Bioactive Secondary Metabolites from an Arctic Marine-Derived Strain, Streptomyces sp. MNP-1, Using the OSMAC Strategy
Abstract
1. Introduction
2. Results
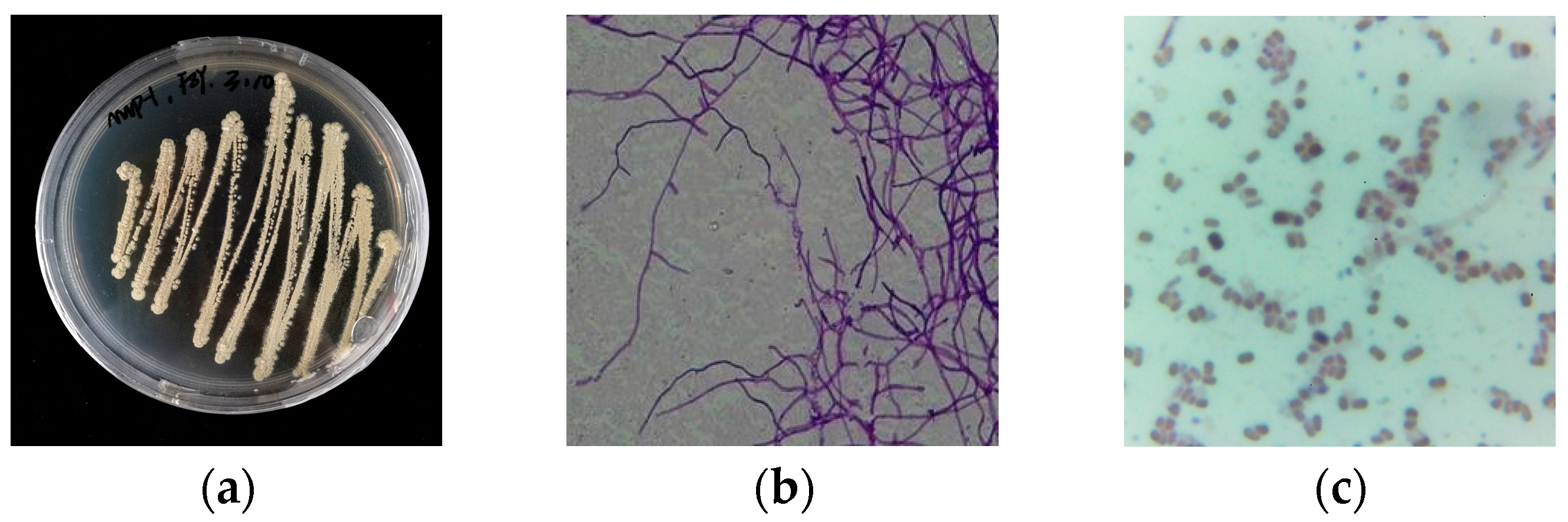
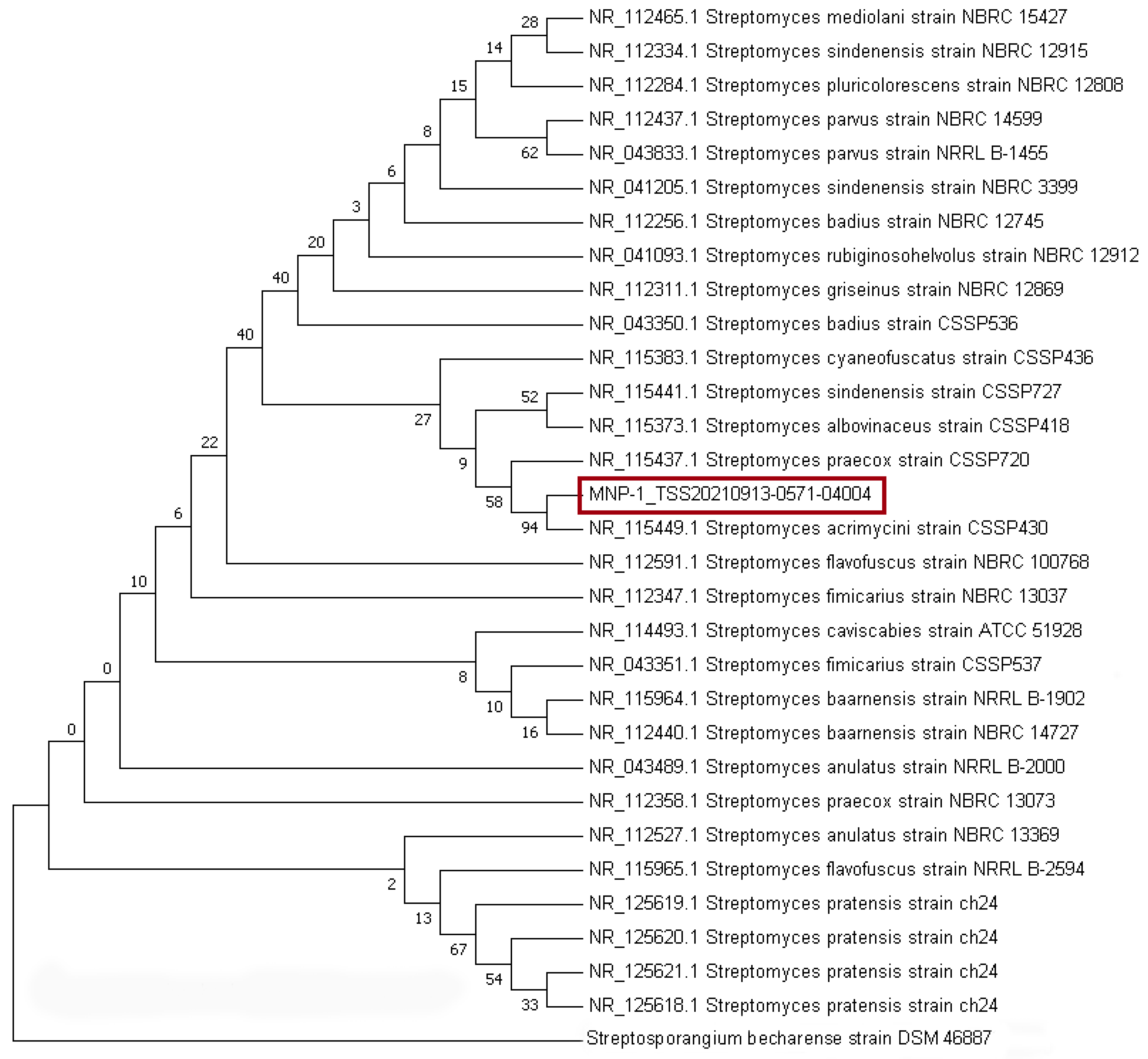
2.1. Strain Classification
2.2. Medium Evaluation
2.3. Structure Elucidation
2.4. Bioactivity
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Biological Materials
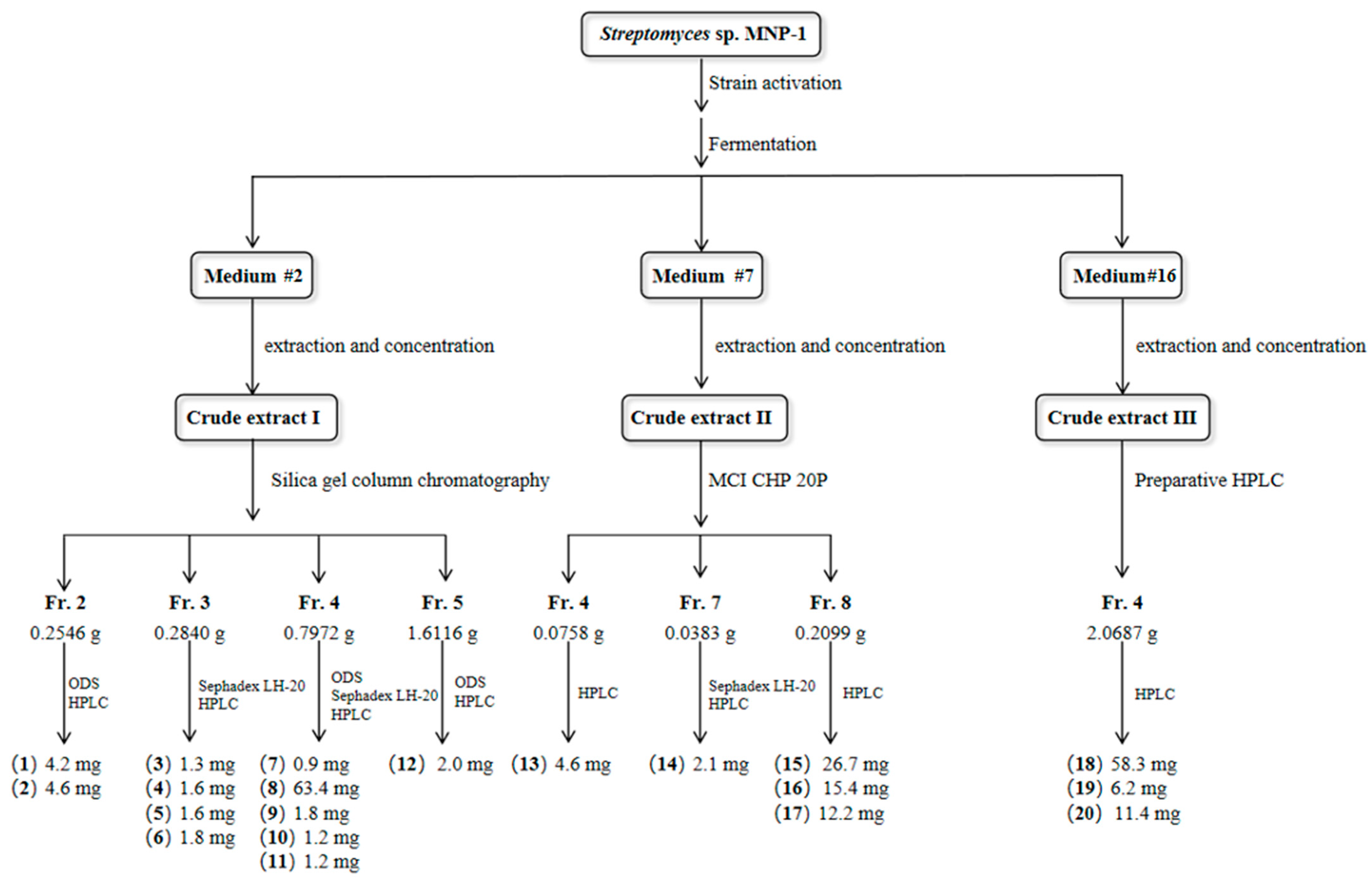
4.3. Fermentation and Extraction
4.4. Isolation and Purification
4.5. Antimicrobial Assay
4.6. Cytotoxicity Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DMSO | Dimethyl sulfoxide |
| ESI-MS | Electrospray ionization mass spectrometry |
| 1H NMR | Proton nuclear magnetic resonance |
| HPLC | High-performance liquid chromatography |
| IC50 | Half maximal inhibitory concentration |
| MIC | Minimal inhibitory concentration |
| MTT | Methylthiazolyldiphenyl-tetrazolium bromide |
| ODS | Octadecylsilyl |
| OSMAC | One strain many compounds |
| ESI-MS | Electrospray ionization mass spectrometry |
References
- Rampelotto, P.H. Extremophiles and extreme environments. Life 2013, 3, 482–485. [Google Scholar] [CrossRef]
- Wilson, Z.E.; Brimble, M.A. Molecules derived from the extremes of life: A decade later. Nat. Prod. Rep. 2021, 38, 24–82. [Google Scholar] [CrossRef]
- Mast, Y.; Stegmann, E. Actinomycetes: The antibiotics producers. Antibiotics 2019, 8, 105. [Google Scholar] [CrossRef]
- Lyudmila, P.T.; Gul, B.B.; Baiken, B.B.; Assya, S.B.; Saule, T.D.; Elmira, R.F.; John, A.B. Beyond traditional screening: Unveiling antibiotic potentials of actinomycetes in extreme environments. Heliyon 2024, 10, e40371. [Google Scholar] [CrossRef]
- Magot, F.; Van Soen, G.; Buedenbender, L.; Li, F.; Soltwedel, T.; Grauso, L.; Mangoni, A.; Blümel, M.; Tasdemir, D. Bioactivity and metabolome mining of deep-sea sediment-derived microorganisms reveal new hybrid PKS-NRPS macrolactone from Aspergillus versicolor PS108-62. Mar. Drugs 2023, 21, 95. [Google Scholar] [CrossRef]
- Martín-Aragón, V.R.; Millán, F.R.; Cuadrado, C.; Daranas, A.H.; Medarde, A.F.; López, J.M.S. Induction of new aromaticpolyketides from the marine actinobacterium Streptomyces griseorubiginosus through an OSMAC approach. Mar. Drugs 2023, 21, 526. [Google Scholar] [CrossRef]
