Screening of Antioxidant Maillard Reaction Products Using HPLC-HRMS and Study of Reaction Conditions for Their Production as Food Preservatives
Abstract
1. Introduction
2. Results and Discussion
2.1. Optimisation of Chromatographic Method
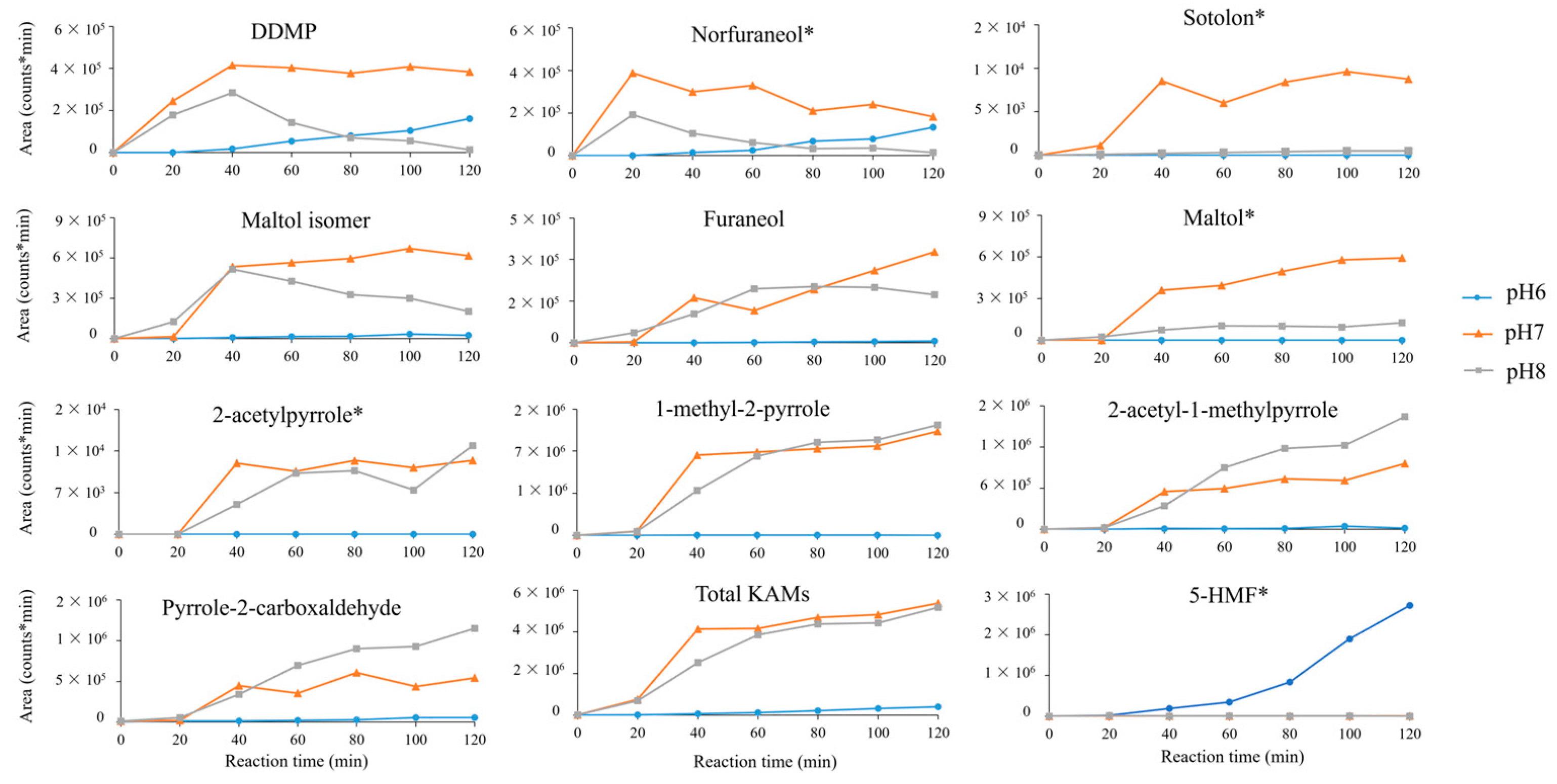
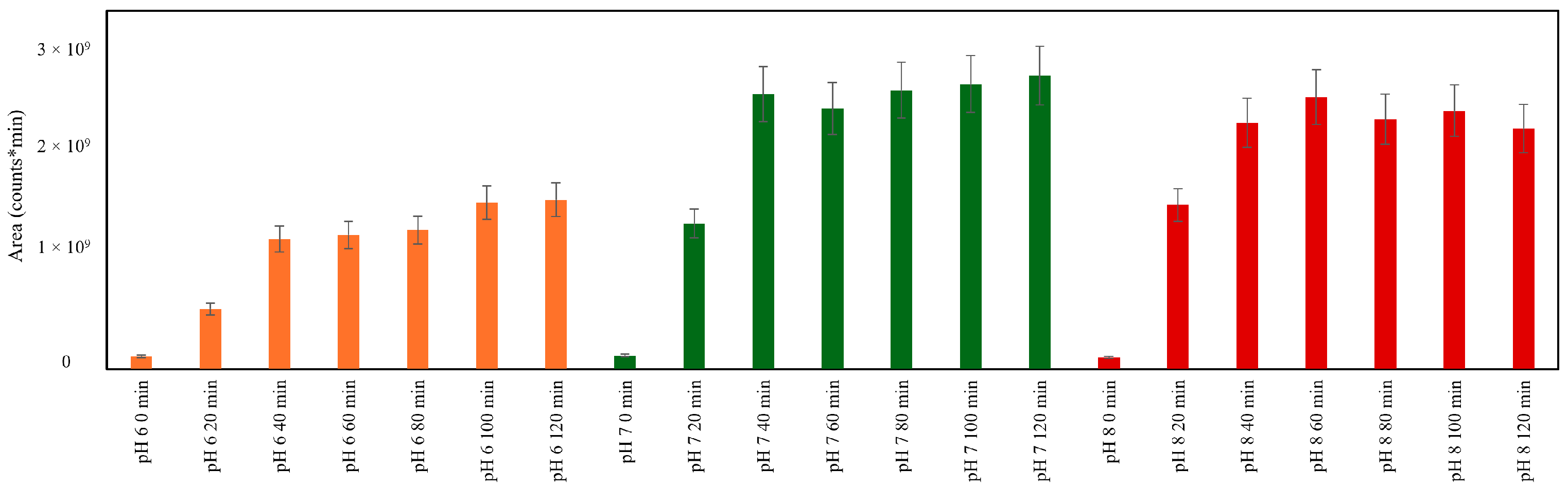
2.2. Influence of Initial pH on Antioxidant Production in MR
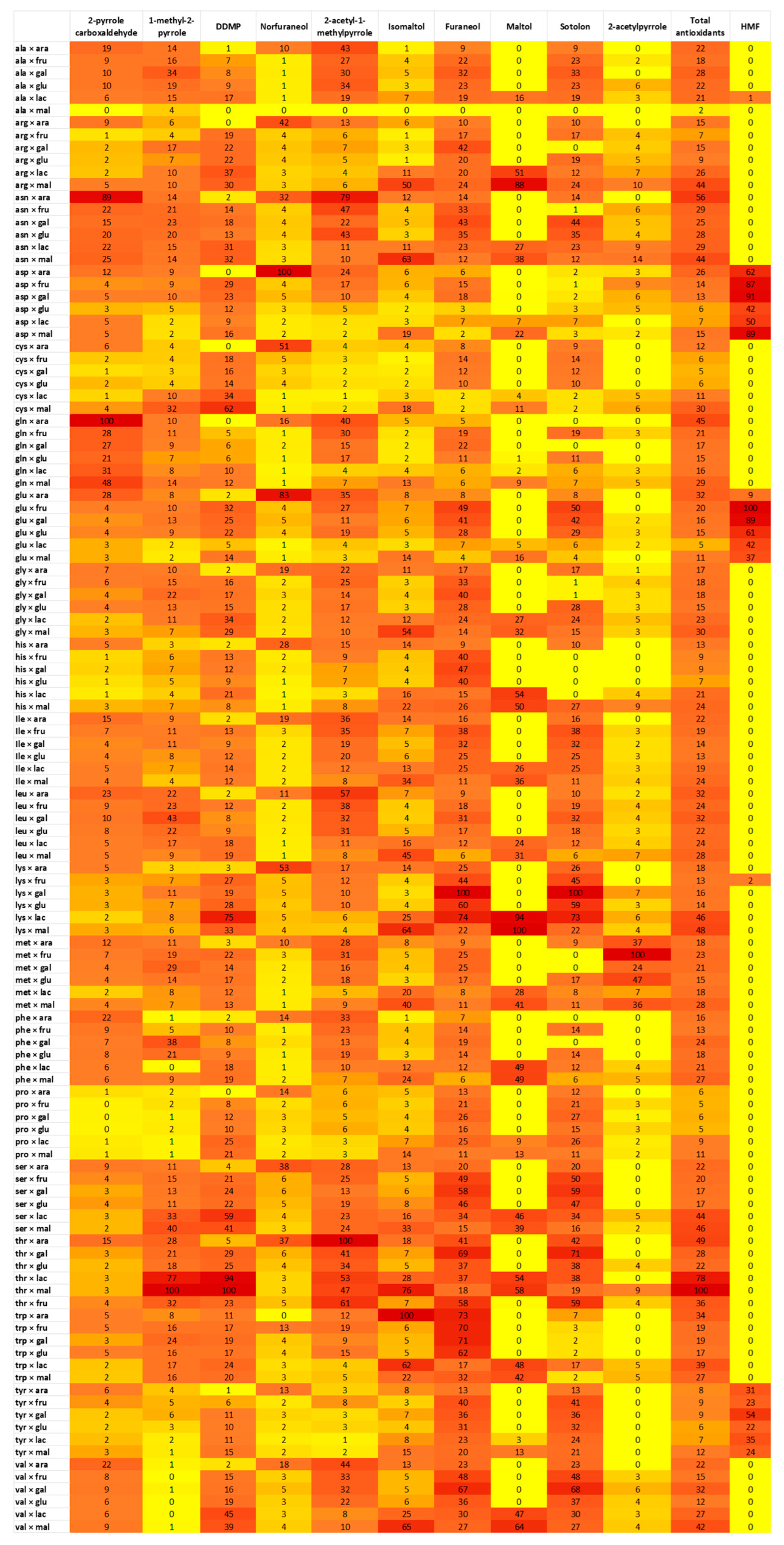
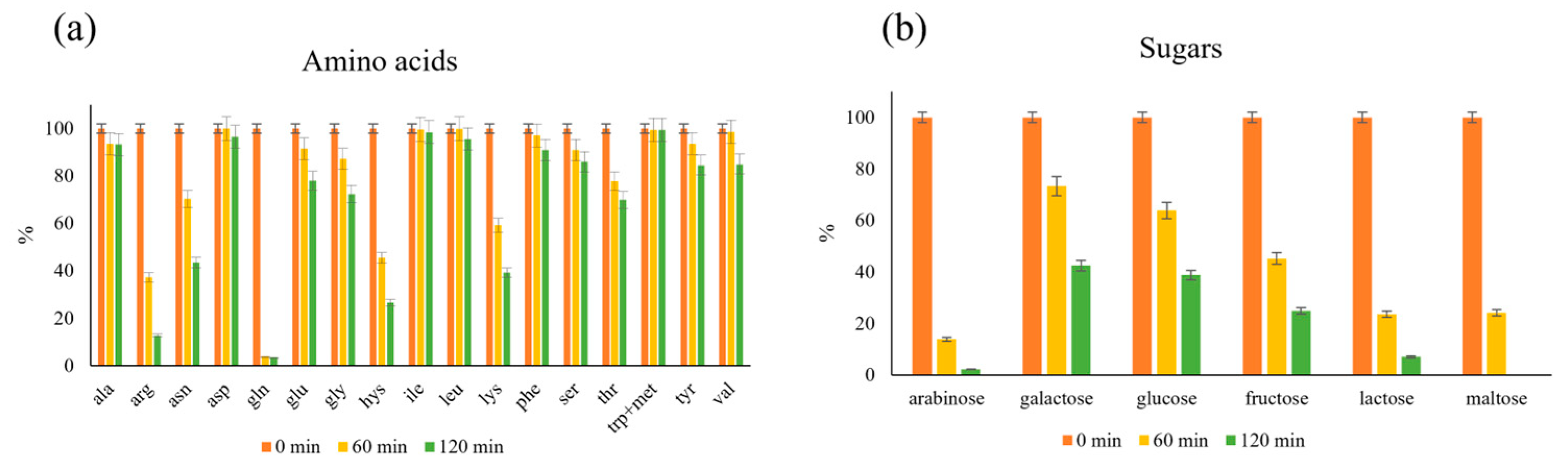
2.3. Influence of Amino Acids and Sugars on Antioxidant Production in MR
2.4. Consumption of Reagents in MR
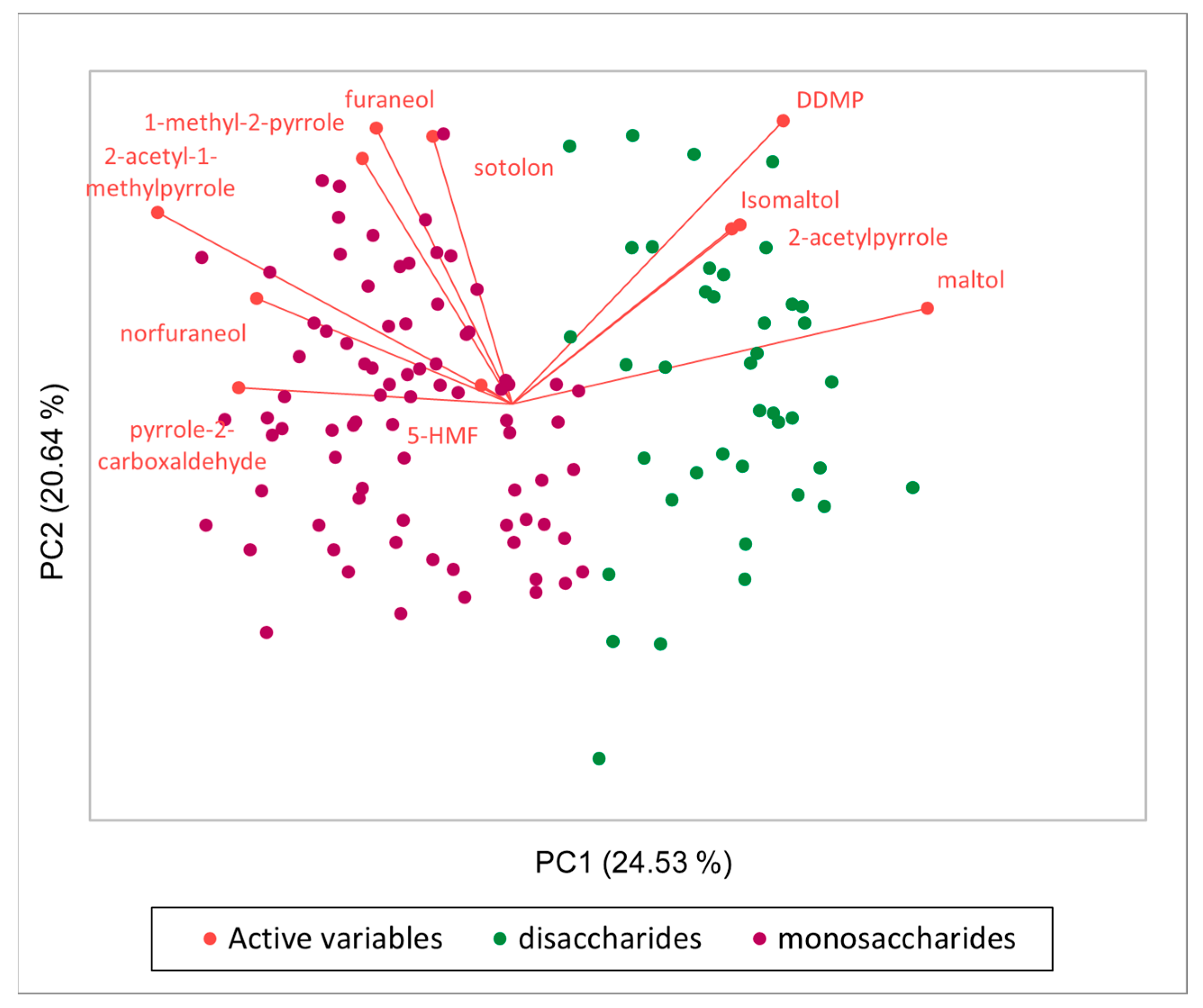
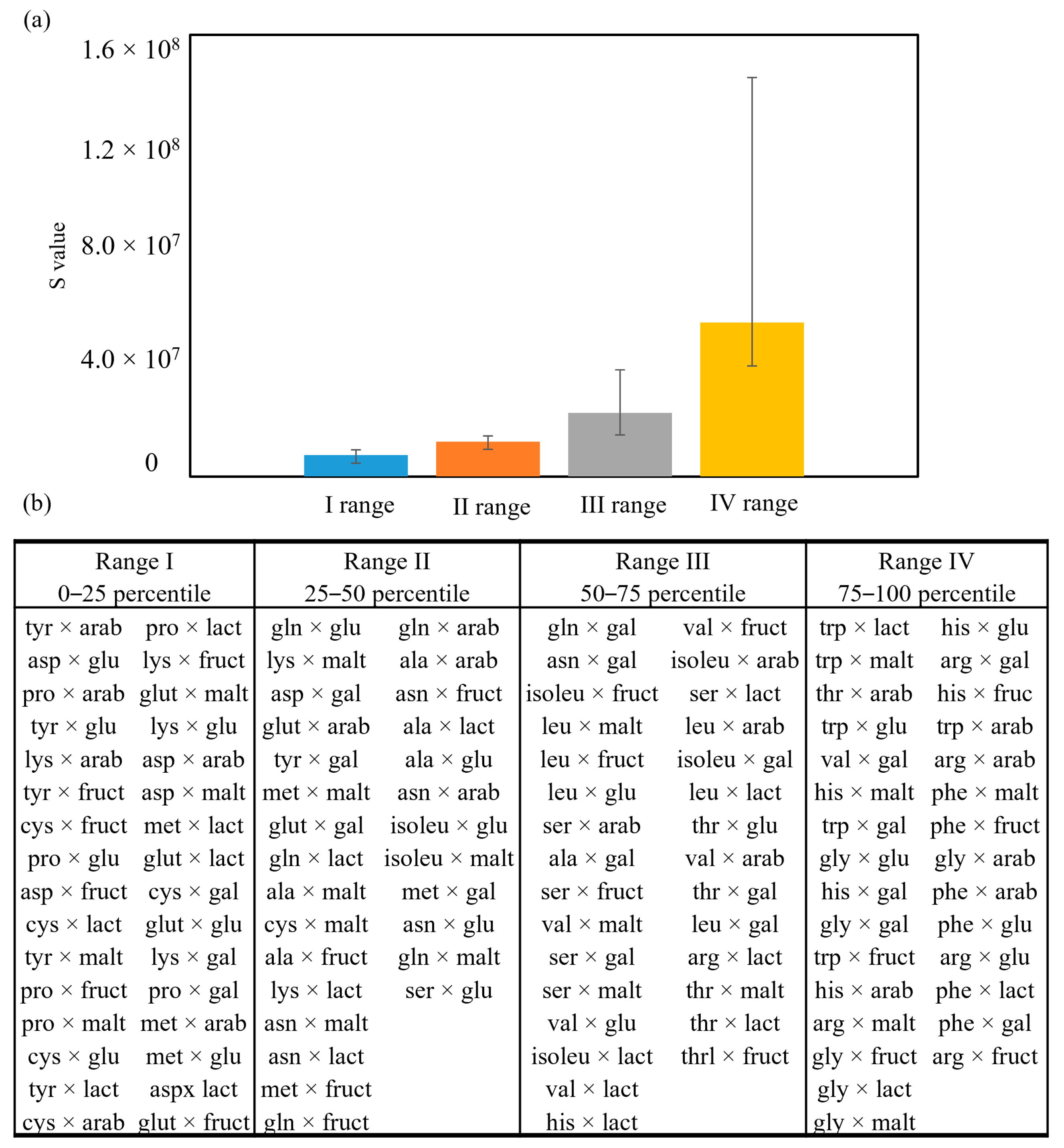
2.5. Untargeted Approach to Create a Library of Potential Antioxidant MRPs
Study of Variable Influence on Antioxidant MRPs Production Using the Library Created
3. Materials and Methods
3.1. Analytical Methods
3.1.1. HPLC-HRMS Analyses
3.1.2. Amino Acids Quantification
3.1.3. Sugars Quantification
3.2. Sample Preparation and Maillard Reaction
3.2.1. Optimisation of Chromatographic Method
3.2.2. Influence of Initial pH on Antioxidants Production in MR
3.2.3. Influence of Amino Acids and Sugar Composition on Antioxidant Production in MR
3.2.4. Consumption of Reagents in MR
3.2.5. Untargeted Approach to Create a Library of Potential Antioxidant MRPs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Nooshkam, M.; Varidi, M.; Bashash, M. The Maillard reaction products as food-born antioxidant and antibrowning agents in model and real food systems. Food Chem. 2019, 275, 644–660. [Google Scholar] [CrossRef] [PubMed]
