Genomics-Driven Discovery of Plantariitin A, a New Lipopeptide in Burkholderia plantarii DSM9509
Abstract
1. Introduction
2. Results and Discussion
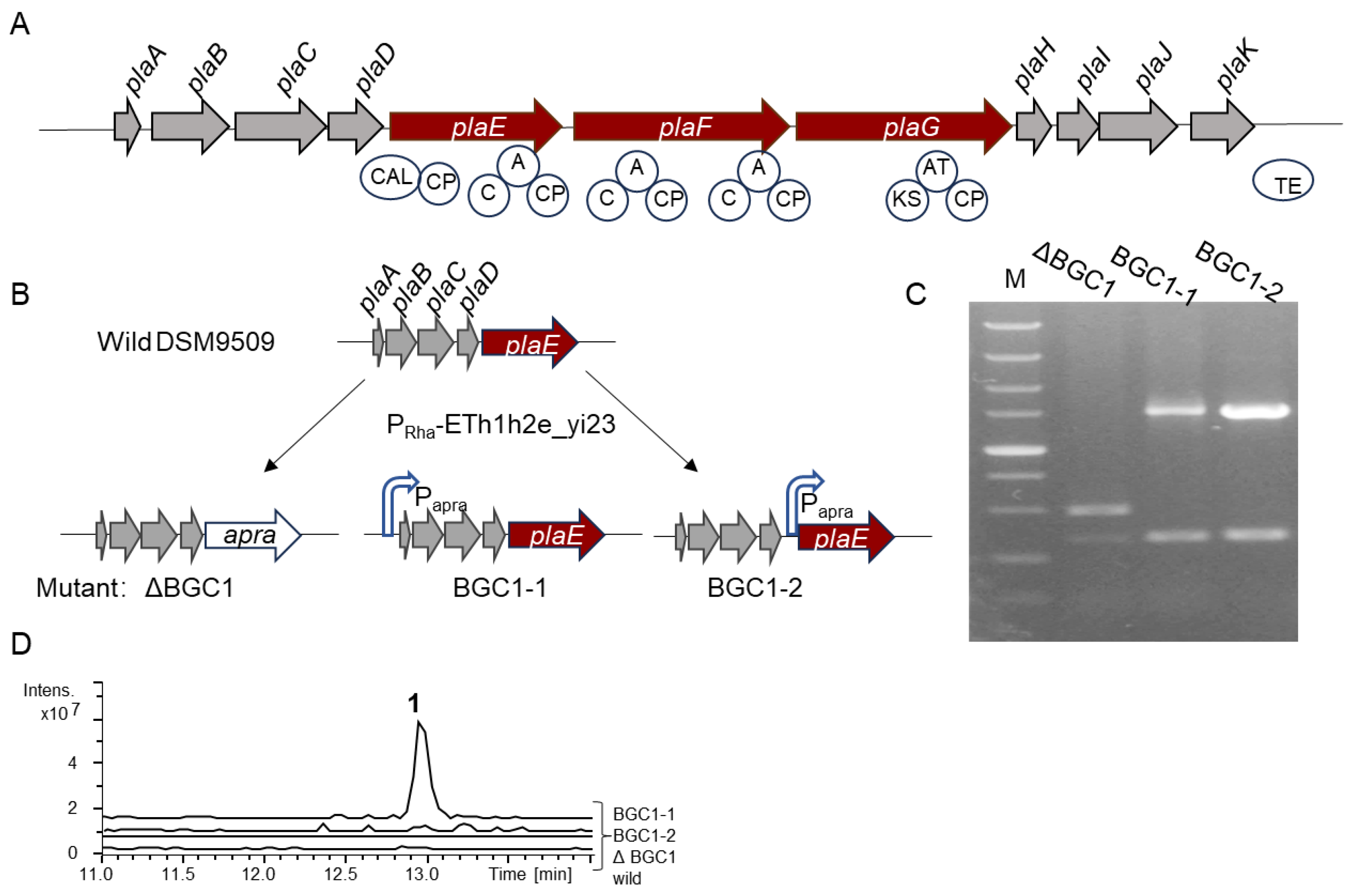
2.1. Bioinformatics Analysis of BGC pla
2.2. In Situ Activation of BGC pla in B. plantarii DSM9509
2.3. Identification of the New Lipopeptide (1)
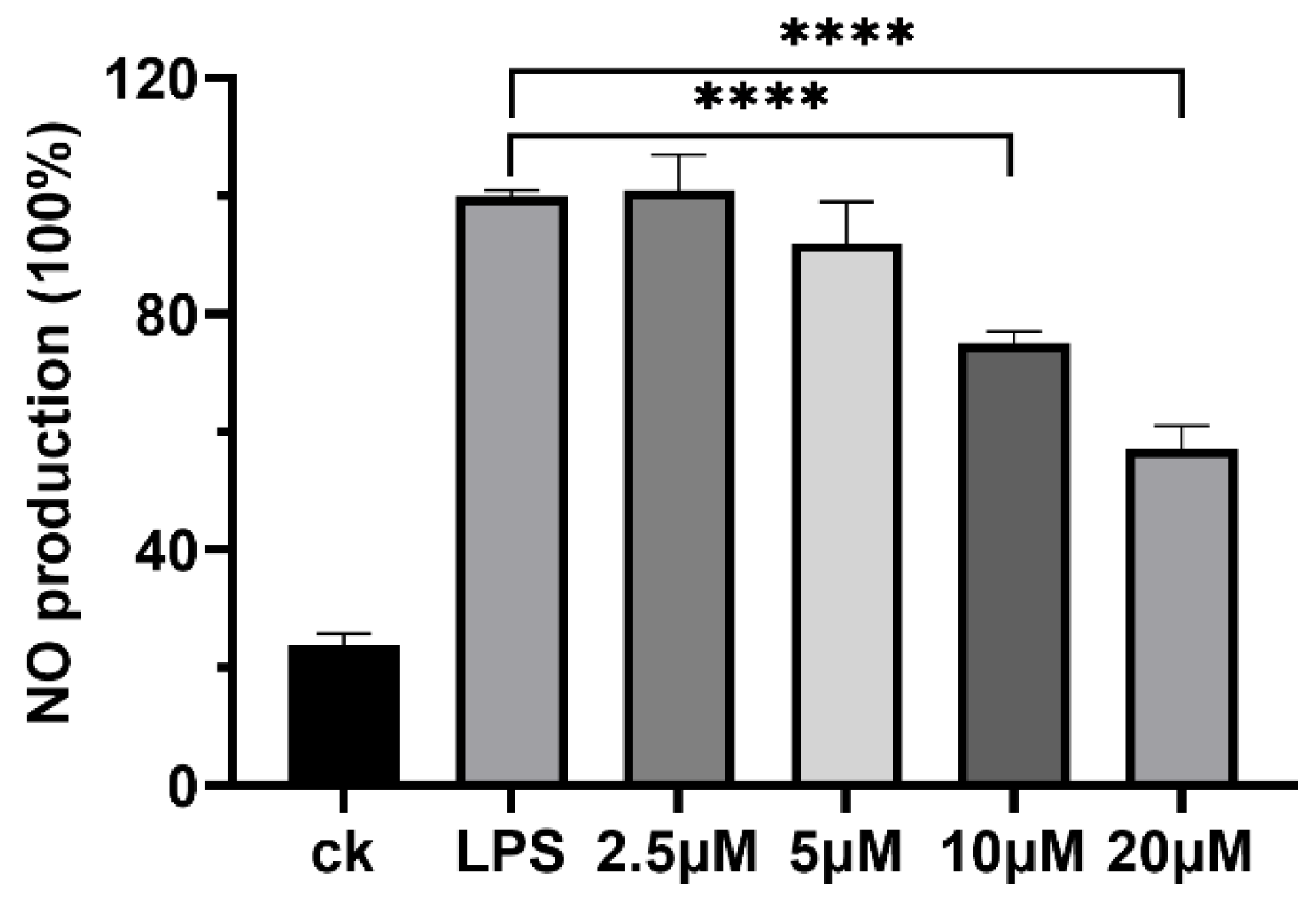
2.4. Anti-Inflammatory Activity of Plantariitin A (1)
3. Materials and Methods
3.1. Materials
3.2. Construction of Mutants
3.3. Fermentation, Extraction, and Isolation
3.4. Determination of the Amino Acid Ala and Hse Configuration
3.5. Determination of the Amino Acid ATDPP Configuration
3.6. Detection of NO Production
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BGCs | biosynthetic gene clusters |
| LPs | lipopeptides |
| NRPSs | non-ribosomal peptide synthetases |
| PKSs | polyketide synthases |
| FA | fatty acid chain |
| Ala | alanine |
| Hse | homoserine |
| ATDPP | amino-1,2,3,6-tetrahydro-2,6-dioxo-4-pyrimidinepropanoic acid |
| NMR | nuclear magnetic resonance |
| HR-ESI-MS | high-resolution electrospray ionization mass spectrometry |
| DEPT | distortionless enhancement via polarization transfer |
| HSQC | heteronuclear single quantum coherence |
| COSY | correlated spectroscopy |
| HMBC | heteronuclear multiple-bond correlation spectroscopy |
References
- Kunakom, S.; Adaikpoh Barbara, I.; Tran Tuan, A.; Eustáquio Alessandra, S. Complete genome sequence of soil bacterium Burkholderia sp. strain FERM BP-3421, a producer of spliceostatins. Microbiol. Resour. Ann. 2023, 12, e00111-23. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Bok, E.Y.; Jung, Y.H.; Hur, T.Y.; Kim, Y.O.; Kong, H.J.; Kim, D.G.; Kim, Y.S.; Oem, J.K. Antifungal activity of aminopyrrolnitrin against Trichophyton verrucosum in a guinea pig model of dermatophytosis. Mycoses 2024, 67, e13748. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, S.M.; Nirmal, A.J.; Rahman, J.; Xu, R.; Drill, E.; Galasso, N.; Ganesan, N.; Davey, T.; Hancock, H.; Perez, L.; et al. Duvelisib plus romidepsin in relapsed/refractory T cell lymphomas: A phase 1b/2a trial. Nat. Med. 2024, 30, 2517–2527. [Google Scholar] [CrossRef]