- Yu, H.-B.; Ning, Z.; Hu, B.; Zhu, Y.-P.; Lu, X.-L.; He, Y.; Jiao, B.-H.; Liu, X.-Y. Cytosporin derivatives from Arctic-derived fungus Eutypella sp. D-1 via the OSMAC approach. Mar. Drugs 2023, 21, 382. [Google Scholar] [CrossRef]
- Hussain, A.; Rather, M.A.; Dar, M.S.; Aga, M.A.; Ahmad, N.; Manzoor, A.; Qayum, A.; Shah, A.; Mushtaq, S.; Ahmad, Z.; et al. Novel bioactive molecules from Lentzea Violacea strain AS 08 using one strain many compounds (OSMAC) approach. Bioorg. Med. Chem. Lett. 2017, 27, 2579–2582. [Google Scholar] [CrossRef]
- Pan, R.; Bai, X.L.; Chen, J.W.; Zhang, H.W.; Wang, H. Exploring structural diversity of microbe secondary metabolites using OSMAC strategy: A literature review. Front. Microbiol. 2019, 10, 294. [Google Scholar] [CrossRef]
- Pinedo-Rivilla, C.; Aleu, J.; Durán-Patrón, R. Cryptic metabolites from marine-derived microorganisms using OSMAC and epigenetic approaches. Mar. Drugs 2022, 20, 84. [Google Scholar] [CrossRef]
- Rateb, M.E.; Houssen, W.E.; Harrison, W.T.A.; Deng, H.; Okoro, C.K.; Asenjo, J.A.; Andrews, B.A.; Bull, A.T.; Goodfellow, M.; Ebel, R.; et al. Diverse metabolic profiles of a Streptomyces strain isolated from a hyper-arid environment. J. Nat. Prod. 2011, 74, 1965–1971. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.H.; Liang, X.; Qi, S.H. Eight new cyclopentenone and cyclohexenone derivatives from the marine-derived fungus Aspergillus sp. SCSIO 41501 by OSMAC strategy. Nat. Prod. Res. 2020, 35, 3810–3819. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, J.; Tatamidani, H.; Fukumoto, Y.; Chatani, N. A new synthesis of aldehydes by the palladium-catalyzed reaction of 2-pyridinyl esters with hydrosilanes. Synlett 2006, 37, 869–872. [Google Scholar] [CrossRef]
- Mehnaz, S.; Saleem, R.S.; Yameen, B.; Pianet, I.; Schnakenburg, G.; Pietraszkiewicz, H.; Valeriote, F.; Josten, M.; Sahl, H.G.; Franzblau, S.G.; et al. Lahorenoic acids A-C, ortho-dialkyl-substituted aromatic acids from the biocontrol strain Pseudomonas aurantiaca PB-St2. J. Nat. Prod. 2013, 76, 135–141. [Google Scholar] [CrossRef]