- Salter, L.J.; Mottram, D.S.; Whitfield, F.B. Volatile compounds produced in maiilard reactions involving glycine, ribose and phospholipid. J. Sci. Food Agric. 1989, 46, 227–242. [Google Scholar] [CrossRef]
- Sun, A.; Wu, W.; Soladoye, O.P.; Aluko, R.E.; Bak, K.H.; Fu, Y.; Zhang, Y.H. Maillard reaction of food-derived peptides as a potential route to generate meat flavor compounds: A review. Food Res. Int. 2022, 151, 110823. [Google Scholar] [CrossRef] [PubMed]
- Michalska, A.; Amigo-Benavent, M.; Zielinski, H.; del Castillo, M.D. Effect of bread making on formation of Maillard reaction products contributing to the overall antioxidant activity of rye bread. J. Cereal Sci. 2008, 48, 123–132. [Google Scholar] [CrossRef]
- Nooshkam, M.; Varidi, M.; Verma, D.K. Functional and biological properties of Maillard conjugates and their potential application in medical and food: A review. Food Res. Int. 2020, 131, 109003. [Google Scholar] [CrossRef]
- Wang, H.Y.; Qian, H.; Yao, W.R. Melanoidins produced by the Maillard reaction: Structure and biological activity. Food Chem. 2011, 128, 573–584. [Google Scholar] [CrossRef]
- Novais, C.; Molina, A.K.; Abreu, R.M.V.; Santo-Buelga, C.; Ferreira, I.C.F.R.; Pereira, C.; Barros, L. Natural Food Colorants and Preservatives: A Review, a Demand, and a Challenge. J. Agric. Food Chem. 2022, 70, 2789–2805. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhang, Y.; Soladoye, O.P.; Aluko, R.E. Maillard reaction products derived from food protein-derived peptides: Insights into flavor and bioactivity. Crit. Rev. Food Sci. Nutr. 2020, 60, 3429–3442. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xia, B.; Hu, L.T.; Ni, Z.J.; Thakur, K.; Wei, Z.J. Maillard conjugates and their potential in food and nutritional industries: A review. Food Front. 2020, 1, 382–397. [Google Scholar] [CrossRef]