- Foxfire, A.; Buhrow, A.R.; Orugunty, R.S.; Smith, L. Drug discovery through the isolation of natural products from Burkholderia. Expert. Opin. Drug Discov. 2021, 16, 807–822. [Google Scholar] [CrossRef] [PubMed]
- Vicente-Garcia, C.; Colomer, I. Lipopeptides as tools in catalysis, supramolecular, materials and medicinal chemistry. Nat. Rev. Chem. 2023, 7, 710–731. [Google Scholar] [CrossRef] [PubMed]
- Pilz, M.; Cavelius, P.; Qoura, F.; Awad, D.; Brück, T. Lipopeptides development in cosmetics and pharmaceutical applications: A comprehensive review. Biotechnol. Adv. 2023, 67, 108210. [Google Scholar] [CrossRef]
- Grünewald, J.; Sieber, S.A.; Mahlert, C.; Linne, U.; Marahiel, M.A. Synthesis and derivatization of daptomycin: A chemoenzymatic route to acidic lipopeptide antibiotics. J. Am. Chem. Soc. 2004, 126, 17025–17031. [Google Scholar] [CrossRef]
- Luhavaya, H.; Sigrist, R.; Chekan, J.R.; McKinnie, S.M.K.; Moore, B.S. Biosynthesis of l-4-chlorokynurenine, an antidepressant prodrug and a non-proteinogenic amino acid found in lipopeptide antibiotics. Angew. Chem. Int. Ed. Engl. 2019, 58, 8394–8399. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Wu, Z.; Ji, W.; Liu, D.; Guo, X.; Yang, D.; Fan, A.; Jia, H.; Ma, M.; Lin, W. Discovery of cyclohexadepsipeptides with anti-Zika virus activities and biosynthesis of the nonproteinogenic building block (3S)-methyl-l-proline. J. Biol. Chem. 2021, 297, 100822. [Google Scholar] [CrossRef] [PubMed]
- Sreedharan, S.M.; Rishi, N.; Singh, R. Microbial lipopeptides: Properties, mechanics and engineering for novel lipopeptides. Microbiol. Res. 2023, 271, 127363. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Diao, X.; Zhang, N.; Li, F.; Zhou, H.; Chen, H.; Bai, X.; Ren, X.; Zhang, Y.; Wu, D.; et al. Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis. Nat. Commun. 2021, 12, 296. [Google Scholar] [CrossRef] [PubMed]
