Design, Synthesis, Biological Evaluation, and In Silico Studies of Novel Multitarget Cinnamic Acid Hybrids †
Abstract
1. Introduction
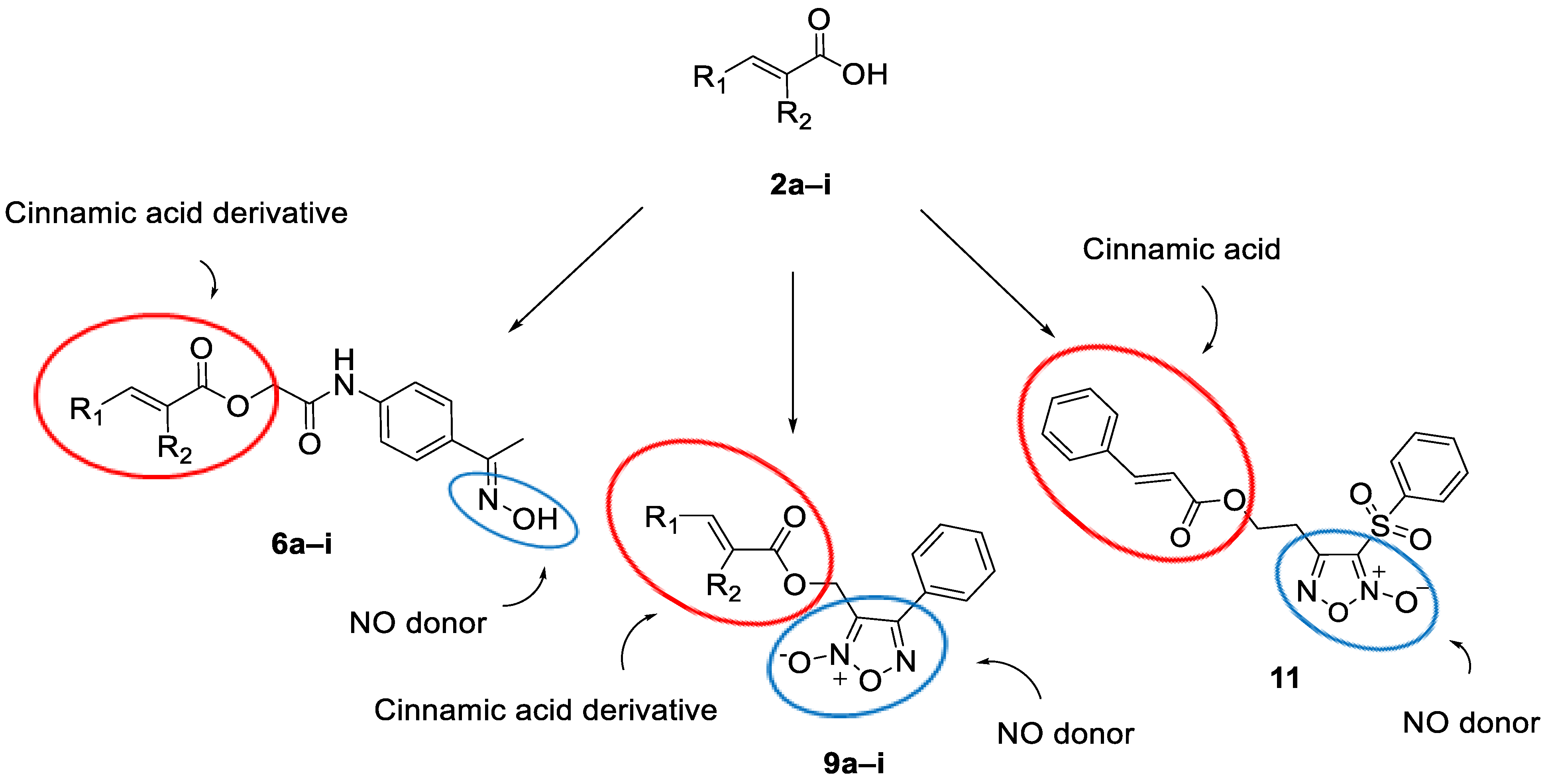
2. Results and Discussion
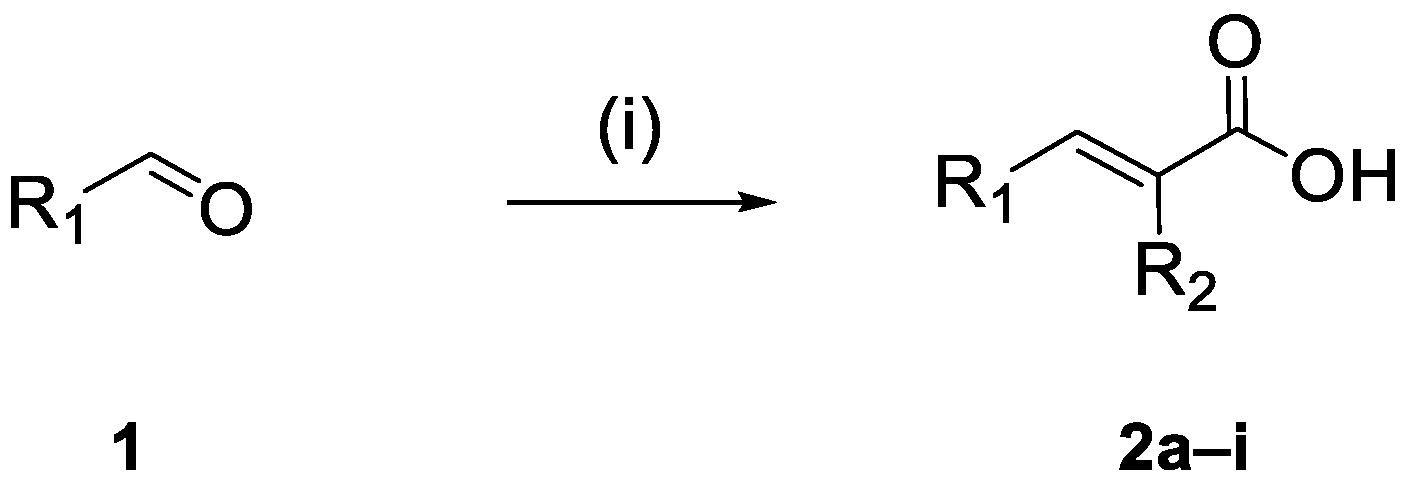
2.1. Chemistry
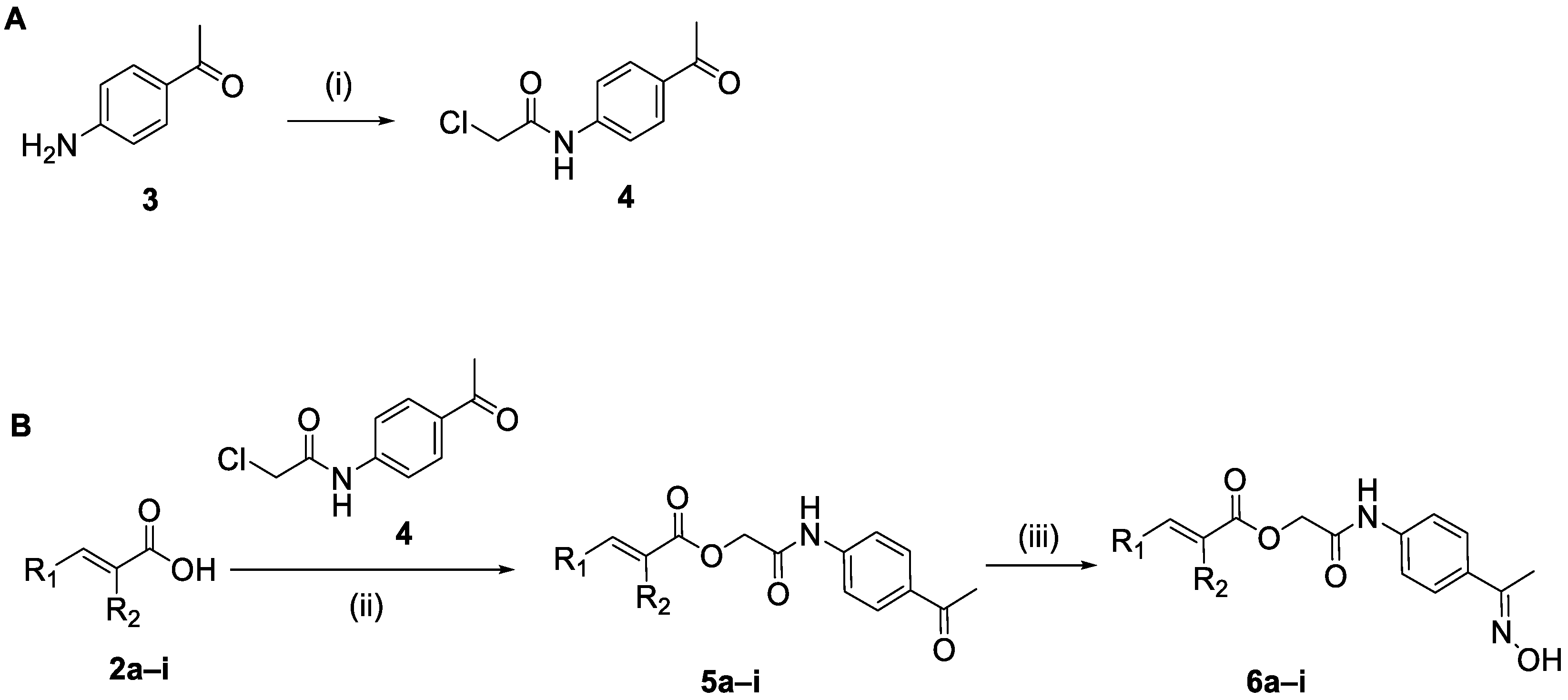
2.1.1. Synthesis of the Cinnamic Acid-Arylacetamide Oxime Hybrids 6a–i
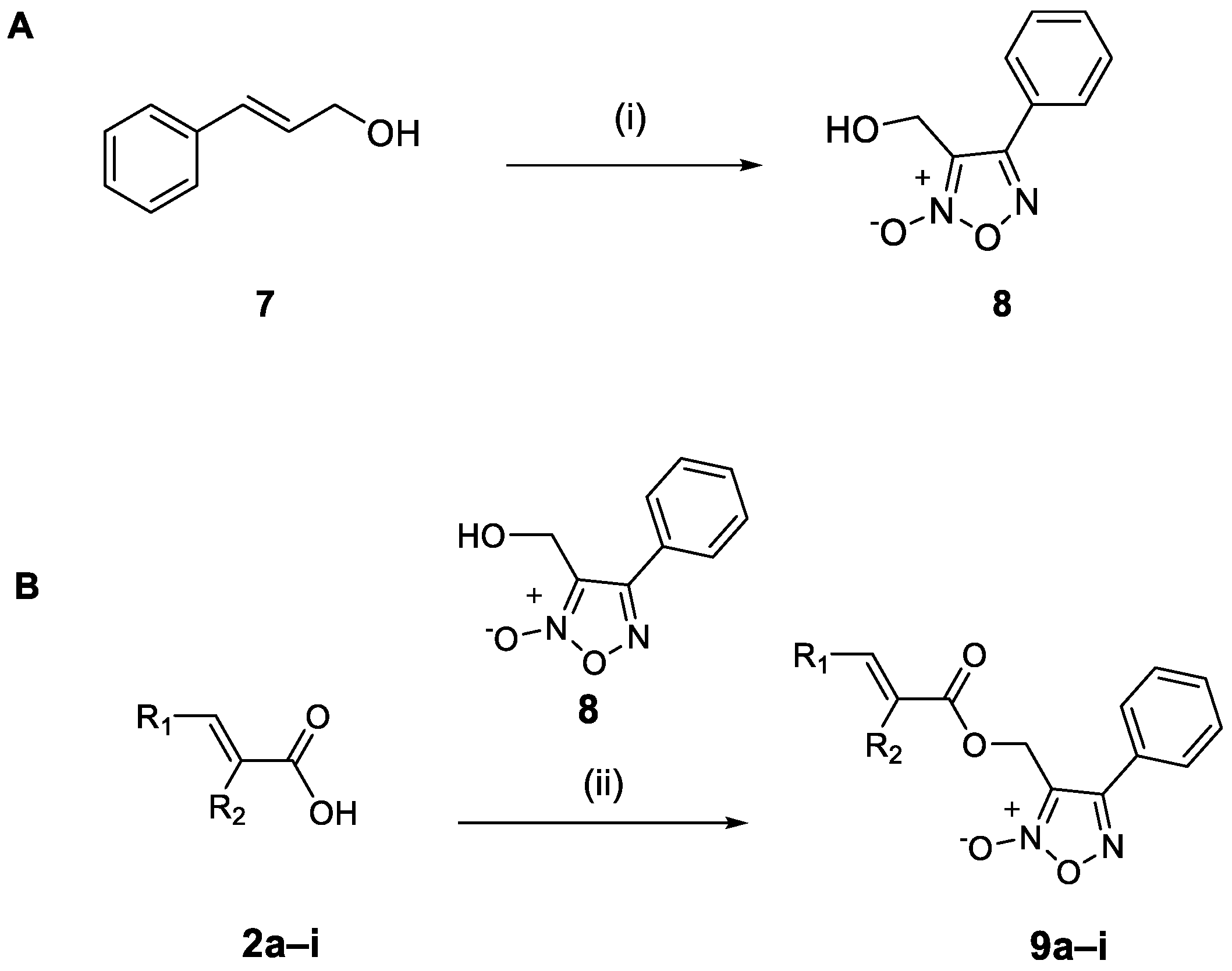
2.1.2. Synthesis of the Cinnamic Acid–Phenyl Furoxan Hybrids 9a–i
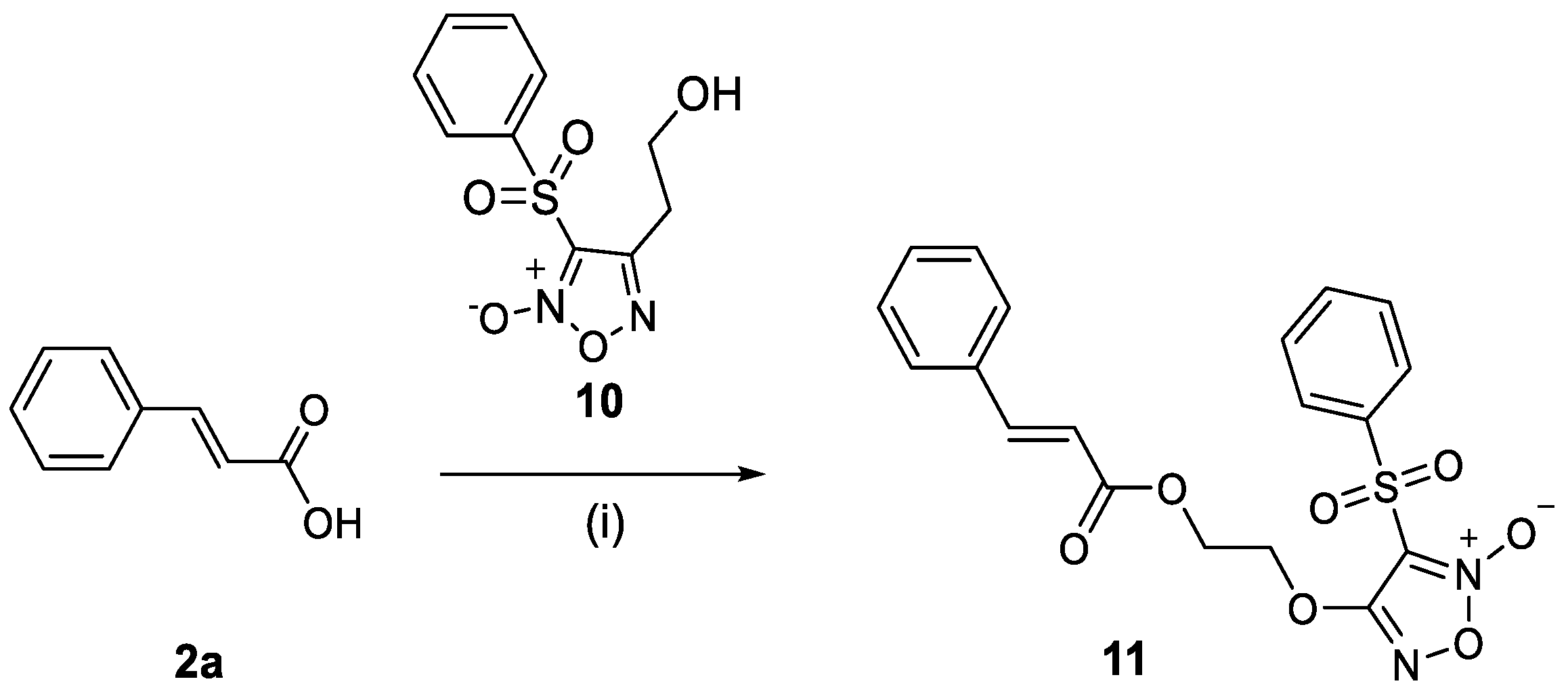
2.1.3. Synthesis of the Cinnamic Acid–Phenylsulfonyl Furoxan Hybrid 11
2.2. Physicochemical Studies
2.2.1. In Silico Determination of Drug-likeness and ADMET Properties
| Compound | Milog P a | TPSA b | No Atoms | No O, N c | No OH, NH d | No Violations | No Rotational Bonds e | Volume f | MW g | logBB h |
|---|---|---|---|---|---|---|---|---|---|---|
| 5a | 3.23 | 72.47 | 24 | 5 | 1 | 0 | 7 | 294.57 | 323.35 | 0.0909 |
| 5b | 4.21 | 72.47 | 28 | 5 | 1 | 0 | 7 | 338.56 | 373.41 | 0.0295 |
| 5c | 2.95 | 72.47 | 23 | 5 | 1 | 0 | 7 | 285.28 | 329.38 | 0.3054 |
| 5d | 2.31 | 85.61 | 23 | 6 | 1 | 0 | 7 | 276.14 | 313.31 | 0.2410 |
| 5e | 4.25 | 72.47 | 27 | 5 | 1 | 0 | 7 | 329.27 | 379.44 | 0.0369 |
| 5f | 3.61 | 85.61 | 27 | 6 | 1 | 0 | 7 | 320.13 | 363.37 | 0.0282 |
| 5g | 4.56 | 72.47 | 30 | 5 | 1 | 0 | 8 | 365.98 | 399.45 | 0.2139 |
| 5h | 5.54 | 72.47 | 34 | 5 | 1 | 1 | 8 | 409.97 | 449.51 | 0.0762 |
| 5i | 4.28 | 72.47 | 29 | 5 | 1 | 0 | 8 | 356.69 | 405.48 | 0.7968 |
| 6a | 3.29 | 88.00 | 25 | 6 | 2 | 0 | 7 | 306.86 | 338.36 | 0.0797 |
| 6b | 4.27 | 88.00 | 29 | 6 | 2 | 0 | 7 | 350.86 | 388.42 | 0.0980 |
| 6c | 3.01 | 88.00 | 24 | 6 | 2 | 0 | 7 | 297.58 | 344.39 | 0.0296 |
| 6d | 2.37 | 101.14 | 24 | 7 | 2 | 0 | 7 | 288.43 | 328.32 | 0.0286 |
| 6e | 4.32 | 88.00 | 28 | 6 | 2 | 0 | 7 | 341.57 | 394.45 | 0.0231 |
| 6f | 3.68 | 101.14 | 28 | 7 | 2 | 0 | 7 | 332.42 | 378.38 | 0.0287 |
| 6g | 4.63 | 88.00 | 31 | 6 | 2 | 0 | 8 | 378.27 | 414.46 | 0.2401 |
| 6h | 5.61 | 88.00 | 35 | 6 | 2 | 1 | 8 | 422.26 | 464.52 | 0.3068 |
| 6i | 4.35 | 88.00 | 30 | 6 | 2 | 0 | 8 | 368.98 | 420.49 | 0.0733 |
| 9a | 4.45 | 77.80 | 24 | 6 | 0 | 0 | 6 | 280.81 | 322.32 | 0.4173 |
| 9b | 5.43 | 77.80 | 28 | 6 | 0 | 1 | 6 | 324.80 | 372.38 | 0.0433 |
| 9c | 4.17 | 77.80 | 23 | 6 | 0 | 0 | 6 | 271.52 | 328.35 | 0.3747 |
| 9d | 3.53 | 90.94 | 23 | 7 | 0 | 0 | 6 | 262.38 | 312.28 | 0.3675 |
| 9e | 5.48 | 77.80 | 27 | 6 | 0 | 1 | 6 | 315.51 | 378.41 | 0.1693 |
| 9f | 4.83 | 90.94 | 27 | 7 | 0 | 0 | 6 | 306.37 | 362.34 | 0.1375 |
| 9g | 5.79 | 77.80 | 30 | 6 | 0 | 1 | 7 | 352.22 | 398.42 | 1.3975 |
| 9h | 6.77 | 77.80 | 34 | 6 | 0 | 1 | 7 | 396.21 | 448.48 | 0.2642 |
| 9i | 5.50 | 77.80 | 29 | 6 | 0 | 1 | 7 | 342.93 | 404.45 | 0.9812 |
| 11 | 3.78 | 121.17 | 29 | 9 | 0 | 0 | 9 | 338.03 | 416.41 | 0.1441 |
hERG Inhibition