- Roemer, A.; Scholl, H.; Budzikiewicz, H.; Korth, H.; Pulverer, G. Bacterial constituents. part II. phenazines from pseudomonads. Org. Chem. 1981, 36B, 1037–1046. [Google Scholar]
- Chen, C.; Pan, Y.; Zhao, H.; Xu, X.; Luo, Z.; Cao, L.; Xi, S.; Li, H.; Xu, L. Ruthenium(II)-catalyzed regioselective C-8 hydroxylation of 1,2,3,4-tetrahydroquinolines. Org. Lett. 2018, 20, 6799–6803. [Google Scholar] [CrossRef]
- Li, C.; Wang, M.; Lu, X.; Zhang, L.; Jiang, J.; Zhang, L. Reusable brønsted acidic ionic liquid efficiently catalyzed N-formylation and N-acylation of amines. ACS Sustain. Chem. Eng. 2020, 8, 4353–4361. [Google Scholar]
- Xu, H.J.; Liang, Y.F.; Cai, Z.Y.; Qi, H.X.; Yang, C.Y.; Feng, Y.S. CuI-nanoparticles-catalyzed selective synthesis of phenols, anilines, and thiophenols from aryl halides in aqueous solution. J. Org. Chem. 2001, 76, 2296–2300. [Google Scholar] [CrossRef]
- Hund, H.K.; de Beyer, A.; Lingens, F. Microbial metabolism of quinoline and related compounds. VI. degradation of quinaldine by Arthrobacter sp. Biol. Chem. Hoppe. Seyler. 1990, 371, 1005–1008. [Google Scholar] [CrossRef]
- Suzuki, H.; Ohnishi, Y.; Furusho, Y.; Sakuda, S.; Horinouchi, S. Novel benzene ring biosynthesis from C(3) and C(4) primary metabolites by two enzymes. J. Biol. Chem. 2006, 281, 36944–36951. [Google Scholar] [CrossRef]
- Toyobo Co., Ltd. 3-Amino-4-hydroxybenzoic acid biosynthetic genes from Streptomyces griseus, and use in production of aminohydroxy aromatic carboxylic acid. JP2004283163. 14 October 2004. [Google Scholar]
- Galm, U.; Dessoy, M.A.; Schmidt, J.; Wessjohann, L.A.; Heide, L. In vitro and in vivo production of new aminocoumarins by a combined biochemical, genetic, and synthetic approach. Chem. Biol. 2004, 11, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, S.; Yasui, K. Interaction of N-6-substituted-9-methyladenines to poly-5-bromouridylic acid. Nucleic Acids Symp. Ser. 1983, 12, 185–188. [Google Scholar]
- Seim, K.L.; Obermeyer, A.C.; Francis, M.B. Oxidative modification of native protein residues using cerium(IV) ammonium nitrate. J. Am. Chem. Soc. 2001, 133, 16970–16976. [Google Scholar] [CrossRef]
