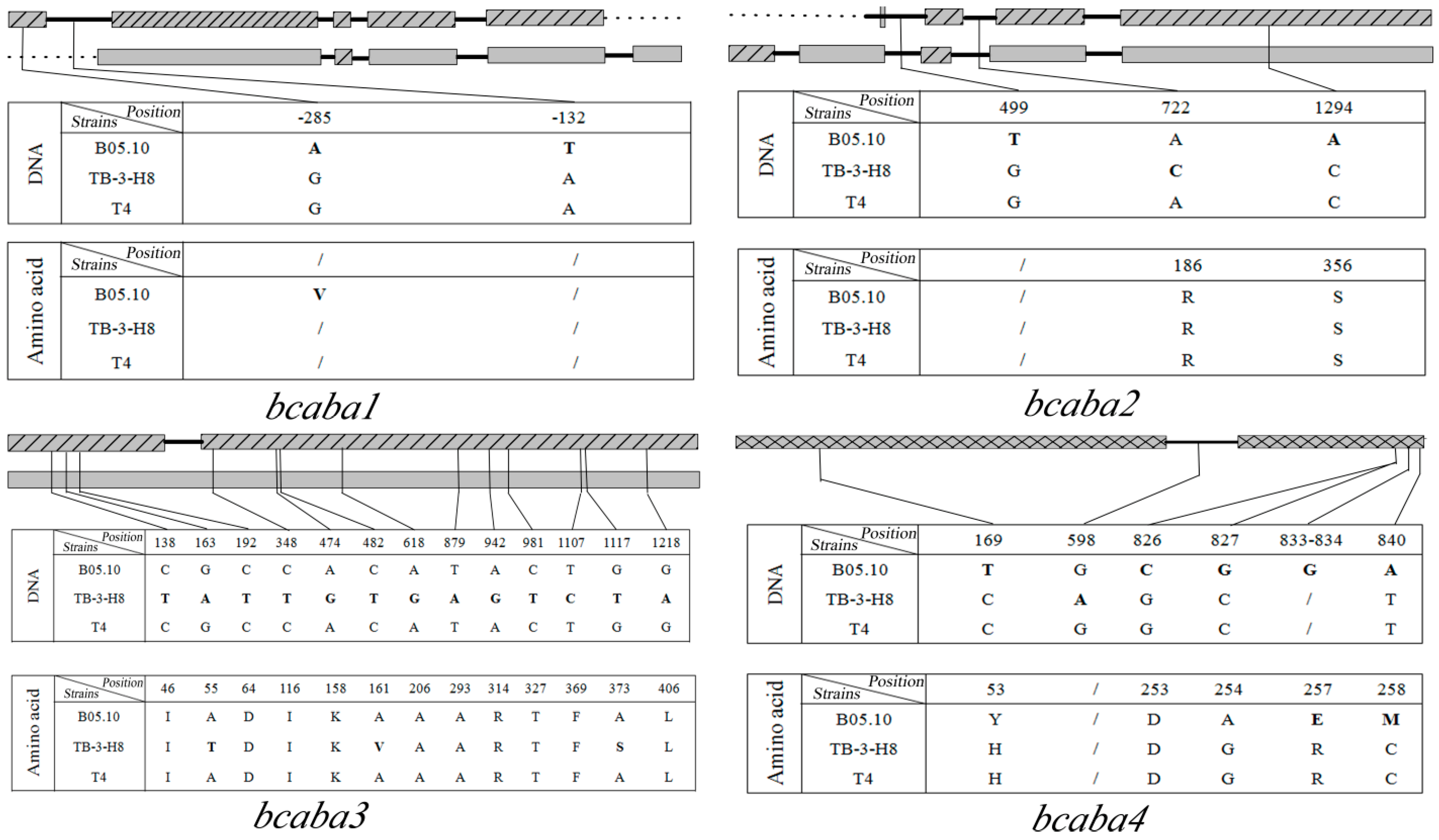
| Gene Name | Strains Names | GenBank Accession No. | DNA Length (bp) | cDNA Length (bp) | Protein Size (Amino Acids) | No. of Exons | No. of Introns |
|---|---|---|---|---|---|---|---|
| bcaba1 | TB-3-H8 | / | 1769 | 1530 | 509 | 5 | 4 |
| B05.10 | XM_001553921 | 1925 | 1482 | 493 | 5 | 4 | |
| T4 | FQ790338.1 | 1769 | 1530 | 509 | 5 | 4 | |
| bcaba2 | TB-3-H8 | / | 1810 | 1584 | 527 | 5 | 4 |
| B05.10 | XM_001553920 | 1507 | 1332 | 443 | 4 | 3 | |
| T4 | FQ790338.1 | 1810 | 1587 | 528 | 5 | 4 | |
| bcaba3 | TB-3-H8 | / | 1323 | 1323 | 440 | 1 | 0 |
| B05.10 | XM_001553924 | 1323 | 1269 | 422 | 2 | 1 | |
| T4 | FQ790338.1 | 1323 | 1323 | 440 | 1 | 0 | |
| bcaba4 | TB-3-H8 | / | 842 | 777 | 258 | 2 | 1 |
| B05.10 | XM_001553919 | 842 | 777 | 258 | 2 | 1 | |
| T4 | FQ790338.1 | 842 | 777 | 258 | 2 | 1 |
2.3. Transcriptional Analysis of the bcaba Cluster
 cell weight,
cell weight,  ABA production,
ABA production,  relative expression level of bcaba1, bcaba2, bcaba3 and bcaba4, respectively.
relative expression level of bcaba1, bcaba2, bcaba3 and bcaba4, respectively.
 cell weight,
cell weight,  ABA production,
ABA production,  relative expression level of bcaba1, bcaba2, bcaba3 and bcaba4, respectively.
relative expression level of bcaba1, bcaba2, bcaba3 and bcaba4, respectively.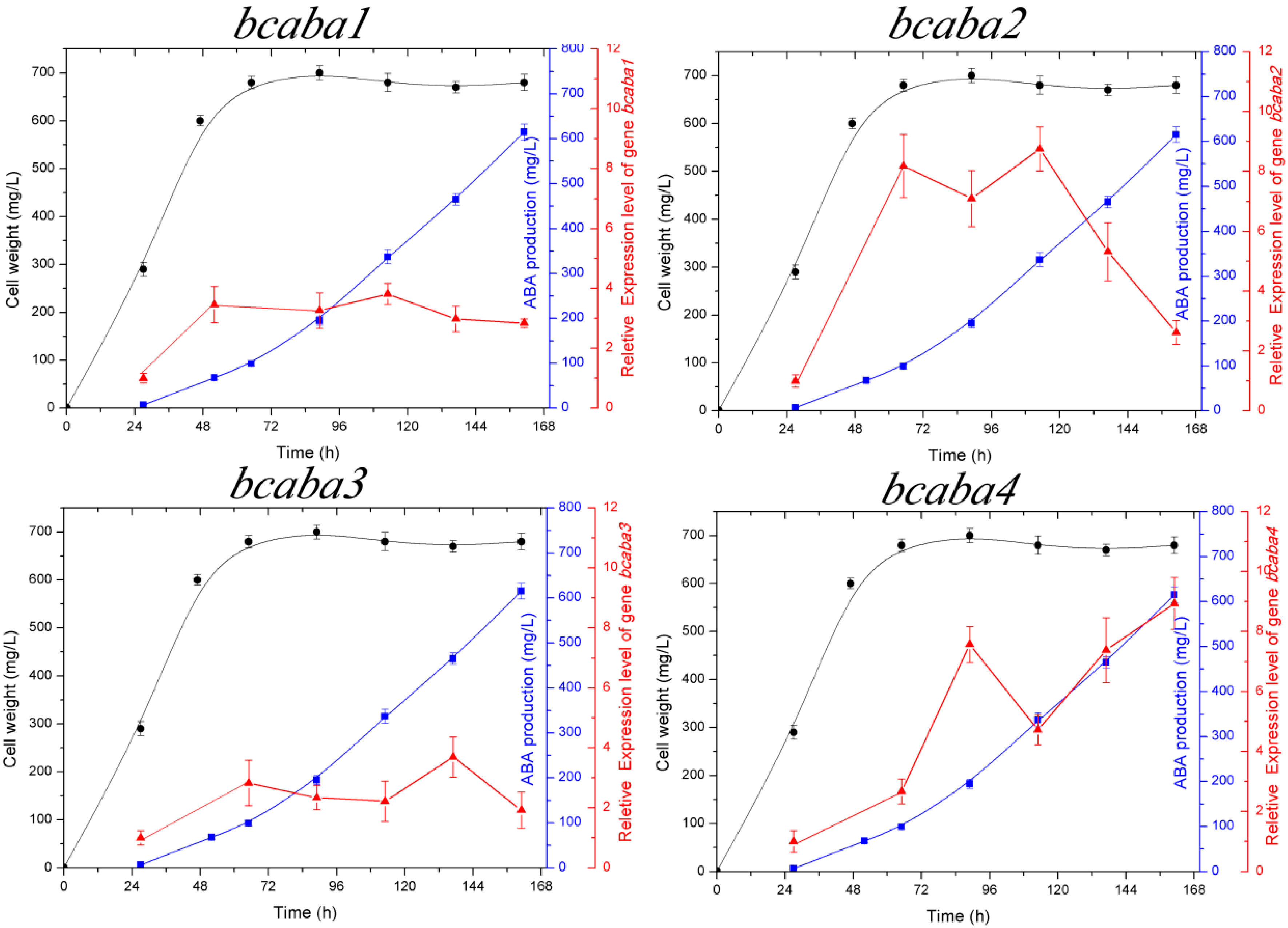
3. Discussions
4. Experimental Section
4.1. Strains, Plasmids and Culture Conditions
4.2. Fed-Batch Fermentation and Determination of ABA
4.3. DNA Manipulation and PCR Amplification
4.4. DNA Sequencing
4.5. RNA Extraction
4.6. Coding Sequence and Intron Identification by Reverse Transcriptase (RT)-PCR