Carcinogenicity in Mouse and Rat
Ames Test
2.3. Biological Evaluation
2.3.1. In Vitro Lipid Peroxidation
2.3.2. In Vitro LOX Inhibition
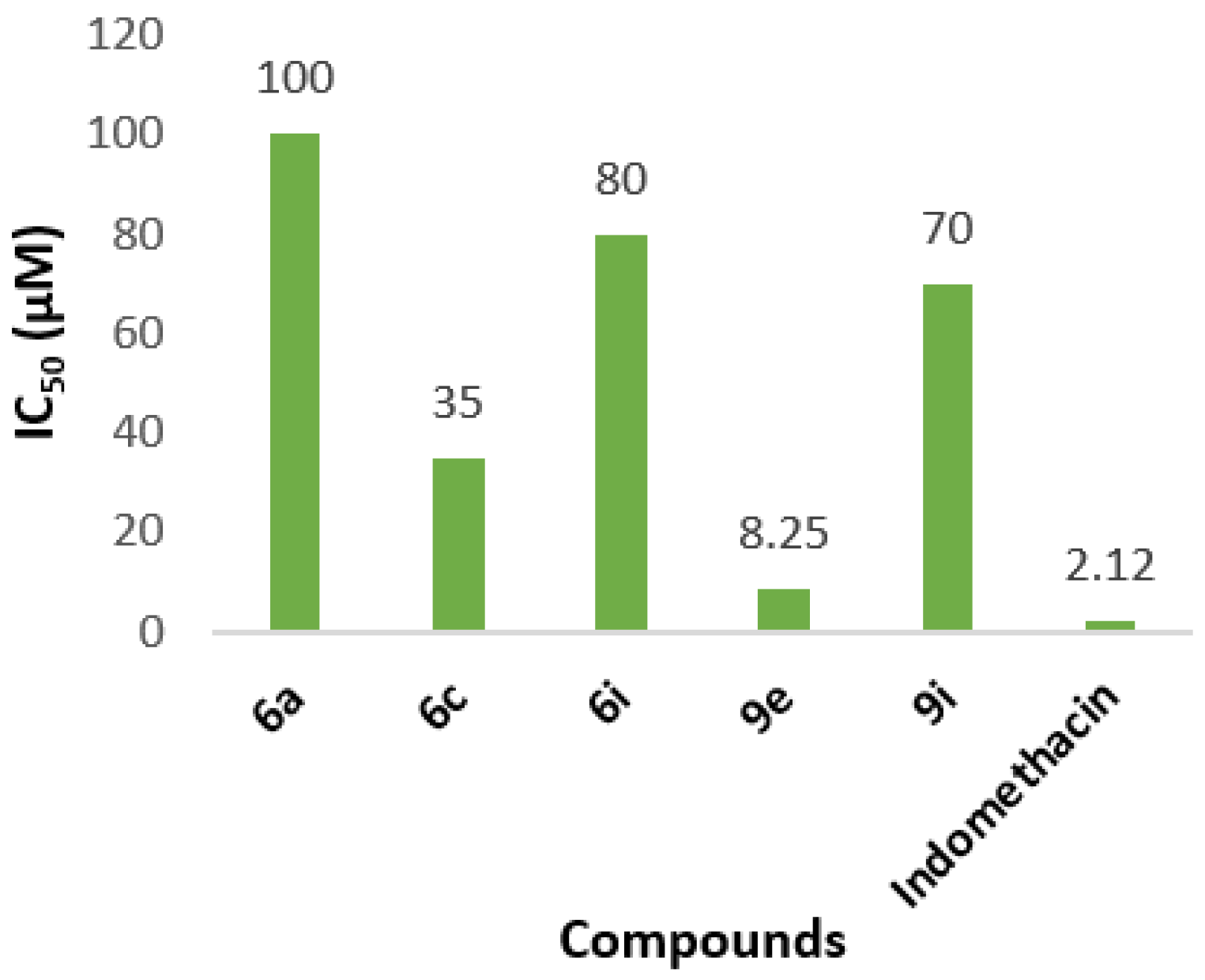
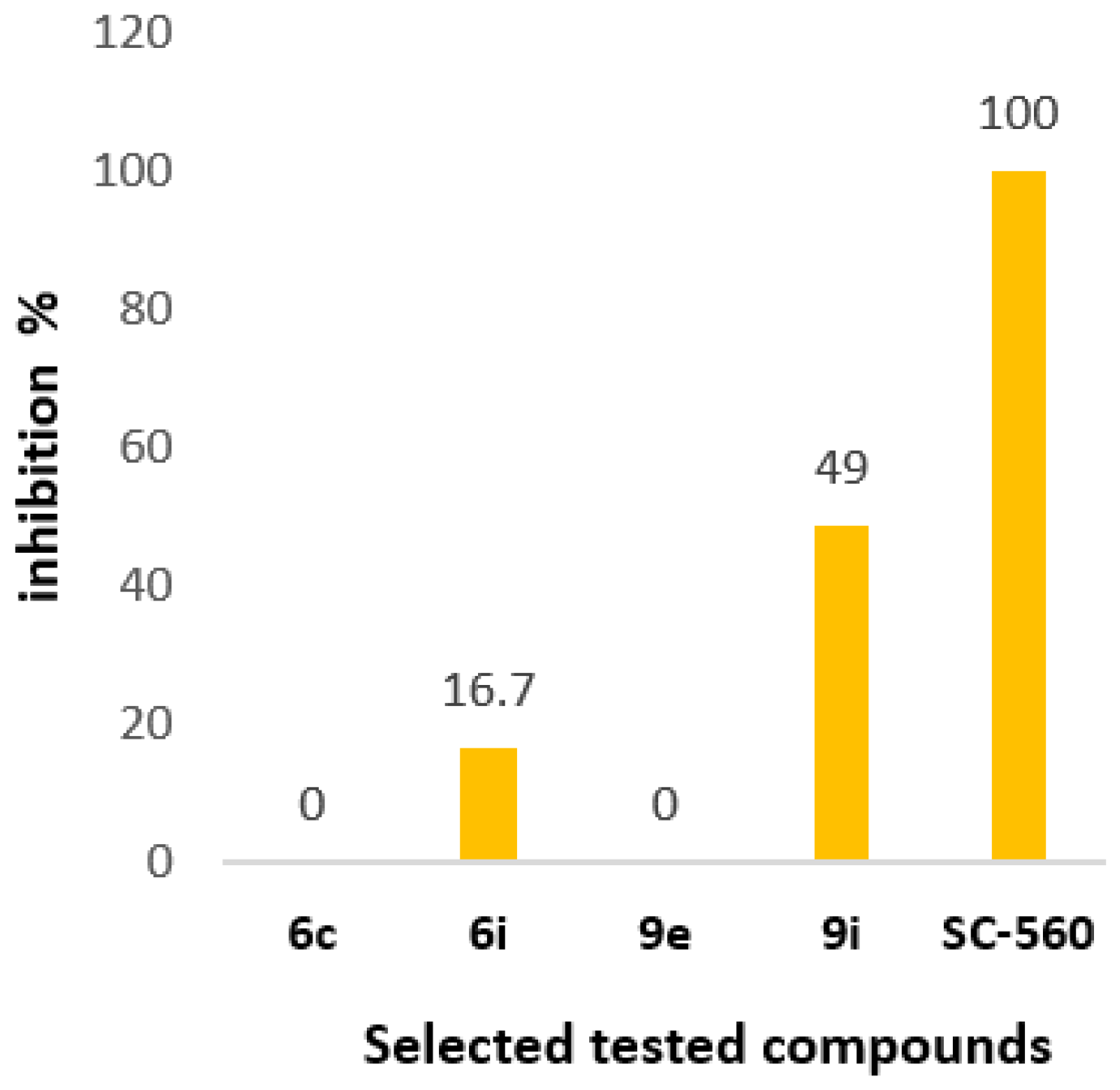
2.3.3. In Vitro COX-2 and COX-1 Inhibition
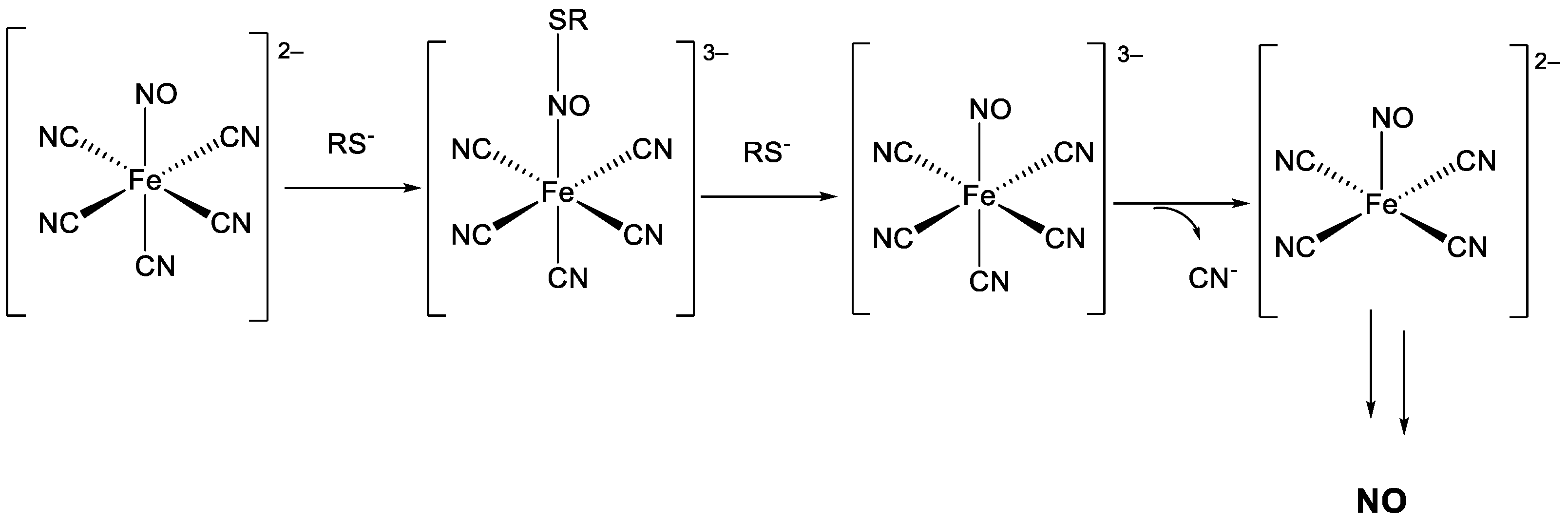
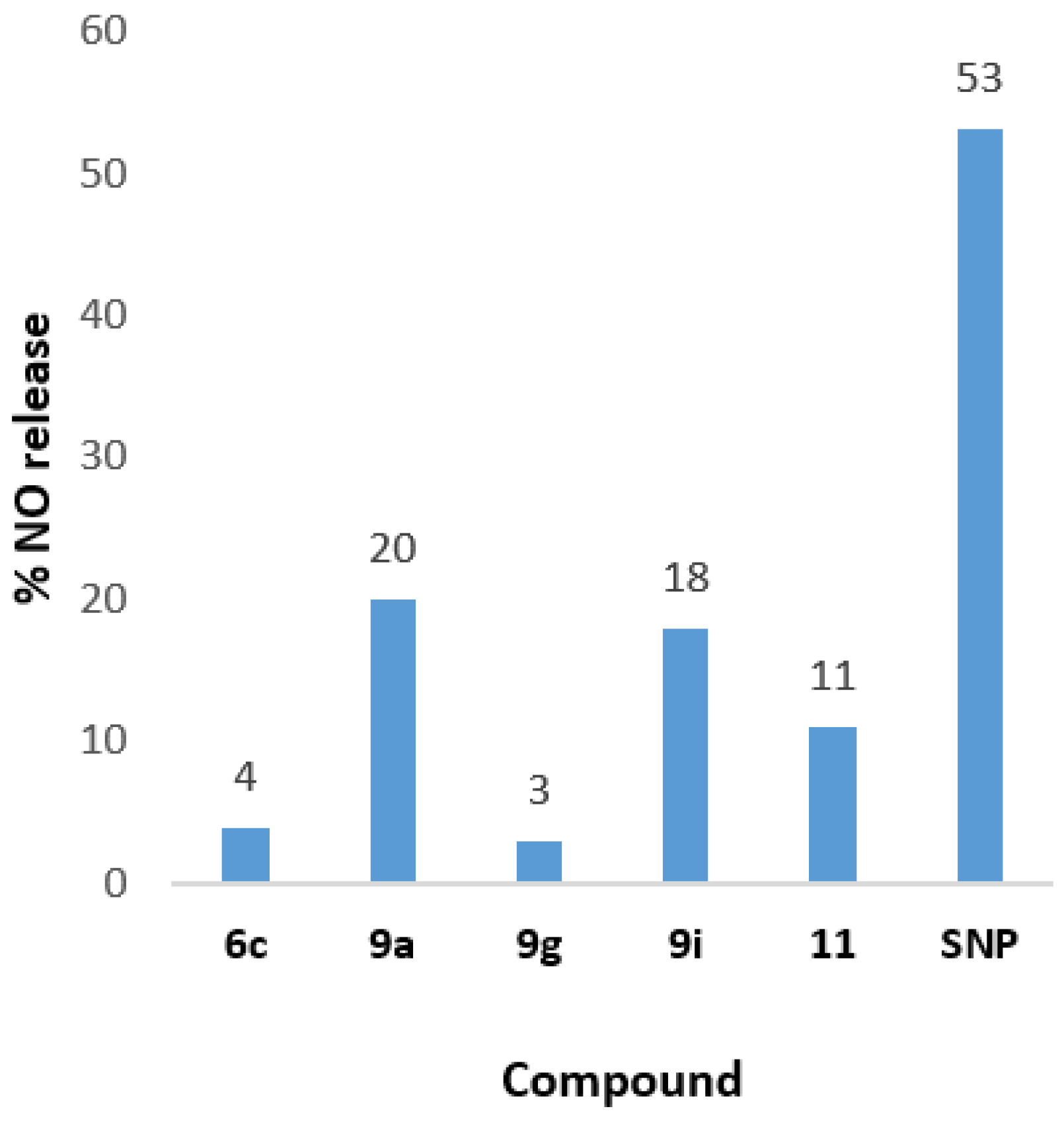
2.3.4. NO Release Ability
2.3.5. In Vitro Inhibition of Albumin Denaturation
2.3.6. MTT Assays on Cancer Cell Lines
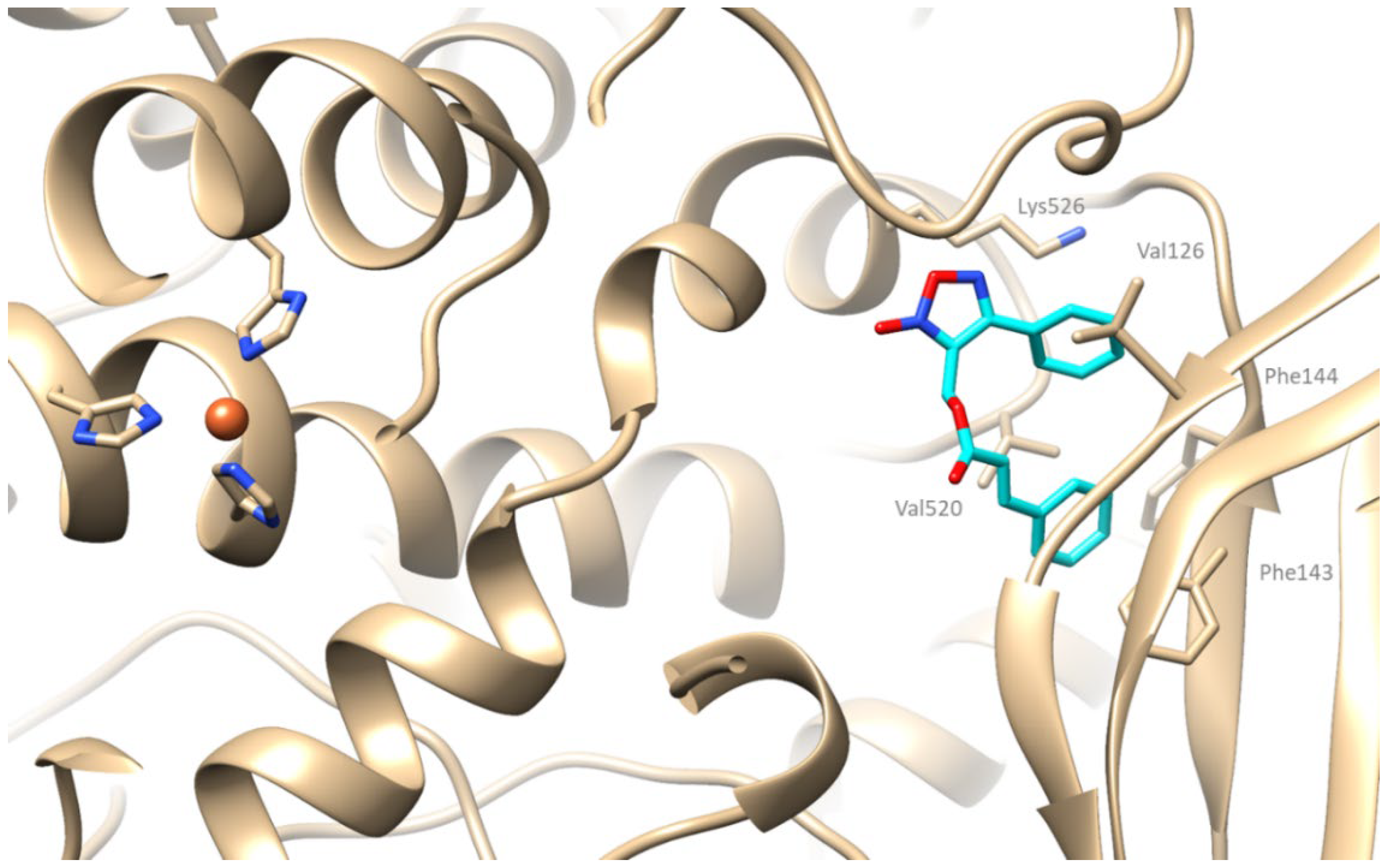
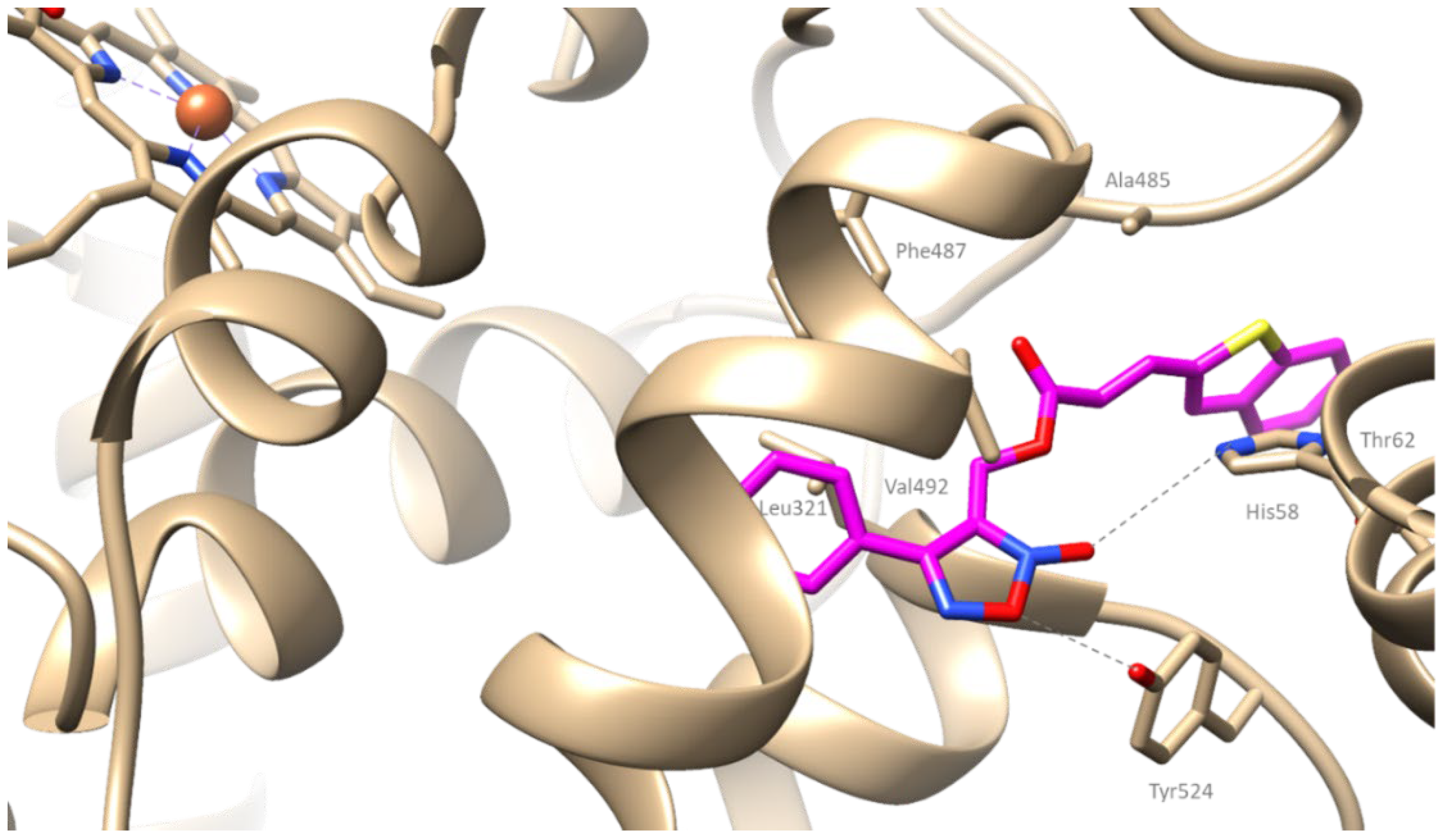
2.4. Computational Studies—Docking Simulation
2.4.1. Docking Studies on Soybean Lipoxygenase
2.4.2. Docking Studies on COX-2
3. Materials and Methods
3.1. General Information
- AutoDockVina v.1.1.1 (The Scripps Research Institute), via the PyRx program [73]
- UCSF Chimera v.1.5.3 (University of California) [74]
- GROMACS toolkit v.4.6.5 [75]
- Modeller [76]
- OpenBabel v.2.2.3 [77]
- AmberTools [78]
- Molinspiration (https://www.molinspiration.com, accessed on 10 May 2025) [47]
- preADMET (https://preadmet.webservice.bmdrc.org/, accessed on 10 April 2025) [46]
3.2. Computational Studies
3.2.1. Molecular Docking Studies on Soybean Lipoxygenase
3.2.2. Molecular Docking Studies on COX-2
3.3. Physicochemical Studies
3.3.1. In Silico Determination of miLogP
3.3.2. In Silico Determination of ADMET Properties and Drug-likeness
3.4. Chemistry
3.4.1. Synthesis of N-(4-Acetylphenyl)-2-chloroacetamide (4) [34]
3.4.2. General Procedure for the Synthesis of Hybrid Compounds 5a–i
2-((4-Acetylphenyl)amino)-2-oxoethyl Cinnamate (5a)
2-((4-Acetylphenyl)amino)-2-oxoethyl (E)-3-(naphthalen-1-yl)acrylate (5b)
2-((4-Acetylphenyl)amino)-2-oxoethyl (E)-3-(thiophen-2-yl)acrylate (5c)
2-((4-Acetylphenyl)amino)-2-oxoethyl (E)-3-(furan-2-yl)acrylate (5d)
2-((4-Acetylphenyl)amino)-2-oxoethyl (E)-3-(benzo[b]thiophen-2-yl)acrylate (5e)
2-((4-Acetylphenyl)amino)-2-oxoethyl (E)-3-(benzofuran-2-yl)acrylate (5f)
2-((4-Acetylphenyl)amino)-2-oxoethyl (E)-2,3-diphenylacrylate (5g)
2-((4-Acetylphenyl)amino)-2-oxoethyl (E)-3-(naphthalen-1-yl)-2-phenylacrylate (5h)
2-((4-Acetylphenyl)amino)-2-oxoethyl (E)-2-phenyl-3-(thiophen-2-yl)acrylate (5i)
3.4.3. General Procedure for the Synthesis of Hybrid Compounds 6a–i
2-((4-((E)-1-(Hydroxyimino)ethyl)phenyl)amino)-2-oxoethyl cinnamate (6a)
2-((4-((E)-1-(Hydroxyimino)ethyl)phenyl)amino)-2-oxoethyl (E)-3-(naphthalen-1-yl)acrylate (6b)
2-((4-((E)-1-(Hydroxyimino)ethyl)phenyl)amino)-2-oxoethyl (E)-3-(thiophen-2-yl)acrylate (6c)
2-((4-((E)-1-(Hydroxyimino)ethyl)phenyl)amino)-2-oxoethyl (E)-3-(furan-2-yl)acrylate (6d)
2-((4-((E)-1-(Hydroxyimino)ethyl)phenyl)amino)-2-oxoethyl (E)-3-(benzo[b]thiophen-2-yl)acrylate (6e)
2-((4-((E)-1-(Hydroxyimino)ethyl)phenyl)amino)-2-oxoethyl (E)-3-(benzofuran-2-yl)acrylate (6f)
2-((4-((E)-1-(Hydroxyimino)ethyl)phenyl)amino)-2-oxoethyl (E)-2,3-diphenylacrylate (6g)