- Kanzler, C.; Haase, P.T.; Schestkowa, H.; Kroh, L.W. Antioxidant Properties of Heterocyclic Intermediates of the Maillard Reaction and Structurally Related Compounds. J. Agric. Food Chem. 2016, 64, 7829–7837. [Google Scholar] [CrossRef] [PubMed]
- Yanagimoto, K.; Lee, K.G.; Ochi, H.; Shibamoto, T. Antioxidative activity of heterocyclic compounds formed in Maillard reaction products. Int. Congr. Ser. 2002, 1245, 335–340. [Google Scholar] [CrossRef]
- Martins, S.I.F.S.; Van Boekel, M.A.J.S. Kinetics of the glucose/glycine Maillard reaction pathways: Influences of pH and reactant initial concentrations. Food Chem. 2005, 92, 437–448. [Google Scholar] [CrossRef]
- Zhang, L.; Kong, X.; Yang, X.; Zhang, Y.Y.; Sun, B.G.; Chen, H.T.; Sun, Y. Kinetics of 5-hydroxymethylfurfural formation in the sugar—Amino acid model of Maillard reaction. J. Sci. Food Agric. 2019, 99, 2340–2347. [Google Scholar] [CrossRef] [PubMed]
- Delgado-andrade, C.; Seiquer, I.; Haro, A.; Castellano, R.; Navarro, M.P. Development of the Maillard reaction in foods cooked by different techniques. Intake of Maillard-derived compounds. Food Chem. 2010, 122, 145–153. [Google Scholar] [CrossRef]
- Yaylayan, V.A.; Mandeville, S. Stereochemical Control of Maltol Formation in Maillard Reaction. J. Agric. Food Chem. 1994, 42, 771–775. [Google Scholar] [CrossRef]
- Amaya-Farfan, J.; Rodriguez-Amaya, D.B. (Eds.) The Mailard Reactions. In Chemical Changes During Processing and Storage of Foods; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar] [CrossRef]