- Jiao, J.; Zhang, X.R.; Chang, N.H.; Wang, J.; Wei, J.F.; Shi, X.Y.; Chen, Z.G. A facile and practical copper powder-catalyzed, organic solvent-and ligand-free ullmann amination of aryl halides. J. Org. Chem. 2011, 76, 1180–1183. [Google Scholar] [CrossRef]
- Senior, M.M.; Williamson, R.T.; Martin, G.E. Using HMBC and adequate NMR data to define and differentiate long-range coupling pathways: Is the crews rule obsolete? J. Nat. Prod. 2013, 76, 2088–2093. [Google Scholar] [CrossRef]
- Choi, J.Y.; Choi, E.H.; Jung, H.W.; Oh, J.S.; Lee, W.H.; Lee, J.G.; Lee, S.H. Melanogenesis inhibitory compounds from saussureae radix. Arch. Pharm. Res. 2008, 31, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Tashiro, T.; Akasaka, K.; Ohrui, H.; Fattorusso, E. Determination of the absolute configuration at the two cyclopropane moieties of plakoside A, an immunosuppressive marine galactosphingolipid. Tetrahedron Lett. 2002, 43, 3719–3722. [Google Scholar] [CrossRef]
- Huang, X.A.; Yang, R.Z. A new hydroquinone diglucoside from Lysimachia fordiana. Chem. Nat. Compd. 2004, 40, 457–459. [Google Scholar] [CrossRef]
- Gutierrez-Lugo, M.T.; Woldemichael, G.M.; Singh, M.P.; Suarez, P.A.; Maiese, W.M.; Montenegro, G.; Timmermann, B.N. Isolation of three new naturally occurring compounds from the culture of Micromonospora sp. P1068. Nat. Prod. Res. 2005, 19, 645–652. [Google Scholar] [CrossRef]
- He, J.; Fan, P.; Feng, S.; Shao, P.; Sun, P. Isolation and purification of two isoflavones from hericium erinaceum mycelium by high-speed counter-current chromatography. Molecules 2018, 23, 560. [Google Scholar] [CrossRef]
- Selepe, M.A.; Drewes, S.E.; Van Heerden, F.R. Total synthesis of the pyranoisoflavone kraussianone 1 and related isoflavones. J. Nat. Prod. 2010, 73, 1680–1685. [Google Scholar] [CrossRef] [PubMed]
- Kol, S.; Merlo, M.E.; Scheltema, R.A.; de Vries, M.; Vonk, R.J.; Kikkert, N.A.; Dijkhuizen, L.; Breitling, R.; Takano, E. Metabolomic characterization of the salt atress response in Streptomyces coelicolor. Appl. Environ. Microbiol. 2010, 76, 2574–2581. [Google Scholar]
- Zhao, F.; Qin, Y.H.; Zheng, X.; Zhao, H.W.; Chai, D.Y.; Li, W.; Pu, M.X.; Zuo, X.S.; Qian, W.; Ni, P. Biogeography and adaptive evolution of Streptomyces strains from saline environments. Sci. Rep. 2016, 6, 32718. [Google Scholar] [CrossRef]