2-((4-((E)-1-(Hydroxyimino)ethyl)phenyl)amino)-2-oxoethyl (E)-3-(naphthalen-1-yl)-2-phenylacrylate (6h)
2-((4-((E)-1-(Hydroxyimino)ethyl)phenyl)amino)-2-oxoethyl (E)-2-phenyl-3-(thiophen-2-yl)acrylate (6i)
3.4.4. Synthesis of 3-(Hydroxymethyl)-4-phenyl-1,2,5-oxadiazole 2-oxide (8)
3.4.5. General Procedure for the Synthesis of Hybrid Compounds 9a–i
3-((Cinnamoyloxy)methyl)-4-phenyl-1,2,5-oxadiazole 2-oxide (9a)
3-(((3-(Naphthalen-1-yl)acryloyl)oxy)methyl)-4-phenyl-1,2,5-oxadiazole 2-oxide (9b)
(E)-4-Phenyl-3-(((3-(thiophen-2-yl)acryloyl)oxy)methyl)-1,2,5-oxadiazole 2-oxide (9c)
(E)-3-(((3-(Furan-2-yl)acryloyl)oxy)methyl)-4-phenyl-1,2,5-oxadiazole 2-oxide (9d)
(E)-3-(((3-(Benzo[b]thiophen-2-yl)acryloyl)oxy)methyl)-4-phenyl-1,2,5-oxadiazole 2-oxide (9e)
(E)-3-(((3-(Benzofuran-2-yl)acryloyl)oxy)methyl)-4-phenyl-1,2,5-oxadiazole 2-oxide (9f)
(E)-3-(((2,3-Diphenylacryloyl)oxy)methyl)-4-phenyl-1,2,5-oxadiazole 2-oxide (9g)
(E)-3-(((3-(Naphthalen-1-yl)-2-phenylacryloyl)oxy)methyl)-4-phenyl-1,2,5-oxadiazole 2-oxide (9h)
(E)-4-Phenyl-3-(((2-phenyl-3-(thiophen-2-yl)acryloyl)oxy)methyl)-1,2,5-oxadiazole 2-oxide (9i)
3.4.6. Synthesis of the 4-(2-(Cinnamoyloxy)ethoxy)-3-(phenylsulfonyl)-1,2,5-oxadiazole 2-oxide (11)
3.5. Biological In Vitro Assays
3.5.1. In Vitro Inhibition of Linoleic Acid Lipid Peroxidation
3.5.2. In Vitro Inhibition of Soybean Lipoxygenase
3.5.3. In Vitro Inhibition of Ovine Cyclooxygenase-2 (COX-2)
3.5.4. In Vitro Inhibition of COX-1
3.5.5. In Vitro Determination of NO Release with Griess Reagent
3.5.6. In Vitro Inhibition of Albumin
3.5.7. MTT Assays on Cancer Cell Lines
Cell Culture
MTT Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fleit, H.B. Chronic Inflammation. Pathobiol. Hum. Dis. Dyn. Encycl. Dis. Mech. 2014, 300–314. [Google Scholar] [CrossRef]
- Punchard, N.A.; Whelan, C.J.; Adcock, I. The Journal of Inflammation. J. Inflamm. 2004, 1, 1. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rådmark, O.; Werz, O.; Steinhilber, D.; Samuelsson, B. 5-Lipoxygenase: Regulation of expression and enzyme activity. Trends Biochem. Sci. 2007, 32, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, F.A. Cyclooxygenase Enzymes: Regulation and Function. Curr. Pharm. Des. 2005, 10, 577–588. [Google Scholar] [CrossRef]
- Chelombitko, M.A. Role of Reactive Oxygen Species in Inflammation: A Minireview. Mosc. Univ. Biol. Sci. Bull. 2018, 73, 199–202. [Google Scholar] [CrossRef]
- Mittal, M.; Siddiqui, M.R.; Tran, K.; Reddy, S.P.; Malik, A.B. Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox Signal. 2014, 20, 1126–1167. [Google Scholar] [CrossRef]
- Hakim, J. Reactive oxygen species and inflammation. Comptes Rendus Seances Soc. Biol. Fil. 1993, 187, 286–295. [Google Scholar] [CrossRef]
- Krumenacker, J.S.; Hanafy, K.A.; Murad, F. Regulation of nitric oxide and soluble guanylyl cyclase. Brain Res. Bull. 2004, 62, 505–515. [Google Scholar] [CrossRef]