- Gallo, A.; Guzzon, R.; Ongaro, M.; Paolini, M.; Nardin, T.; Malacarne, M.; Roman, T.; Larcher, R. Biological acidification of ‘Vino Santo di Gambellara’ by mixed fermentation of L. thermotolerans and S. cerevisiae. Role of nitrogen in the evolution of fermentation and aroma profile. Oeno. One 2023, 57, 205–217. [Google Scholar] [CrossRef]
- Di Lella, S.; Tognetti, R.; La Porta, N.; Lombardi, F.; Nardin, T.; Larcher, R. Characterization of Silver fir Wood Decay Classes Using Sugar Metabolites Detected with Ion Chromatography. J. Wood Chem. Technol. 2019, 39, 90–110. [Google Scholar] [CrossRef]
- Van Boekel, M.A.J.S. Kinetic aspects of the Maillard reaction: A critical review. Food/Nahrung 2001, 45, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Hodge, J.E. Browning Reaction Theories Integrated in Review Chemistry of Browning Reactions in Model Systems. Agric. Food Chem. 1953, 1, 928–943. [Google Scholar] [CrossRef]
- Zhuang, G.D.; Gu, W.T.; Xu, S.H.; Cao, D.M.; Deng, S.M.; Chen, Y.S.; Wang, S.M.; Tang, D. Rapid screening of antioxidant from natural products by AAPH-Incubating HPLC-DAD-HR MS/MS method: A case study of Gardenia jasminoides fruit. Food Chem. 2023, 401, 134091. [Google Scholar] [CrossRef] [PubMed]
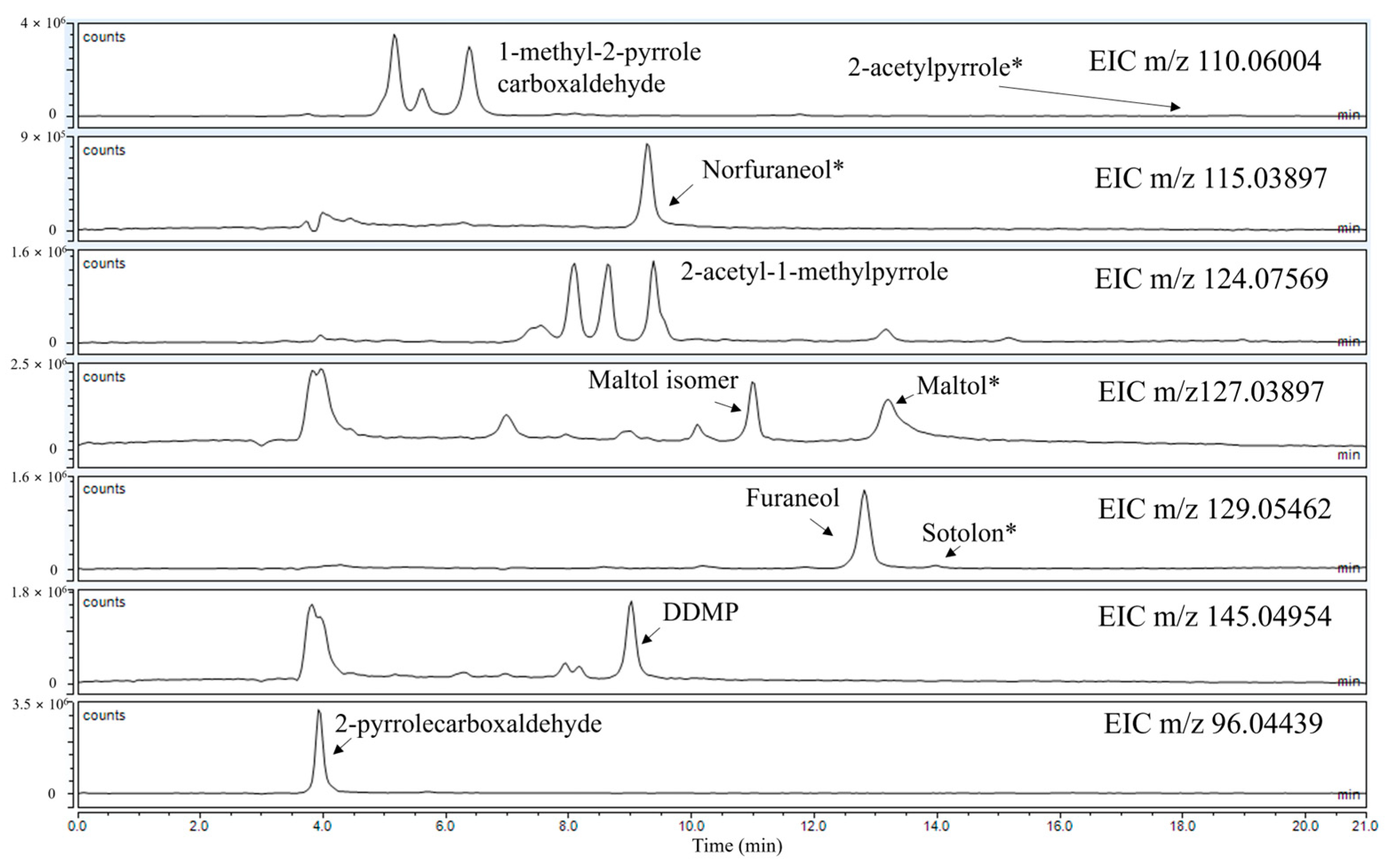

| Compound | Theoretical m/z | Measured m/z | Δppm | Retention Time | Ionisation | Reference |
|---|---|---|---|---|---|---|
| Maltol * | 127.03897 | 127.03859 | 0.00038 | 13.27 | [M + H]+ | [10] |
| Maltol isomer | 127.03897 | 127.03905 | 0.00008 | 10.98 | [M + H]+ | [10] |
| 2-acetylpyrrole * | 110.06004 | 110.06005 | 0.00001 | 18.79 | [M + H]+ | [10] |
