Metabolism, a Blossoming Target for Small-Molecule Anticancer Drugs
Abstract
1. Introduction
2. Developing Therapeutic Agents Based on Metabolism
2.1. Aerobic Glycolysis
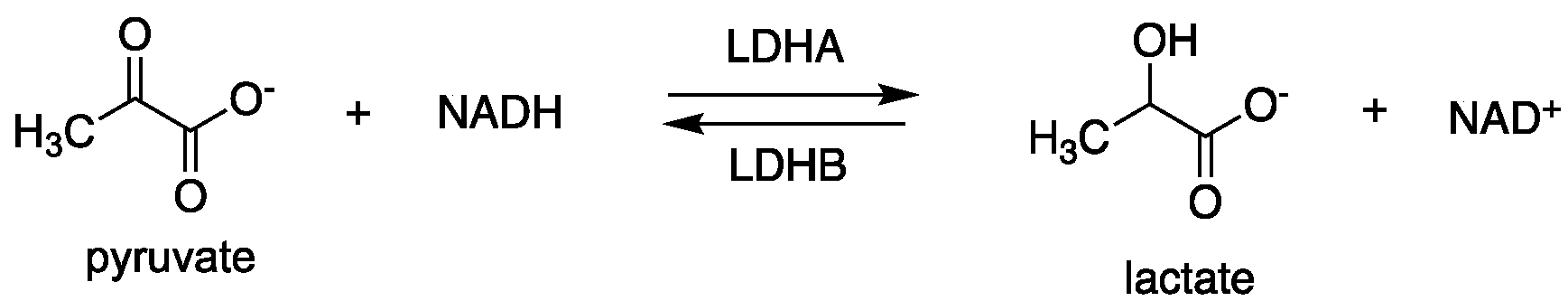
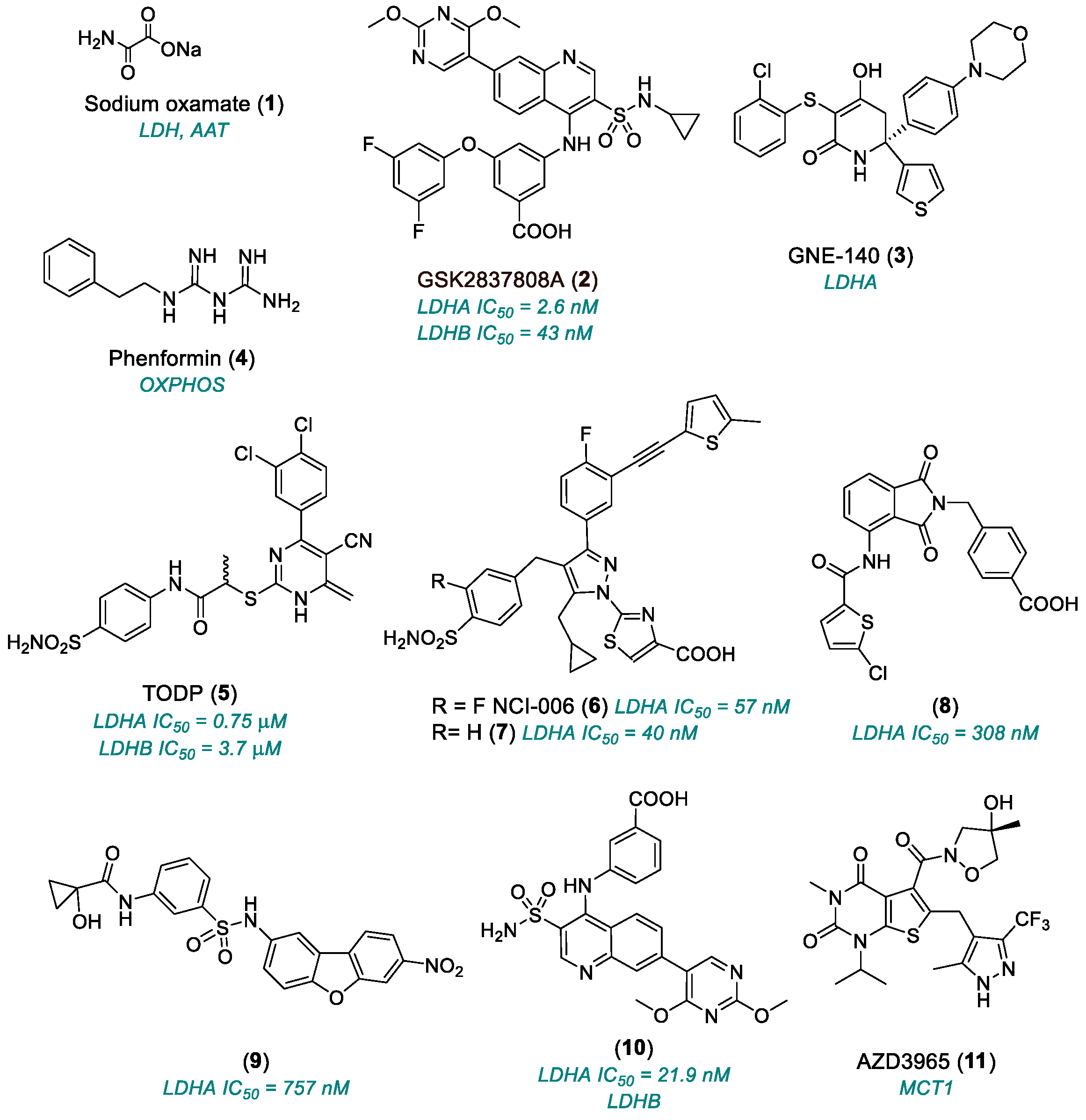
2.1.1. Lactate Dehydrogenase (LDH)