- Calcott, M.J.; Owen, J.G.; Ackerley, D.F. Efficient rational modification of non-ribosomal peptides by adenylation domain substitution. Nat. Commun. 2020, 11, 4554. [Google Scholar] [CrossRef]
- Zhang, M.; Peng, Z.; Huang, Z.; Fang, J.; Li, X.; Qiu, X. functional diversity and engineering of the adenylation domains in nonribosomal peptide synthetases. Mar. Drugs 2024, 22, 349. [Google Scholar] [CrossRef]
- Chen, H.; Zhong, L.; Zhou, H.; Bai, X.; Sun, T.; Wang, X.; Zhao, Y.; Ji, X.; Tu, Q.; Zhang, Y.; et al. Biosynthesis and engineering of the nonribosomal peptides with a C-terminal putrescine. Nat. Commun. 2023, 14, 6619. [Google Scholar] [CrossRef] [PubMed]
- Clements-Decker, T.; Kode, M.; Khan, S.; Khan, W. Underexplored bacteria as reservoirs of novel antimicrobial lipopeptides. Front. Chem. 2022, 10, 1025979. [Google Scholar] [CrossRef]
- Esmaeel, Q.; Pupin, M.; Jacques, P.; Leclère, V. Nonribosomal peptides and polyketides of Burkholderia: New compounds potentially implicated in biocontrol and pharmaceuticals. Environ. Sci. Pollut. Res. Int. 2018, 25, 29794–29807. [Google Scholar] [CrossRef] [PubMed]
- Gavriilidou, A.; Kautsar, S.A.; Zaburannyi, N.; Krug, D.; Müller, R.; Medema, M.H.; Ziemert, N. Compendium of specialized metabolite biosynthetic diversity encoded in bacterial genomes. Nat. Microbiol. 2022, 7, 726–735. [Google Scholar] [CrossRef]
- Chen, H.; Zhou, H.; Sun, T.; Xu, J.; Tu, Q.; Yang, J.; Zhang, Y.; Bian, X. Identification of holrhizins E–Q reveals the diversity of nonribosomal lipopeptides in Paraburkholderia rhizoxinica. J. Nat. Prod. 2020, 83, 537–541. [Google Scholar] [CrossRef]
- Dose, B.; Ross, C.; Niehs, S.P.; Scherlach, K.; Bauer, J.P.; Hertweck, C. Food-poisoning bacteria employ a citrate synthase and a Type II NRPS to synthesize bolaamphiphilic lipopeptide antibiotics. Angew. Chem. Int. Ed. Engl. 2020, 59, 21535–21540. [Google Scholar] [CrossRef]
- Bach, E.; Passaglia, L.M.P.; Jiao, J.; Gross, H. Burkholderia in the genomic era: From taxonomy to the discovery of new antimicrobial secondary metabolites. Crit. Rev. Microbiol. 2022, 48, 121–160. [Google Scholar] [CrossRef] [PubMed]