| Sotolon * | 129.05462 | 129.04928 | 0.00534 | 14.56 | [M + H]+ | [10] |
| Norfuraneol * | 115.03897 | 115.03938 | 0.00041 | 9.25 | [M + H]+ | [10] |
| Furaneol | 129.05462 | 129.06004 | 0.00542 | 12.62 | [M + H]+ | [10] |
| DDMP | 145.04954 | 145.05029 | 0.00075 | 10.50 | [M + H]+ | [10] |
| 2-pyrrolecarboxaldehyde | 96.04439 | 96.04450 | 0.00011 | 4.00 | [M + H]+ | [11] |
| 1-methyl-2-pyrrole carboxaldehyde | 110.06004 | 110.06099 | 0.00095 | 6.00 | [M + H]+ | [11] |
| 2-acetyl-1-methylpyrrole | 124.07569 | 124.07662 | 0.00093 | 9.30 | [M + H]+ | [11] |
| Compound | % of Degradation after 1 h with Radical Initiator | Theoretical m/z | Retention Time (min) | Best AA × Sugar |
|---|---|---|---|---|
| 2-pyrrolecarboxaldehyde | 88% | 96.0444 | 4 | Glutamine × arabinose |
| Norfuraneol | 70% | 115.039 | 9.25 | Aspartic acid × arabinose |
| Furaneol | 69% | 129.055 | 12.62 | Lysine × galactose |
| 1-methyl-2-pyrrole carboxaldehyde | 60% | 110.06 | 6 | Threonine × maltose |
| DDMP | 42% | 145.05 | 10.5 | Threonine × maltose |
| 2-acetyl-1-methylpyrrole | 38% | 124.076 | 9.3 | Threonine × arabinose |
| Maltol | 35% | 127.039 | 13.27 | Lysine × maltose |
| Maltol isomer | 31% | 127.039 | 10.98 | Tryptophan × arabinose |
| 2-acetylpyrrole | 29% | 110.06 | 18.79 | Methionine × fructose |
| Sotolon | 13% | 129.055 | 14.56 | Lysine × galactose |
| Name | % Degradation after 1 h with AAPH | m/z | RT [min] | Best AA × Sugar |
|---|---|---|---|---|
| Unknown 1 | 96.2 | 230.1140 | 8.41 | Arginine × fructose |
| Unknown 2 | 93.0 | 199.1077 | 10.36 | Asparagine × fructose |
| Unknown 3 | 92.6 | 154.0497 | 4.39 | Glycine × arabinose |