- Cirino, G.; Distrutti, E.; Wallace, J.L. Nitric oxide and inflammation. Inflamm. Allergy-Drug Targets 2006, 5, 115–119. [Google Scholar] [CrossRef]
- Laroux, F.S.; Pavlick, K.P.; Hines, I.N.; Kawachi, S.; Harada, H.; Bharwani, S.; Hoffman, J.M.; Grisham, M.B. Role of nitric oxide in inflammation. Acta. Physiol. Scand. 2001, 173, 113–118. [Google Scholar] [CrossRef]
- Ruwizhi, N.; Aderibigbe, B.A. Cinnamic Acid Derivatives and Their Biological Efficacy. Int. J. Mol. Sci. 2020, 21, 5712. [Google Scholar] [CrossRef] [PubMed]
- Pontiki, E.; Hadjipavlou-Litina, D. Multi-Target Cinnamic Acids for Oxidative Stress and Inflammation: Design, Synthesis, Biological Evaluation and Modeling Studies. Molecules 2018, 24, 12. [Google Scholar] [CrossRef]
- Ivasiv, V.; Albertini, C.; Gonçalves, A.E.; Rossi, M.; Bolognesi, M.L. Molecular Hybridization as a Tool for Designing Multitarget Drug Candidates for Complex Diseases. Curr. Top. Med. Chem. 2019, 19, 1694–1711. [Google Scholar] [CrossRef]
- Bosquesi, P.L.; Melo, T.R.F.; Vizioli, E.O.; Santos, J.L.D.; Chung, M.C. Anti-Inflammatory Drug Design Using a Molecular Hybridization Approach. Pharmaceuticals 2011, 4, 1450–1474. [Google Scholar] [CrossRef]
- Sharma, R.; Joubert, J.; Malan, S.F. Recent Developments in Drug Design of NO-donor Hybrid Compounds. Mini-Rev. Med. Chem. 2018, 18, 1175–1198. [Google Scholar] [CrossRef]
- Wasfi, A.A.M.; Al-Masoudi, N.A.; Saeed, B.A.; Winter, R.; Pannecouque, C. Synthesis, In Vitro Anti-HIV Activity, Cytotoxicity, and Computational Studies of Some New Steroids and Their Pyrazoline and Oxime Analogues. Russ. J. Bioorg. Chem. 2020, 46, 822–836. [Google Scholar] [CrossRef]
- Yang, Y.; Pannecouque, C.; De Clercq, E.; Zhuang, C.; Chen, F.E. Privileged scaffold inspired design of novel oxime-biphenyl-DAPYs in treatment of HIV-1. Bioorg. Chem. 2020, 99, 103825. [Google Scholar] [CrossRef]
- Abd-Ellah, H.S.; Abdel-Aziz, M.; Shoman, M.E.; Beshr, E.A.M.; Kaoud, T.S.; Ahmed, A.S.F.F. New 1,3,4-oxadiazole/oxime hybrids: Design, synthesis, anti-inflammatory, COX inhibitory activities and ulcerogenic liability. Bioorg. Chem. 2017, 74, 15–29. [Google Scholar] [CrossRef]
- Abd-Ellah, H.S.; Abdel-Aziz, M.; Shoman, M.E.; Beshr, E.A.M.; Kaoud, T.S.; Ahmed, A.S.F.F. Novel 1,3,4-oxadiazole/oxime hybrids: Synthesis, docking studies and investigation of anti-inflammatory, ulcerogenic liability and analgesic activities. Bioorg. Chem. 2016, 69, 48–63. [Google Scholar] [CrossRef]
- Koikov, L.N.; Alekseeva, N.V.; Lisitza, E.A.; Krichevsky, E.S.; Grigoriev, N.B.; Danilov, A.V.; Severina, I.S.; Pyatakova, N.V.; Granik, V.G. Oximes, amidoximes and hydroxamic acids as nitric oxide donors. Mendeleev Commun. 1998, 8, 165–168. [Google Scholar] [CrossRef]