| Genes | Forward Primers (5'-3') | Reverse Primers (5'-3') |
|---|---|---|
| bcababa1 | 5'-ATGTCTAATTCTATATTGAAC-3' | 5'-CTATTTGTATTCTGTTCCC-3' |
| bcababa2 | 5'- ATGCTGCTTAGCATTAAAGA-3' | 5'-CTATCTAGGAACCTCTTTTA-3' |
| bcababa3 | 5'- ATGCAGCAAGTTATTACTCA-3' | 5'-CTAGGTACTTTCTCCACGAT-3' |
| bcababa4 | 5'-ATGTCCTCTCAACCATTCAC-3' | 5'-CTAACATCTCCATCCGCCAT-3' |
4.7. Sequence Analysis
4.8. Analysis of Gene Expression Profiling by SYBR Green Real-Time PCR Assays
| Genes | Forward Primers (5'-3') | Reverse Primers (5'-3') | Product Size (bp) |
|---|---|---|---|
| bcaba1 | 5'-GCCCAAAGCCTACTGATAAA-3' | 5'-TGTCGAATGAATGACCCAAG-3' | 124 |
| bcaba2 | 5'-CTTATTACTTCCCGTTTACTC-3' | 5'-CTTATTACTTCCCGTTTACTC-3' | 101 |
| bcaba3 | 5'-CAAGGAACTCAGCAAGCCC-3' | 5'-AGTCGATGCCAACAAAAGG-3' | 102 |
| bcaba4 | 5'-CTTGGACGAGTGGGAGTT-3' | 5'-GCCGTTGTTAGCCATTAC-3' | 91 |
| 18S rRNA | 5'-GAAACTCACCAGGTCCAGA-3' | 5'-CAAATCACTCCACCAACTAAG-3' | 104 |
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rabbani, M.A.; Maruyama, K.; Abe, H.; Khan, M.A.; Katsura, K.; Ito, Y.; Yoshiwara, K.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 2003, 133, 1755–1767. [Google Scholar] [CrossRef] [PubMed]
- Tsavkelova, E.; Klimova, S.Y.; Cherdyntseva, T.; Netrusov, A. Microbial producers of plant growth stimulators and their practical use: A review. Appl. Biochem. Microbiol. 2006, 42, 117–126. [Google Scholar] [CrossRef]
- Secchi, F.; Perrone, I.; Chitarra, W.; Zwieniecka, A.K.; Lovisolo, C.; Zwieniecki, M.A. The dynamics of embolism refilling in abscisic acid (ABA)-deficient tomato plants. Int. J. Mol. Sci. 2012, 14, 359–377. [Google Scholar] [CrossRef] [PubMed]