- Romanowski, S.B.; Lee, S.; Kunakom, S.; Paulo, B.S.; Recchia, M.J.J.; Liu, D.Y.; Cavanagh, H.; Linington, R.G.; Eustáquio, A.S. Identification of the lipodepsipeptide selethramide encoded in a giant nonribosomal peptide synthetase from a Burkholderia bacterium. Proc. Natl. Acad. Sci. USA 2023, 120, e2304668120. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Sun, T.; Bai, X.; Yang, J.; Yan, F.; Yu, L.; Tu, Q.; Li, A.; Tang, Y.; Zhang, Y.; et al. Genomics-driven activation of silent biosynthetic gene clusters in Burkholderia gladioli by screening recombineering system. Molecules 2021, 26, 700. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Shi, H.; Zhao, X.; Liu, X.; Duan, Q.; Song, C.; Chen, H.; Zheng, W.; Shen, Q.; Wang, M.; et al. Development and application of an efficient recombineering system for Burkholderia glumae and Burkholderia plantarii. Microb. Biotechnol. 2021, 14, 1809–1826. [Google Scholar] [CrossRef]
- Gong, K.; Wang, M.; Duan, Q.; Li, G.; Yong, D.; Ren, C.; Li, Y.; Zhang, Q.; Wang, Z.; Sun, T.; et al. High-yield production of FK228 and new derivatives in a Burkholderia chassis. Metab. Eng. 2023, 75, 131–142. [Google Scholar] [CrossRef]
- Kunakom S, Eustáquio AS: Burkholderia as a source of natural products. J. Nat. Prod. 2019, 82, 2018–2037. [CrossRef]
- Paulo, B.S.; Recchia, M.J.J.; Lee, S.; Fergusson, C.H.; Romanowski, S.B.; Hernandez, A.; Krull, N.; Liu, D.Y.; Cavanagh, H.; Bos, A.; et al. Discovery of megapolipeptins by genome mining of a Burkholderiales bacteria collection. Chem. Sci. 2024, 15, 16567–16581. [Google Scholar] [CrossRef]
- Bajsa-Hirschel, J.; Pan, Z.; Pandey, P.; Asolkar, R.N.; Chittiboyina, A.G.; Boddy, L.; Machingura, M.C.; Duke, S.O. Spliceostatin C, a component of a microbial bioherbicide, is a potent phytotoxin that inhibits the spliceosome. Front. Plant Sci. 2022, 13, 1019938. [Google Scholar] [CrossRef]