- Ferioli, R.; Folco, G.C.; Ferretti, C.; Gasco, A.M.; Medana, C.; Fruttero, R.; Civelli, M.; Gasco, A. A new class of furoxan derivatives as NO donors: Mechanism of action and biological activity. Br. J. Pharmacol. 1995, 114, 816. [Google Scholar] [CrossRef]
- Gasco, A.; Fruttero, R.; Sorba, G.; Di Stilo, A.; Calvino, R. NO donors: Focus on furoxan derivatives. Pure Appl. Chem. 2004, 76, 973–981. [Google Scholar] [CrossRef]
- Pontiki, E.; Hadjipavlou-Litina, D.; Geromichalos, G.; Papageorgiou, A. Anticancer activity and quantitative-structure activity relationship (QSAR) studies of a series of antioxidant/anti-inflammatory aryl-acetic and hydroxamic acids. Chem. Biol. Drug Des. 2009, 74, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Heravi, M.M.; Asadi, S.; Azarakhshi, F. Recent Applications of Doebner, Doebner-von Miller and Knoevenagel-Doebner Reactions in Organic Syntheses. Curr. Org. Synth. 2014, 11, 701–731. [Google Scholar] [CrossRef]
- Klein, J.; Bergmann, E.D. The Reaction of Acetals with Malonic Acid and its Derivatives. A Contribution to the Knowledge of the Knoevenagel–Doebner Reaction. J. Am. Chem. Soc. 1957, 79, 3452–3454. [Google Scholar] [CrossRef]
- Fotopoulos, I.; Pontiki, E.; Litina, D.H. Targeting Inflammation with Conjugated Cinnamic Amides, Ethers and Esters. Lett. Drug Des. Discov. 2018, 17, 3–11. [Google Scholar] [CrossRef]
- Zhou, H.; Gao, S.J.; Zhang, M.T.; Jia, J.; Chen, F.X.; Chen, C.L.; Yang, P.F.; Mao, J.L. Synthesis, configurational analysis and antiviral activities of novel diphenylacrylic acids with caffeic acid as the lead compound. J. Mol. Struct. 2023, 1291, 136016. [Google Scholar] [CrossRef]
- Csankó, K.; Illés, L.; Felföldi, K.; Kiss, J.T.; Sipos, P.; Pálinkó, I. CH⋯S hydrogen bonds as the organising force in 2,3-thienyl- and phenyl- or 2,3-dithienyl-substituted propenoic acid aggregates studied by the combination of FT-IR spectroscopy and computations. J. Mol. Struct. 2011, 993, 259–263. [Google Scholar] [CrossRef]
- Zhu, L.; Lei, N.; Miao, Z.; Sheng, C.; Zhuang, C.; Yao, J.; Zhang, W. β-alanine-DBU: A highly efficient catalytic system for knoevenagel-doebner reaction under mild conditions. Chin. J. Chem. 2012, 30, 139–143. [Google Scholar] [CrossRef]
- Obushak, M.D.; Pokhodylo, N.T.; Ostapiuk, Y.V.; Matiychuk, V.S. Synthesis of 3-Substituted (6-[(E)-2-(1-Benzofuran-2-yl)ethenyl][1,2,4]triazolo [3,4-b][1,3,4]thiadiazoles. Phosphorus Sulfur Silicon Relat. Elem. 2007, 183, 136–143. [Google Scholar] [CrossRef]
- Qin, J.; Li, H.; Wang, X.; Zhang, Y.; Duan, Y.; Yao, Y.; Yang, H.; Sun, M. Discovery of a novel piperlongumine analogue as a microtubule polymerization inhibitor with potent anti-angiogenic and anti-metastatic efficacy. Eur. J. Med. Chem. 2022, 243, 114738. [Google Scholar] [CrossRef]
- Bellassoued, M.; Lensen, N.; Bakasse, M.; Mouelhi, S. Two-carbon homologation of aldehydes via silyl ketene acetals: A new stereoselective approach to (E)-alkenoic acids. J. Org. Chem. 1998, 63, 8785–8789. [Google Scholar] [CrossRef]