- Dorffling, K.; Petersen, W.; Sprecher, E.; Urbasch, I.; Hanssen, H. Abscisic acid in phytopathogenic fungi of the genera Botrytis, Ceratocystis, Fusarium, and Rhizoctoma. Zeitschrift fur Naturforschung. Sect. C Biosci. 1984, 39, 683–684. [Google Scholar]
- Marumo, S.; Katayama, M.; Komori, E.; Ozaki, Y.; Natsume, M.; Kondo, S. Microbial production of abscisic acid by Botrytis cinerea. Agric. Biol. Chem. 1982, 46, 1967–1968. [Google Scholar]
- Zhou, J.; Wu, K.; Lei, B.; Yang, J.; Tan, H. Isolation method and biological activity of metabolites from Botrytis cinerea. Chin. J. Appl. Environ. Biol. 2001, 8, 532–534. [Google Scholar]
- Wang, H.; Niu, X.; Zhang, J.; Dong, J.; Shang, H. Isolation and identification of abscisic acid from BC4 isolate of Botrytis cinerea. J. Northwest. Sci.-Tech. Univ. Agric. For. 2003, 32, 34–36. [Google Scholar]
- Zhang, H.; Liu, J.; He, W. Screening of abscisic acid producing fungi and optimization of its fermentation conditions. Ind. Microbiol. 2008, 1, 49–52. [Google Scholar]
- Hirai, N.; Yoshida, R.; Todoroki, Y.; Ohigashi, H. Biosynthesis of abscisic acid by the non-mevalonate pathway in plants, and by the mevalonate pathway in fungi. Biosci. Biotechnol. Bbiochem. 2000, 64, 1448–1458. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Oritani, T.; Kiyota, H. Biosynthesis and metabolism of abscisic acid and related compounds. Nat. Prod. Rep. 2003, 20, 414–425. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, L.M. Plant Growth and Development: Hormones and Environment; Academic Press: London, UK, 2002; pp. 217–231. [Google Scholar]
- Inomata, M.; Hirai, N.; Yoshida, R.; Ohigashi, H. The biosynthetic pathway to abscisic acid via ionylideneethane in the fungu Botrytis cinerea. Phytochemistry 2004, 65, 2667–2678. [Google Scholar]