- Novella, I.S.; Sánchez, J. Effects of 5-Azacytidine on physiological differentiation of Streptomyces antibioticus. Res. Microbiol. 1995, 146, 721–728. [Google Scholar] [CrossRef]
- Kumar, J.; Sharma, V.K.; Singh, D.K.; Mishra, A.; Gond, S.K.; Verma, S.K.; Kumar, A.; Kharwar, R.N. Epigenetic activation of antibacterial property of an endophytic Streptomyces coelicolor strain AZRA 37 and identification of the induced protein using MALDI TOF MS/MS. PLoS ONE 2016, 11, e0147876. [Google Scholar] [CrossRef][Green Version]
- Karuppiah, V.; Alagappan, K.; Sivakumar, K.; Kannan, L. Phenazine-1-Carboxylic Acid-Induced Programmed Cell Death in Human Prostate Cancer Cells Is Mediated by Reactive Oxygen Species Generation and Mitochondrial-Related Apoptotic Pathway. J. Appl. Biomed. 2016, 14, 199–209. [Google Scholar] [CrossRef]
- Cimmino, A.; Bahmani, Z.; Castaldi, S.; Masi, M.; Isticato, R.; Abdollahzadeh, J.; Amini, J.; Evidente, A. Phenazine-1-Carboxylic Acid (PCA), produced for the first time as an antifungal metabolite by truncatella angustata, a causal agent of Grapevine Trunk Diseases (GTDs) in iran. J. Agric. Food Chem. 2021, 69, 12143–12147. [Google Scholar] [CrossRef]
- Zhang, L.L.; Tian, X.Y.; Kuang, S.; Liu, G.; Zhang, C.S.; Sun, C.M. Antagonistic activity and mode of action of phenazine-1-carboxylic acid, produced by marine bacterium Pseudomonas aeruginosa PA31x, against Vibrio anguillarum in vitro and in a zebrafish in vivo model. Front. Microbiol. 2017, 8, 289. [Google Scholar] [CrossRef]
- Nishida, A.; Oda, A.; Takeuchi, A.; Lee, T.; Awano, H.; Hashimoto, N.; Takeshima, Y.; Matsuo, M. Staurosporine allows dystrophin expression by skipping of nonsense-encoding exon. Brain Dev. 2016, 38, 738–745. [Google Scholar] [CrossRef]
- Yadav, S.S.; Prasad, C.B.; Prasad, S.B.; Pandey, L.K.; Singh, S.; Pradhan, S.; Narayan, G. Anti-Tumor Activity of Staurosporine in the Tumor Microenvironment of Cervical Cancer: An in Vitro Study. Life Sci. 2015, 133, 21–28. [Google Scholar] [CrossRef]