- Owens, D.K.; Bajsa-Hirschel, J.; Duke, S.O.; Carbonari, C.A.; Gomes, G.; Asolkar, R.; Boddy, L.; Dayan, F.E. The contribution of romidepsin to the herbicidal activity of Burkholderia rinojensis biopesticide. J. Nat. Prod. 2020, 83, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Wang, X.; Zhou, H.; Zhang, Y.; Li, A.; Bian, X. Establishment of recombineering genome editing system in Paraburkholderia megapolitana empowers activation of silent biosynthetic gene clusters. Microb. Biotechnol. 2020, 13, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.H.; Huang, E. Novel linear lipopeptide paenipeptin C′ binds to lipopolysaccharides and lipoteichoic acid and exerts bactericidal activity by the disruption of cytoplasmic membrane. BMC Microbiol. 2019, 19, 6. [Google Scholar] [CrossRef]
- Wagenlehner, F.; Lucenteforte, E.; Pea, F.; Soriano, A.; Tavoschi, L.; Steele, V.R.; Henriksen, A.S.; Longshaw, C.; Manissero, D.; Pecini, R.; et al. Systematic review on estimated rates of nephrotoxicity and neurotoxicity in patients treated with polymyxins. Clin. Microbiol. Infect. 2021, 27, 671–686. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, W.; Wu, Y.; Ma, X.; Zhou, W.; Lai, Y. Lipopeptide 78 from Staphylococcus epidermidis activates β-catenin to inhibit skin inflammation. J. Immunol. 2019, 202, 1219–1228. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, C.; Shi, Y.; Huang, M.; Song, Z.; Simal-Gandara, J.; Li, N.; Shi, J. Classification, application, multifarious activities and production improvement of lipopeptides produced by Bacillus. Crit. Rev. Food Sci. Nutr. 2024, 64, 7451–7464. [Google Scholar] [CrossRef]
- Rodríguez-Cisneros, M.; Morales-Ruíz, L.M.; Salazar-Gómez, A.; Rojas-Rojas, F.U.; Estrada-de Los Santos, P. Compilation of the antimicrobial compounds produced by Burkholderia Sensu Stricto. Molecules 2023, 28, 1646. [Google Scholar] [CrossRef]
- Lodewyk, M.W.; Siebert, M.R.; Tantillo, D.J. Computational prediction of 1H and 13C chemical shifts: A useful tool for natural product, mechanistic, and synthetic organic chemistry. Chem. Rev. 2012, 112, 1839–1862. [Google Scholar] [CrossRef]
- Zanardi, M.M.; Sarotti, A.M. Sensitivity analysis of DP4+ with the probability distribution terms: Development of a universal and customizable method. J. Org. Chem. 2021, 86, 8544–8548. [Google Scholar] [CrossRef] [PubMed]