- Yamamoto, H.; Inomata, M.; Tsuchiya, S.; Nakamura, M.; Oritani, T. Metabolism of chiral ionylideneacetic acids on the abscisic acid biosynthetic pathway in Cercospora. Biosci. Biotechnol. Bbiochem. 2000, 64, 2644–2650. [Google Scholar] [CrossRef]
- Bennett, R.D.; Norman, S.M.; Maier, V. Intermediate steps in the biosynthesis of abscisic acid from farnesyl pyrophosphate in Cercospora rosicola. Phytochemistry 1990, 29, 3473–3477. [Google Scholar]
- Inomata, M.; Hirai, N.; Yoshida, R.; Ohigashi, H. Biosynthesis of abscisic acid by the direct pathway via ionylideneethane in a fungus, Cercospora cruenta. Biosci. Biotechnol. Biochem. 2004, 68, 2571–2580. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Oritani, T.; Yamashita, K. Metabolism of (2Z,4E)-γ-ionylideneethanol and (2Z,4E)-γ-ionylideneacetic acid in Cercospora cruenta. Agric. Biol. Chem. 1987, 51, 2695–2699. [Google Scholar] [CrossRef]
- Okamoto, M.; Hirai, N.; Koshimizu, K. Biosynthesis of abscisic acid from α-ionylideneethanol in Cercospora pini-densiflorae. Phytochemistry 1988, 27, 3465–3469. [Google Scholar]
- Siewers, V.; Kokkelink, L.; Smedsgaard, J.; Tudzynski, P. Identification of an abscisic acid gene cluster in the grey mold Botrytis cinerea. Appl. Environ. Microbiol. 2006, 72, 4619–4626. [Google Scholar] [CrossRef] [PubMed]
- Siewers, V.; Smedsgaard, J.; Tudzynski, P. The P450 monooxygenase BcABA1 is essential for abscisic acid biosynthesis in Botrytis cinerea. Appl. Environ. Microbiol. 2004, 70, 3868–3876. [Google Scholar] [CrossRef] [PubMed]