- Zambrano, J.N.; Williams, C.J.; Williams, C.B.; Hedgepeth, L.; Burger, P.; Dilday, T.; Eblen, S.T.; Armeson, K.; Hill, E.G.; Yeh, E.S. Staurosporine, an inhibitor of hormonally up-regulated neu-associated kinase. Oncotarget 2018, 9, 35962–35973. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.D.; Gillespie, S.K.; Hersey, P. Staurosporine Induces Apoptosis of Melanoma by Both Caspase-Dependent and -Independent Apoptotic Pathways. Mol. Cancer Ther. 2004, 3, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Rajeswari, P.; Jose, P.A.; Amiya, R.; Jebakumar, S.R.D. Characterization of saltern based Streptomyces sp. and statistical media optimization for its improved antibacterial activity. Front. Microbiol. 2015, 5, 2014. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Chen, S.; Shen, Y.; Wei, D.; Deng, Z. Effect of copper sulfate on biosynthesis of FR-008/candicidin complex production in Streptomyces sp. World J. Microbiol. Biotechnol. 2011, 27, 2033–2039. [Google Scholar] [CrossRef]
- Bind, S.; Bind, S.; Sharma, A.K.; Chaturvedi, P. Epigenetic modification: A key tool for secondary metabolite production in microorganisms. Front. Microbiol. 2022, 13, 784109. [Google Scholar] [CrossRef]
- Zdouc, M.M.; Iorio, M.; Maffioli, S.I.; Crüsemann, M.; Donadio, S.; Sosio, M. Planomonospora: A metabolomics perspective on an underexplored Actinobacteria genus. J. Nat. Prod. 2021, 84, 204–219. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, L.; Han, G.; Zhou, J.; Zhao, Y. Determination of antibacterial activity of aucubigenin and aucubin. Asian J. Chem. 2014, 26, 559–561. [Google Scholar] [CrossRef]
- Erkuş, B.; Özcan, H.; Zaim, Ö. Synthesis, antimicrobial activity, and ion transportation investigation of four new [1 + 1] condensed furan and thiophene-based cycloheterophane amides. J. Heterocycl. Chem. 2020, 57, 1956–1962. [Google Scholar] [CrossRef]
- Shehabeldine, A.M.; Doghish, A.S.; El-Dakroury, W.A.; Hassanin, M.M.H.; Al-Askar, A.A.; AbdElgawad, H.; Hashem, A.H. Antimicrobial, antibiofilm, and anticancer activities of syzygium aromaticum essential oil nanoemulsion. Molecules 2023, 28, 5812. [Google Scholar] [CrossRef]

| Compound | MIC Value (μg/mL) | IC50 Value (µM) | ||||
|---|---|---|---|---|---|---|