| Unknown 4 | 91.1 | 124.0757 | 10.12 | Asparagine × glucose |
| Unknown 5 | 91.0 | 164.0818 | 5.07 | Arginine × arabinose |
| Unknown 6 | 86.8 | 170.0811 | 4.59 | Leucine × galactose |
| Unknown 7 | 86.0 | 135.0552 | 9.21 | Histidine × maltose |
| Unknown 8 | 83.3 | 230.1140 | 6.76 | Arginine × fructose |
| Unknown 9 | 82.5 | 212.1028 | 3.10 | Histidine × glucose |
| Unknown 10 | 79.7 | 230.1140 | 4.13 | Arginine × fructose |
| Unknown 11 | 79.6 | 127.0390 | 6.97 | Tryptophan × arabinose |
| Unknown 12 | 79.2 | 151.123 | 6.65 | Leucine × lactose |
| Unknown 13 | 77.1 | 124.0757 | 8.39 | Threonine × fructose |
| Unknown 14 | 74.8 | 140.0706 | 4.61 | Threonine × arabinose |
| Unknown 15 | 74.5 | 119.0350 | 4.73 | Valine × galactose |
| Unknown 16 | 73.8 | 127.0389 | 3.93 | Aspartic acid × lactose |
| Unknown 17 | 72.1 | 154.0498 | 4.72 | Glycine × arabinose |
| Unknown 18 | 69.1 | 212.1034 | 8.03 | Arginine × fructose |
| Unknown 19 | 65.0 | 184.1085 | 4.72 | Arginine × fructose |
| Unknown 20 | 64.5 | 123.0914 | 3.48 | Glycine × fructose |
| Unknown 21 | 63.7 | 135.0553 | 8.77 | Histidine × maltose |
| Unknown 22 | 59.7 | 110.0600 | 5.15 | threonine × maltose |
| Unknown 23 | 58.1 | 166.0861 | 10.08 | Phenylalanine × lactose |
| Unknown 24 | 56.2 | 124.0756 | 10.56 | Asparagine × arabinose |
| Unknown 25 | 53.8 | 127.0389 | 10.12 | Tryptophan × maltose |
| Unknown 26 | 53.6 | 123.0915 | 3.71 | Glycine × fructose |
| Unknown 27 | 53.1 | 110.0600 | 5.61 | Threonine × arabinose |
| Unknown 28 | 51.7 | 230.1141 | 5.53 | Arginine × galactose |
| Unknown 29 | 51.6 | 196.0224 | 7.01 | Glutamine × lactose |
| Unknown 30 | 49.9 | 110.0600 | 6.38 | Leucine × galactose |