- Law, Y.-S.; Gudimella, R.; Song, B.-K.; Ratnam, W.; Harikrishna, J.A. Molecular characterization and comparative sequence analysis of defense-related gene, oryza rufipogon receptor-like protein kinase 1. Int. J. Mol. Sci. 2012, 13, 9343–9362. [Google Scholar] [CrossRef] [PubMed]
- Botrytis cinerea B05.10 genome. Available online: http://www.broadinstitute.org/annotation/genome/botrytis_cinerea/GenomeDescriptions.html#BC1 (accessed on 16 August 2014).
- Botrytis cinerea T4genome. Available online: http://www.broadinstitute.org/annotation/genome/botrytis_cinerea/GenomeDescriptions.html#Botcin_T4_vankan (accessed on 16 August 2014).
- Botrytis cinerea T4genome. Available online: https://urgi.versailles.inra.fr/Species/Botrytis (accessed on 16 August 2014).
- Amselem, J.; Cuomo, C.A.; van Kan, J.A.; Viaud, M.; Benito, E.P.; Couloux, A.; Coutinho, P.M.; de Vries, R.P.; Dyer, P.S.; Fillinger, S. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 2011, 7, e1002230. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.A.; Staats, M.; van Kan, J.A.; Rokas, A.; Slot, J.C. Repeated loss of an anciently horizontally transferred gene cluster in Botrytis. Mycologia 2013, 105, 1126–1134. [Google Scholar] [PubMed]
- Tan, H.; Gong, G.; Li, Z.; Peng, S.; Lei, B.; Liu, D.; Ding, L. High-yield ABA producing strain obtained by UV irradiation of protoplasts. Chin. J. Appl. Envrion. Biol. 1998, 3, 281–285. [Google Scholar]
- Gong, T.; Shu, D.; Zhao, M.; Zhong, J.; Deng, H.Y.; Tan, H. Isolation of genes related to abscisic acid production in Botrytis cinerea TB-3-H8 by cDNA-AFLP. J. Basic Microbiol. 2014, 54, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Conserved Domains and Protein Classification. Available online: http://www.ncbi.nlm.nih.gov/cdd/ (accessed on 18 October 2013).