- Upare, A.A.; Gadekar, P.K.; Sivaramakrishnan, H.; Naik, N.; Khedkar, V.M.; Sarkar, D.; Choudhari, A.; Roopan, S.M. Design, synthesis and biological evaluation of (E)-5-styryl-1,2,4-oxadiazoles as anti-tubercular agents. Bioorg. Chem. 2019, 86, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Anthwal, A.; Thakur, B.K.; Rawat, M.S.M.; Rawat, D.S.; Tyagi, A.K.; Aggarwal, B.B. Synthesis, Characterization and In Vitro Anticancer Activity of C-5 Curcumin Analogues with Potential to Inhibit TNF-α-Induced NF-B Activation. BioMed Res. Int. 2014, 2014, 524161. [Google Scholar] [CrossRef]
- Tsopka, I.C.; Hadjipavlou-Litina, D. 2-((4-((E)-1-(Hydroxyimino)ethyl)phenyl)amino)-2-oxoethyl Cinnamate. Molbank 2021, 2021, M1239. [Google Scholar] [CrossRef]
- Rafiq, M.; Nazir, Y.; Ashraf, Z.; Rafique, H.; Afzal, S.; Mumtaz, A.; Hassan, M.; Ali, A.; Afzal, K.; Yousuf, M.R.; et al. Synthesis, computational studies, tyrosinase inhibitory kinetics and antimelanogenic activity of hydroxy substituted 2-[(4-acetylphenyl)amino]-2-oxoethyl derivatives. J. Enzym. Inhib. Med. Chem. 2019, 34, 1562–1572. [Google Scholar] [CrossRef]
- Horton, A.; Nash, K.; Tackie-Yarboi, E.; Kostrevski, A.; Novak, A.; Raghavan, A.; Tulsulkar, J.; Alhadidi, Q.; Wamer, N.; Langenderfer, B.; et al. Furoxans (Oxadiazole-4 N-oxides) with Attenuated Reactivity are Neuroprotective, Cross the Blood Brain Barrier, and Improve Passive Avoidance Memory. J. Med. Chem. 2018, 61, 4593–4607. [Google Scholar] [CrossRef]
- Ling, Y.; Ye, X.; Zhang, Z.; Zhang, Y.; Lai, Y.; Ji, H.; Peng, S.; Tian, J. Novel nitric oxide-releasing derivatives of farnesylthiosalicylic acid: Synthesis and evaluation of antihepatocellular carcinoma activity. J. Med. Chem. 2011, 54, 3251–3259. [Google Scholar] [CrossRef]
- Xie, Y.D.; Shao, L.H.; Wang, Q.T.; Bai, Y.; Li, N.; Yang, G.; Li, Y.P.; Bian, X.L. Design, synthesis and evaluation of phenylfuroxan nitric oxide-donor phenols as potential anti-diabetic agents. Bioorg. Chem. 2019, 89, 103000. [Google Scholar] [CrossRef]
- Norinder, U.; Bergström, C.A.S. Prediction of ADMET Properties. ChemMedChem 2006, 1, 920–937. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods 2000, 44, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Di, L.; Kerns, E.H.; Carter, G.T. Strategies to assess blood-brain barrier penetration. Expert Opin. Drug Discov. 2008, 3, 677–687. [Google Scholar] [CrossRef]
- Martins, I.F.; Teixeira, A.L.; Pinheiro, L.; Falcao, A.O. A Bayesian approach to in Silico blood-brain barrier penetration modeling. J. Chem. Inf. Model. 2012, 52, 1686–1697. [Google Scholar] [CrossRef]
- PreADMET|Prediction of ADME/Tox—Just Another BMDRC Sites Site. Available online: https://preadmet.webservice.bmdrc.org/ (accessed on 8 May 2025).