| Unknown 31 | 47.3 | 164.0817 | 6.20 | Histidine × fructose |
| Unknown 32 | 47.1 | 143.0349 | 3.78 | Glutamic acid × lactose |
| Unknown 33 | 42.0 | 144.0807 | 15.71 | Tryptophan × galactose |
| Unknown 34 | 39.5 | 124.0757 | 8.06 | Threonine × arabinose |
| Unknown 35 | 39.3 | 124.0756 | 4.34 | Alanine × maltose |
| Unknown 36 | 38.2 | 210.1127 | 8.29 | Threonine × maltose |
| Unknown 37 | 37.6 | 124.0756 | 15.82 | Alanine × fructose |
| Unknown 38 | 35.6 | 169.0972 | 3.27 | Serine × galactose |
| Unknown 39 | 34.4 | 127.039 | 3.76 | Serine × lactose |
| Unknown 40 | 31.3 | 124.0757 | 7.49 | Glutamine × fructose |
| Unknown 41 | 30.0 | 212.1035 | 6.82 | Arginine × fructose |
| Unknown 42 | 28.3 | 217.0971 | 17.69 | Tryptophan × arabinose |
| Unknown 43 | 25.5 | 144.0807 | 13.62 | Tryptophan × galactose |
| Unknown 44 | 25.0 | 144.0807 | 17.69 | Tryptophan × arabinose |
| Unknown 45 | 22.0 | 124.0757 | 8.63 | Asparagine × arabinose |
| Unknown 46 | 21.0 | 143.0350 | 6.95 | Tryptophan × arabinose |
| Unknown 47 | 20.1 | 230.1139 | 8.12 | Arginine × fructose |
| Unknown 48 | 14.9 | 95.06033 | 3.11 | Glycine × galactose |
| Unknown 49 | 12.2 | 151.1229 | 6.06 | Valine × galactose |
| Unknown 50 | 10.0 | 143.035 | 5.25 | Methionine × fructose |
| Time (s) | Potential (V vs. Ag/AgCl) | Integration |
|---|---|---|
| 0.00 | 1.35 | Off |
| 0.20 | 1.35 | On |
| 0.40 | 1.35 | Off |
| 0.41 | −1.15 | Off |
| 0.42 | −1.15 | Off |
| 0.43 | 1.45 | Off |
| 0.44 | 1.15 | Off |
| 0.50 | 1.15 | Off |
| Amino Acids | Reducing Sugars | ||
|---|---|---|---|
| Arginine (arg) | Histidine (his) | Lysine (lys) | Glucose (glu) |