- Spiering, M.J.; Moon, C.D.; Wilkinson, H.H.; Schardl, C.L. Gene clusters for insecticidal loline alkaloids in the grass-endophytic fungus Neotyphodium uncinatum. Genetics 2005, 169, 1403–1414. [Google Scholar] [PubMed]
- Zhao, M.; Zhou, J.Y.; Li, Z.D.; Song, W.W.; Gong, T.; Tan, H. Boty-like retrotransposons in the filamentous fungus Botrytis cinerea contain the additional antisense gene brtn. Virology 2011, 417, 248–252. [Google Scholar]
- Choquer, M.; Fournier, E.; Kunz, C.; Levis, C.; Pradier, J.M.; Simon, A.; Viaud, M. Botrytis cinerea virulence factors: New insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol. Lett. 2007, 277, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tudzynski, B.; Sharon, A. Biosynthesis, biological role and application of fungal phytohormones. The Mycota 2002, 10, 183–211. [Google Scholar]
- Tokugawa, N.; Shirai, M.; Yonehara, T. Production of Nature-Type Abscisic Acid. JP6247927A, 1994. [Google Scholar]
- Shu, D.; Xiao, L. Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu, China. Unpublished work. 2013. [Google Scholar]
- Malonek, S.; Rojas, M.; Hedden, P.; Gaskin, P.; Hopkins, P.; Tudzynski, B. Functional characterization of two cytochrome P450 monooxygenase genes, P450–1 and P450–4, of the gibberellic acid gene cluster in Fusarium proliferatum (Gibberella fujikuroi MP-D). Appl. Environ. Microbiol. 2005, 71, 1462–1472. [Google Scholar] [CrossRef] [PubMed]
- Shu, D.; Zhong, J. Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu, China. Unpublished work. 2013. [Google Scholar]
- Ladero, V.; Rattray, F.P.; Mayo, B.; Martín, M.C.; Fernández, M.; Alvarez, M.A. Sequencing and transcriptional analysis of the biosynthesis gene cluster of putrescine-producing Lactococcus lactis. Appl. Environ. Microbiol. 2011, 77, 6409–6418. [Google Scholar] [CrossRef] [PubMed]
- Möller, E.; Bahnweg, G.; Sandermann, H.; Geiger, H. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 1992, 20, 6115. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. ClustalW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, S. Promoter2. 0: For the recognition of PolII promoter sequences. Bioinformatics 1999, 15, 356–361. [Google Scholar] [PubMed]
- Schwartz, S.; Zhang, Z.; Frazer, K.A.; Smit, A.; Riemer, C.; Bouck, J.; Gibbs, R.; Hardison, R.; Miller, W. PipMaker-a web server for aligning two genomic DNA sequences. Genome Res. 2000, 10, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Suarez, M.B.; Walsh, K.; Boonham, N.; O’Neill, T.; Pearson, S.; Barker, I. Development of real-time PCR (TaqMan) assays for the detection and quantification of Botrytis cinerea in planta. Plant Physiol. Biochem. 2005, 43, 890–899. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Huang, Z.; Chen, Y.; Zhang, Y.; Rong, G.; Mu, C.; Xu, Q.; Chen, G. Molecular cloning and functional analysis of the duck TLR4 Gene. Int. J. Mol. Sci. 2013, 14, 18615–18628. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Ma, J.; Xu, G.; Wang, D.; Wang, N. Molecular cloning, structural analysis and tissue expression of protein phosphatase 3 catalytic subunit alpha isoform (PPP3CA) gene in Tianfu goat muscle. Int. J. Mol. Sci. 2014, 15, 2346–2358. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, T.; Shu, D.; Yang, J.; Ding, Z.-T.; Tan, H. Sequencing and Transcriptional Analysis of the Biosynthesis Gene Cluster of Abscisic Acid-Producing Botrytis cinerea. Int. J. Mol. Sci. 2014, 15, 17396-17410. https://doi.org/10.3390/ijms151017396
Gong T, Shu D, Yang J, Ding Z-T, Tan H. Sequencing and Transcriptional Analysis of the Biosynthesis Gene Cluster of Abscisic Acid-Producing Botrytis cinerea. International Journal of Molecular Sciences. 2014; 15(10):17396-17410. https://doi.org/10.3390/ijms151017396
Chicago/Turabian StyleGong, Tao, Dan Shu, Jie Yang, Zhong-Tao Ding, and Hong Tan. 2014. "Sequencing and Transcriptional Analysis of the Biosynthesis Gene Cluster of Abscisic Acid-Producing Botrytis cinerea" International Journal of Molecular Sciences 15, no. 10: 17396-17410. https://doi.org/10.3390/ijms151017396
APA StyleGong, T., Shu, D., Yang, J., Ding, Z.-T., & Tan, H. (2014). Sequencing and Transcriptional Analysis of the Biosynthesis Gene Cluster of Abscisic Acid-Producing Botrytis cinerea. International Journal of Molecular Sciences, 15(10), 17396-17410. https://doi.org/10.3390/ijms151017396