- Molinspiration Cheminformatics. Available online: https://molinspiration.com/ (accessed on 9 July 2025).
- Dmitriev, A.V.; Rudik, A.V.; Karasev, D.A.; Pogodin, P.V.; Lagunin, A.A.; Filimonov, D.A.; Poroikov, V.V. In Silico Prediction of Drug-Drug Interactions Mediated by Cytochrome P450 Isoforms. Pharmaceutics 2021, 13, 538. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- De Graaf, C.; Vermeulen, N.P.E.; Feenstra, K.A. Cytochrome P450 in silico: An integrative modeling approach. J. Med. Chem. 2005, 48, 2725–2755. [Google Scholar] [CrossRef]
- Pelkonen, O.; Mäenpääj, J.; Taavitsainen, P.; Rautio, A.; Raunio, H. Inhibition and induction of human cytochrome P450 (CYP) enzymes. Xenobiotica 1998, 28, 1203–1253. [Google Scholar] [CrossRef]
- Shao, C.-Y.; Su, B.-H.; Tu, Y.-S.; Lin, C.; Lin, O.A.; Tseng, Y.J. CypRules: A rule-based P450 inhibition pre-diction server. Bioinformatics 2015, 31, 1869–1871. [Google Scholar] [CrossRef]
- Hillebrecht, A.; Muster, W.; Brigo, A.; Kansy, M.; Weiser, T.; Singer, T. Comparative evaluation of in silico systems for ames test mutagenicity prediction: Scope and limitations. Chem. Res. Toxicol. 2011, 24, 843–854. [Google Scholar] [CrossRef]
- Modi, S.; Li, J.; Malcomber, S.; Moore, C.; Scott, A.; White, A.; Carmichael, P. Integrated in silico approaches for the prediction of Ames test mutagenicity. J. Comput. Aided Mol. Des. 2012, 26, 1017–1033. [Google Scholar] [CrossRef]
- Ertl, P.; Rohde, B.; Selzer, P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J. Med. Chem. 2000, 43, 3714–3717. [Google Scholar] [CrossRef]
- Prasanna, S.; Doerksen, R.J. Topological polar surface area: A useful descriptor in 2D-QSAR. Curr. Med. Chem. 2009, 16, 21–41. [Google Scholar] [CrossRef] [PubMed]
- Hadjipavlou-Litina, D.; Samadi, A.; Unzeta, M.; Marco-Contelles, J. Analysis of the antioxidant properties of differently substituted 2- and 3-indolyl carbohydrazides and related derivatives. Eur. J. Med. Chem. 2013, 63, 670–674. [Google Scholar] [CrossRef]
- Mankovska, I.M.; Klymenko, O.O.; Gonchar, O.O.; Karasevich, N.V.; Karaban, I.M. Interplay of oxidative stress and mitochondrial dysfunction in Alzheimer’s and Parkinson’s diseases: Mechanisms and treatment strategies. Neurophysiology 2025, 1–11. [Google Scholar] [CrossRef]
- Chong, Z.Z.; Souayah, N. Oxidative Stress: Pathological Driver in Chronic Neurodegenerative Diseases. Antioxidants 2025, 14, 696. [Google Scholar] [CrossRef]
- Wisastra, R.; Dekker, F.J. Inflammation, Cancer and Oxidative Lipoxygenase Activity are Intimately Linked. Cancers 2014, 6, 1500. [Google Scholar] [CrossRef]
- Prigge, S.T.; Boyington, J.C.; Gaffney, B.J.; Amzel, L.M. Structure conservation in lipoxygenases: Structural analysis of soybean lipoxygenase-1 and modeling of human lipoxygenases. Proteins Struct. Funct. Genet. 1996, 24, 275–291. [Google Scholar] [CrossRef]
- Pontiki, E.; Hadjipavlou-Litina, D. Synthesis and pharmacochemical evaluation of novel aryl-acetic acid inhibitors of lipoxygenase, antioxidants, and anti-inflammatory agents. Bioorg. Med. Chem. 2007, 15, 5819–5827. [Google Scholar] [CrossRef]
- Vane, J.R.; Botting, R.M. Anti-inflammatory drugs and their mechanism of action. Inflamm. Res. 1998, 47 (Suppl. S2), 78–87. [Google Scholar] [CrossRef]
- Noti, V.; Pontiki, E.; Hadjipavlou-Litina, D. Development of Novel Pyrrole Derivatives and Their Cinnamic Hybrids as Dual COX-2/LOX Inhibitors. Molecules 2023, 28, 7958. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.N.; Dasgupta, T.P. Mechanism of Nitric Oxide Release. I. Two-electron Reduction of Sodium Nitroprusside by l-cysteine in Aqueous Solution. Inorg. React. Mech. 2002, 3, 181–195. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, X.; Broderick, M.; Fein, H. Measurement of Nitric Oxide Production in Biological Systems by Using Griess Reaction Assay. Sensors 2003, 3, 276–284. [Google Scholar] [CrossRef]
- Tariq, S.; Kamboj, P.; Alam, O.; Amir, M. 1,2,4-Triazole-based benzothiazole/benzoxazole derivatives: Design, synthesis, p38α MAP kinase inhibition, anti-inflammatory activity and molecular docking studies. Bioorg. Chem. 2018, 81, 630–641. [Google Scholar] [CrossRef]
- Sigala, I.; Tsamis, K.I.; Gousia, A.; Alexiou, G.; Voulgaris, S.; Giannakouros, T.; Kyritsis, A.P.; Nikolakaki, E. Expression of SRPK1 in gliomas and its role in glioma cell lines viability. Tumor Biol. 2016, 37, 8699–8707. [Google Scholar] [CrossRef]
- Kostopoulou, I.; Tzani, A.; Polyzos, N.I.; Karadendrou, M.A.; Kritsi, E.; Pontiki, E.; Liargkova, T.; Hadjipavlou-Litina, D.; Zoumpoulakis, P.; Detsi, A. Exploring the 2′-Hydroxy-Chalcone Framework for the Development of Dual Antioxidant and Soybean Lipoxygenase Inhibitory Agents. Molecules 2021, 26, 2777. [Google Scholar] [CrossRef]
- El Khatabi, K.; El-Mernissi, R.; Aanouz, I.; Ajana, M.A.; Lakhlifi, T.; Khan, A.; Wei, D.Q.; Bouachrine, M. Identification of novel acetylcholinesterase inhibitors through 3D-QSAR, molecular docking, and molecular dynamics simulation targeting Alzheimer’s disease. J. Mol. Model. 2021, 27, 302. [Google Scholar] [CrossRef]
- Pontiki, E.; Hadjipavlou-Litina, D. Lipoxygenase inhibitors: A comparative QSAR study review and evaluation of new QSARs. Med. Res. Rev. 2008, 28, 39–117. [Google Scholar] [CrossRef]
- Mavridis, E.; Bermperoglou, E.; Pontiki, E.; Hadjipavlou-Litina, D. 5-(4H)-Oxazolones and Their Benzamides as Potential Bioactive Small Molecules. Molecules 2020, 25, 3173. [Google Scholar] [CrossRef]
- Kostopoulou, I.; Diassakou, A.; Kavetsou, E.; Kritsi, E.; Zoumpoulakis, P.; Pontiki, E.; Hadjipavlou-Litina, D.; Detsi, A. Novel quinolinone–pyrazoline hybrids: Synthesis and evaluation of antioxidant and lipoxygenase inhibitory activity. Mol. Divers. 2021, 25, 723–740. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- UCSF Chimera Home Page. Available online: https://www.cgl.ucsf.edu/chimera/ (accessed on 8 May 2025).
- GROMACS 4.6.5 Online Reference. Available online: https://manual.gromacs.org/archive/4.6.6/online.html (accessed on 8 May 2025).
- About MODELLER. Available online: https://salilab.org/modeller/ (accessed on 8 May 2025).
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- AmberTools25. Available online: https://ambermd.org/AmberTools.php (accessed on 27 August 2025).