| Aspartic acid (asp) | Glutamic acid (glu) | Serine (ser) | Galactose (gal) |
| Threonine (thr) | Asparagine (asn) | Glutamine (gln) | Fructose (fru) |
| Cysteine (cys) | Glycine (gly) | Proline (pro) | Arabinose (ara) |
| Alanine (ala) | Valine (val) | Isoleucine (Ile) | Maltose (mal) |
| Leucine (leu) | Methionine (met) | Phenylalanine (phe) | Lactose (lac) |
| Tyrosine (tyr) | Tryptophan (trp) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bolchini, S.; Larcher, R.; Morozova, K.; Scampicchio, M.; Nardin, T. Screening of Antioxidant Maillard Reaction Products Using HPLC-HRMS and Study of Reaction Conditions for Their Production as Food Preservatives. Molecules 2024, 29, 4820. https://doi.org/10.3390/molecules29204820
Bolchini S, Larcher R, Morozova K, Scampicchio M, Nardin T. Screening of Antioxidant Maillard Reaction Products Using HPLC-HRMS and Study of Reaction Conditions for Their Production as Food Preservatives. Molecules. 2024; 29(20):4820. https://doi.org/10.3390/molecules29204820
Chicago/Turabian StyleBolchini, Sara, Roberto Larcher, Ksenia Morozova, Matteo Scampicchio, and Tiziana Nardin. 2024. "Screening of Antioxidant Maillard Reaction Products Using HPLC-HRMS and Study of Reaction Conditions for Their Production as Food Preservatives" Molecules 29, no. 20: 4820. https://doi.org/10.3390/molecules29204820
APA StyleBolchini, S., Larcher, R., Morozova, K., Scampicchio, M., & Nardin, T. (2024). Screening of Antioxidant Maillard Reaction Products Using HPLC-HRMS and Study of Reaction Conditions for Their Production as Food Preservatives. Molecules, 29(20), 4820. https://doi.org/10.3390/molecules29204820