| S. aureus ATCC 25923 | E. coli ATCC 25922 | C. albicans ATCC 10231 | A549 | MCF-7 | HepG2 | |
| 1 | >64 | >64 | >64 | >100 | >100 | >100 |
| 2 | 16 | 8 | >64 | >100 | >100 | >100 |
| 3 | 16 | >64 | 4 | 21.52 ± 4.36 | 19.88 ± 1.65 | 35.82 ± 2.70 |
| 4 | >64 | >64 | >64 | >100 | >100 | >100 |
| 5 | >64 | >64 | 32 | >100 | >100 | >100 |
| 6 | >64 | >64 | >64 | >100 | >100 | >100 |
| 7 | >64 | >64 | >64 | >100 | >100 | >100 |
| 8 | >64 | >64 | >64 | >100 | >100 | >100 |
| 9 | 32 | >64 | >64 | >100 | >100 | >100 |
| 10 | >64 | >64 | >64 | >100 | >100 | >100 |
| 11 | >64 | >64 | >64 | >100 | >100 | >100 |
| 12 | >64 | >64 | >64 | >100 | >100 | >100 |
| 13 | >64 | >64 | >64 | >100 | >100 | >100 |
| 14 | >64 | >64 | 8 | 27.79 ± 6.70 | 35.57 ± 2.84 | 23.71 ± 2.89 |
| 15 | 32 | 32 | >64 | >100 | >100 | >100 |
| 16 | >64 | >64 | >64 | >100 | >100 | >100 |
| 17 | >64 | >64 | >64 | >100 | >100 | >100 |
| 18 | >64 | >64 | >64 | >100 | >100 | >100 |
| 19 | >64 | >64 | >64 | >100 | >100 | >100 |
| 20 | >64 | 16 | 16 | 90.37 ± 2.46 | >100 | >100 |
| Positive Control | 0.25 | 1 | 0.25 | 14.86 ± 0.00 | 12.34 ± 0.01 | 15.30 ± 0.01 |
| Negative Control | - | - | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, M.; Liu, Z.; Wang, J.; Hu, W.; Zhang, H. Bioactive Secondary Metabolites from an Arctic Marine-Derived Strain, Streptomyces sp. MNP-1, Using the OSMAC Strategy. Molecules 2025, 30, 1657. https://doi.org/10.3390/molecules30081657
Wu M, Liu Z, Wang J, Hu W, Zhang H. Bioactive Secondary Metabolites from an Arctic Marine-Derived Strain, Streptomyces sp. MNP-1, Using the OSMAC Strategy. Molecules. 2025; 30(8):1657. https://doi.org/10.3390/molecules30081657
Chicago/Turabian StyleWu, Mengna, Zijun Liu, Jiahui Wang, Wentao Hu, and Huawei Zhang. 2025. "Bioactive Secondary Metabolites from an Arctic Marine-Derived Strain, Streptomyces sp. MNP-1, Using the OSMAC Strategy" Molecules 30, no. 8: 1657. https://doi.org/10.3390/molecules30081657
APA StyleWu, M., Liu, Z., Wang, J., Hu, W., & Zhang, H. (2025). Bioactive Secondary Metabolites from an Arctic Marine-Derived Strain, Streptomyces sp. MNP-1, Using the OSMAC Strategy. Molecules, 30(8), 1657. https://doi.org/10.3390/molecules30081657