| No. | δC | δH (J in Hz) | ||
|---|---|---|---|---|
| dodecanoic acid | 1 | 172.3 | C | |
| 2 | 35.1 | CH2 | 2.08 (td, 10.2, 2.4) | |
| 3 | 25.2 | CH2 | 1.46 (m) | |
| 4 | 28.8 | CH2 | 1.23 (m a) | |
| 5 | 29.1 | CH2 | 1.23 (m a) | |
| 6 | 29.1 | CH2 | 1.23 (m a) | |
| 7 | 29.0 | CH2 | 1.23 (m a) | |
| 8 | 28.9 | CH2 | 1.23 (m a) | |
| 9 | 28.7 | CH2 | 1.23 (m a) | |
| 10 | 31.3 | CH2 | 1.22 (m a) | |
| 11 | 22.1 | CH2 | 1.26 (m) | |
| 12 | 14.0 | CH3 | 0.85 (t, 7.2) | |
| L-Ala | 1 | 172.4 | C | |
| 2 | 48.1 | CH | 4.24 (p, 7.2) | |
| 3 | 18.0 | CH3 | 1.15 (d, 7.2) | |
| NH | 7.94 (d, 7.8) | |||
| L-Hse | 1 | 171.6 | C | |
| 2 | 50.0 | CH | 4.27 (m) | |
| 3 | 35.1 | CH2 | 1.80 (m); 1.63 (m) | |
| 4 | 57.5 | CH2 | 3.38 (m a) | |
| NH | 7.91 (d, 9.0) | |||
| D-ATDPP | 1 | 171.9 | C | |
| 2 | 49.9 | CH | 4.50 (m) | |
| 3 | 33.7 | CH2 | 2.78 (dd, 14.4, 5.4); 2.61 (dd, 14.4, 8.4) | |
| 4 | 152.3 | C | ||
| 5 | 99.7 | CH | 5.28 (s) | |
| 6 | 164.0 | C | ||
| 6a | NH | 10.89 (brs) | ||
| 8 | 151.6 | C | ||
| 8a | NH | 10.89 (brs) | ||
| NH | 8.09 (m) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Zhang, Z.; Fu, J.; Li, R. Genomics-Driven Discovery of Plantariitin A, a New Lipopeptide in Burkholderia plantarii DSM9509. Molecules 2025, 30, 868. https://doi.org/10.3390/molecules30040868
Wang X, Zhang Z, Fu J, Li R. Genomics-Driven Discovery of Plantariitin A, a New Lipopeptide in Burkholderia plantarii DSM9509. Molecules. 2025; 30(4):868. https://doi.org/10.3390/molecules30040868
Chicago/Turabian StyleWang, Xiuling, Zhuo Zhang, Jun Fu, and Ruijuan Li. 2025. "Genomics-Driven Discovery of Plantariitin A, a New Lipopeptide in Burkholderia plantarii DSM9509" Molecules 30, no. 4: 868. https://doi.org/10.3390/molecules30040868
APA StyleWang, X., Zhang, Z., Fu, J., & Li, R. (2025). Genomics-Driven Discovery of Plantariitin A, a New Lipopeptide in Burkholderia plantarii DSM9509. Molecules, 30(4), 868. https://doi.org/10.3390/molecules30040868