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Fiser, A.; Šali, A. MODELLER: Generation and Refinement of Homology-Based Protein Structure Models. Methods Enzym. 2003, 374, 461–491. [Google Scholar] [CrossRef]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Cerutti, D.S.; Cisneros, G.A.; Cruzeiro, V.W.D.; Forouzesh, N.; Giese, T.J.; Gotz, A.W.; Gohlke, H.; et al. AmberTools. J. Chem. Inf. Model. 2023, 63, 6183–6191. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Struct. Funct. Bioinform. 2010, 78, 1950–1958. [Google Scholar] [CrossRef]
- Li, P.; Roberts, B.P.; Chakravorty, D.K.; Merz, K.M. Rational design of particle mesh ewald compatible lennard-jones parameters for +2 metal cations in explicit solvent. J. Chem. Theory Comput. 2013, 9, 2733–2748. [Google Scholar] [CrossRef]
- Halgren, T.A. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 1996, 17, 490–519. [Google Scholar] [CrossRef]
- El-Ashrey, M.K.; Bakr, R.O.; Fayed, M.A.A.; Refaey, R.H.; Nissan, Y.M. Pharmacophore based virtual screening for natural product database revealed possible inhibitors for SARS-CoV-2 main protease. Virology 2022, 570, 18. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef]
- Hess, B.; Kutzner, C.; Van Der Spoel, D.; Lindahl, E. GRGMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455. [Google Scholar] [CrossRef]
- Peperidou, A.; Kapoukranidou, D.; Kontogiorgis, C.; Hadjipavlou-Litina, D. Multitarget Molecular Hybrids of Cinnamic Acids. Molecules 2014, 19, 20197–20226. [Google Scholar] [CrossRef]
- Pontiki, E.; Hadjipavlou-Litina, D.; Litinas, K.; Nicolotti, O.; Carotti, A. Design, synthesis and pharmacobiological evaluation of novel acrylic acid derivatives acting as lipoxygenase and cyclooxygenase-1 inhibitors with antioxidant and anti-inflammatory activities. Eur. J. Med. Chem. 2011, 46, 191–200. [Google Scholar] [CrossRef]
| Compound 2, 5, 6, and 9 | R1 | R2 |
|---|---|---|
| a | 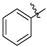 | H |
| b | 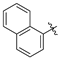 | H |
| c | 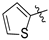 | H |
| d | 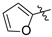 | H |
| e | 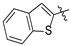 | H |
| f | 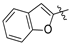 | H |
| g | 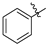 | 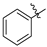 |
| h | 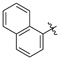 | 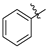 |
| i | 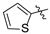 | 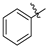 |
| Compound | CYP2C19 Inhibition | CYP2C9 Inhibition | CYP2D6 Inhibition | CYP3A4 Inhibition |
|---|---|---|---|---|
| 5a | no | no | no | yes |
| 5b | no | no | no | yes |
| 5c | no | no | no | yes |
| 5d | no | no | no | yes |
| 5e | no | no | no | yes |
| 5f | no | no | no | yes |
| 5g | no | yes | no | yes |
| 5h | no | yes | no | yes |
| 5i | no | no | no | yes |
| 6a | no | no | no | yes |
| 6b | no | no | no | yes |
| 6c | no | no | no | yes |
| 6d | no | no | no | yes |
| 6e | no | no | no | yes |
| 6f | no | no | no | yes |
| 6g | no | yes | no | yes |
| 6h | no | yes | no | yes |
| 6i | no | no | no | yes |
| 9a | yes | yes | no | yes |
| 9b | yes | yes | no | yes |
| 9c | yes | yes | no | yes |
| 9d | yes | yes | no | yes |
| 9e | yes | yes | no | yes |
| 9f | yes | yes | no | yes |
| 9g | yes | yes | no | yes |
| 9h | yes | yes | no | yes |
| 9i | yes | yes | no | yes |
| 11 | yes | yes | no | yes |
| Compound | hERG Inhibition | Carcinogenicity in Mouse | Carcinogenicity in Rat | Ames Test | TA100_NA | TA100_10RLI | TA1535_NA | TA1535_10RLI |
|---|---|---|---|---|---|---|---|---|
| 5a | Medium risk | negative | negative | mutagen | negative | positive | negative | negative |
| 5b | Medium risk | negative | negative | mutagen | negative | positive | negative | negative |
| 5c | Medium risk | negative | negative | mutagen | negative | positive | negative | positive |
| 5d | Medium risk | negative | negative | mutagen | positive | positive | negative | positive |
| 5e | Medium risk | negative | negative | mutagen | negative | positive | negative | negative |
| 5f | Medium risk | negative | negative | mutagen | positive | positive | negative | negative |
| 5g | Low risk | negative | negative | Non-mutagen | negative | negative | negative | negative |
| 5h | Low risk | negative | negative | mutagen | negative | negative | negative | negative |
| 5i | Medium risk | negative | negative | mutagen | negative | negative | negative | positive |
| 6a | Medium risk | positive | negative | mutagen | negative | positive | negative | negative |
| 6b | high risk | negative | negative | mutagen | negative | positive | negative | negative |
| 6c | Medium risk | negative | negative | mutagen | negative | positive | negative | positive |
| 6d | Medium risk | negative | negative | mutagen | positive | positive | negative | positive |
| 6e | Medium risk | negative | negative | mutagen | negative | positive | negative | negative |
| 6f | Medium risk | negative | negative | mutagen | positive | positive | negative | negative |
| 6g | ambiguous | negative | negative | mutagen | negative | positive | negative | negative |
| 6h | ambiguous | negative | negative | mutagen | negative | positive | negative | negative |
| 6i | high risk | negative | negative | mutagen | negative | positive | negative | positive |
| 9a | low risk | negative | positive | mutagen | positive | positive | positive | positive |
| 9b | low risk | negative | positive | mutagen | negative | positive | positive | negative |
| 9c | Medium risk | negative | positive | mutagen | positive | positive | positive | positive |
| 9d | Medium risk | negative | positive | mutagen | positive | positive | positive | positive |
| 9e | low risk | negative | positive | mutagen | positive | positive | positive | negative |
| 9f | low risk | negative | negative | mutagen | positive | positive | positive | negative |
| 9g | low risk | negative | positive | mutagen | positive | positive | positive | positive |
| 9h | low risk | negative | negative | mutagen | negative | negative | negative | negative |
| 9i | low risk | negative | positive | mutagen | negative | positive | positive | positive |
| 11 | low risk | negative | negative | mutagen | positive | positive | positive | positive |
| Compound | %ABS | Compound | %ABS |
|---|---|---|---|
| 5a | 84 | 6f | 74.11 |
| 5b | 84 | 6g | 78.64 |
| 5c | 84 | 6h | 78.64 |
| 5d | 79.46 | 6i | 78.64 |
| 5e | 84 | 9a | 82.16 |
| 5f | 79.46 | 9b | 82.16 |
| 5g | 84 | 9c | 82.16 |
| 5h | 84 | 9d | 77.63 |
| 5i | 84 | 9e | 82.16 |
| 6a | 78.64 | 9f | 77.63 |
| 6b | 78.64 | 9g | 82.16 |
| 6c | 78.64 | 9h | 82.16 |
| 6d | 74.11 | 9i | 82.16 |
| 6e | 78.64 | 11 | 67.20 |
| Compound | AAPH Inh. at 100 μΜ | LOX Inh. IC50 (μΜ) | COX-2 Inh. IC50 (μΜ) | COX-1 at 100 μM | %NO2− Release mol/mol at 100 μΜ | Albumin |
|---|---|---|---|---|---|---|
| 5a | 31.9% | no | nt | nt | nt | nt |
| 5b | no | 57.5 | nt | nt | nt | nt |
| 5c | 29% | no | nt | nt | nt | nt |
| 5d | no | no | nt | nt | nt | nt |
| 5e | no | no | nt | nt | nt | nt |
| 5f | 39% | 80 | nt | nt | nt | nt |
| 5g | 94.7% | no | nt | nt | nt | nt |
| 5h | 75.8% | 33 | nt | nt | nt | nt |
| 5i | 77% | no | nt | nt | nt | nt |
| 6a | 50.2% | 50 | 100 | nt | no | no |
| 6b | 94% | 60 | no | nt | 1.1% | no |
| 6c | 4% | 10 | 35 | no | 4% | no |
| 6d | 54% | no | no | nt | 0.2% | 10% |
| 6e | no | no | no | nt | 1.1% | 63% |
| 6f | no | 61 | no | nt | 0.3% | no |
| 6g | 89% | 44 | nt | nt | no | no |
| 6h | 97.7% | 62.5 | nt | nt | no | no |
| 6i | 28% | 60 | 80 | 16.7% | no | no |
| 9a | 91.6% | 3.5 | no | nt | 20% | no |
| 9b | 84% | no | no | nt | 1.2% | no |
| 9c | 73.3% | no | no | nt | 1.2% | no |
| 9d | 78% | 63 | no | nt | no | no |
| 9e | 67% | 65 | 8.25 | no | no | no |
| 9f | 97% | 70 | no | nt | no | no |
| 9g | 90% | 55.5 | no | nt | 3% | no |
| 9h | 94.5% | 10 | no | nt | no | no |
| 9i | no | no | 70 | 49% | 1% | no |
| 11 | 90% | no | no | nt | 11% | no |
| Cinnamic acid | 78% | 56 | - | - | - | - |
| Trolox | 93% | - | - | - | - | - |
| NDGA | - | 0.45 | - | - | - | - |
| Indomethacin | - | - | 2.12 | - | - | - |
| SC-560 | - | - | - | 100% | - | - |
| SNP | - | - | - | - | 53% | - |
| Acetylsalicylic acid | 31% |
| Compound | HeLa EC50 (μΜ) | MCF-7 EC50 (μΜ) |
|---|---|---|
| 6c | 83.04 | >100 |
| 6i | 36.69 | 38.34 |
| 9e | 55.46 | 58.53 |
| 9g | 46.59 | >100 |
| 9i | 55.96 | >100 |
| Cell Line | Compound 6i EC50 (μM) |
|---|---|
| HeLa | 36.69 |
| MCF-7 | 38.34 |
| U251 | 40.94 |
| U87 | 45.44 |
| MDA-MB-231 | 42.82 |
| MDA-MB-435 | 52.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsopka, I.-C.; Pontiki, E.; Sigala, I.; Nikolakaki, E.; Prousis, K.C.; Hadjipavlou-Litina, D. Design, Synthesis, Biological Evaluation, and In Silico Studies of Novel Multitarget Cinnamic Acid Hybrids. Molecules 2025, 30, 4582. https://doi.org/10.3390/molecules30234582
Tsopka I-C, Pontiki E, Sigala I, Nikolakaki E, Prousis KC, Hadjipavlou-Litina D. Design, Synthesis, Biological Evaluation, and In Silico Studies of Novel Multitarget Cinnamic Acid Hybrids. Molecules. 2025; 30(23):4582. https://doi.org/10.3390/molecules30234582
Chicago/Turabian StyleTsopka, Ioanna-Chrysoula, Eleni Pontiki, Ioanna Sigala, Eleni Nikolakaki, Kyriakos C. Prousis, and Dimitra Hadjipavlou-Litina. 2025. "Design, Synthesis, Biological Evaluation, and In Silico Studies of Novel Multitarget Cinnamic Acid Hybrids" Molecules 30, no. 23: 4582. https://doi.org/10.3390/molecules30234582
APA StyleTsopka, I.-C., Pontiki, E., Sigala, I., Nikolakaki, E., Prousis, K. C., & Hadjipavlou-Litina, D. (2025). Design, Synthesis, Biological Evaluation, and In Silico Studies of Novel Multitarget Cinnamic Acid Hybrids. Molecules, 30(23), 4582. https://doi.org/10.3390/molecules30234582