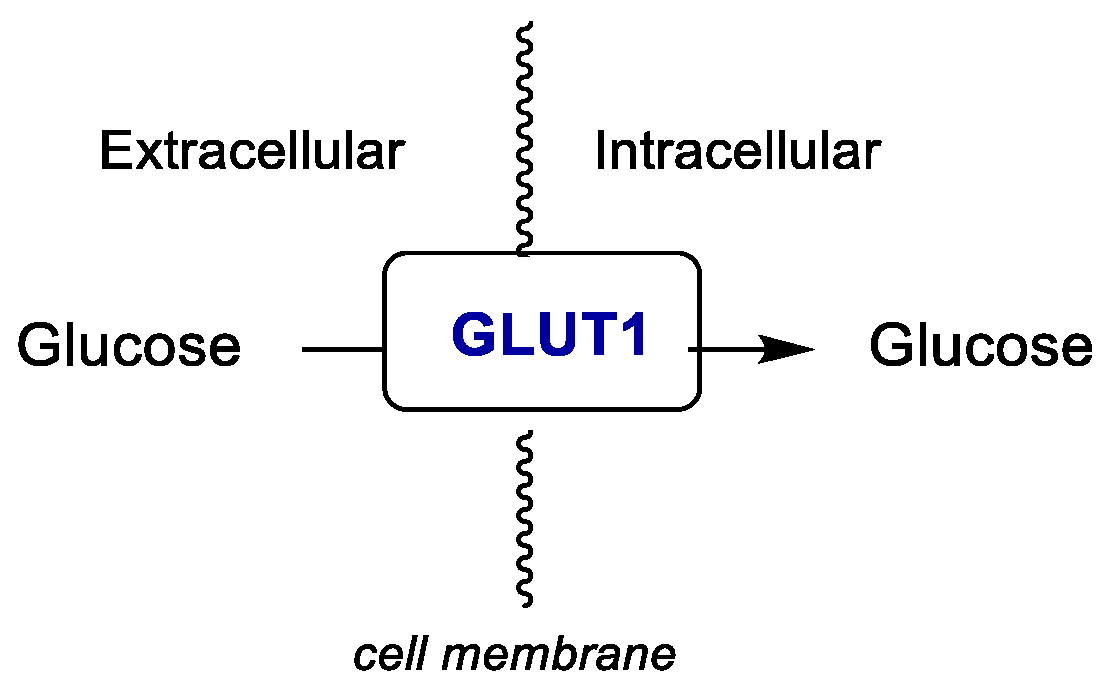
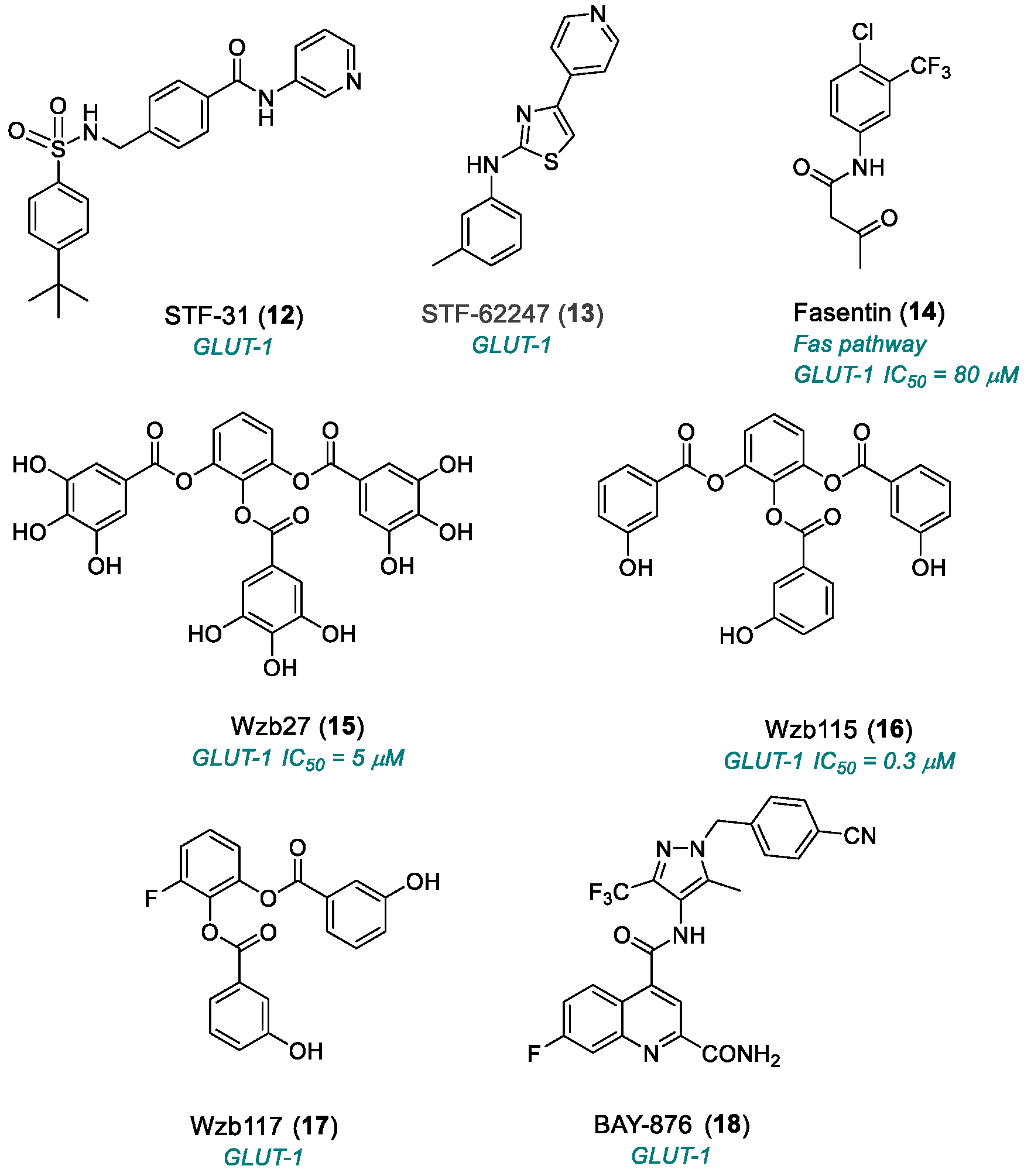
2.1.2. Glucose Transporter 1 (GLUT1)
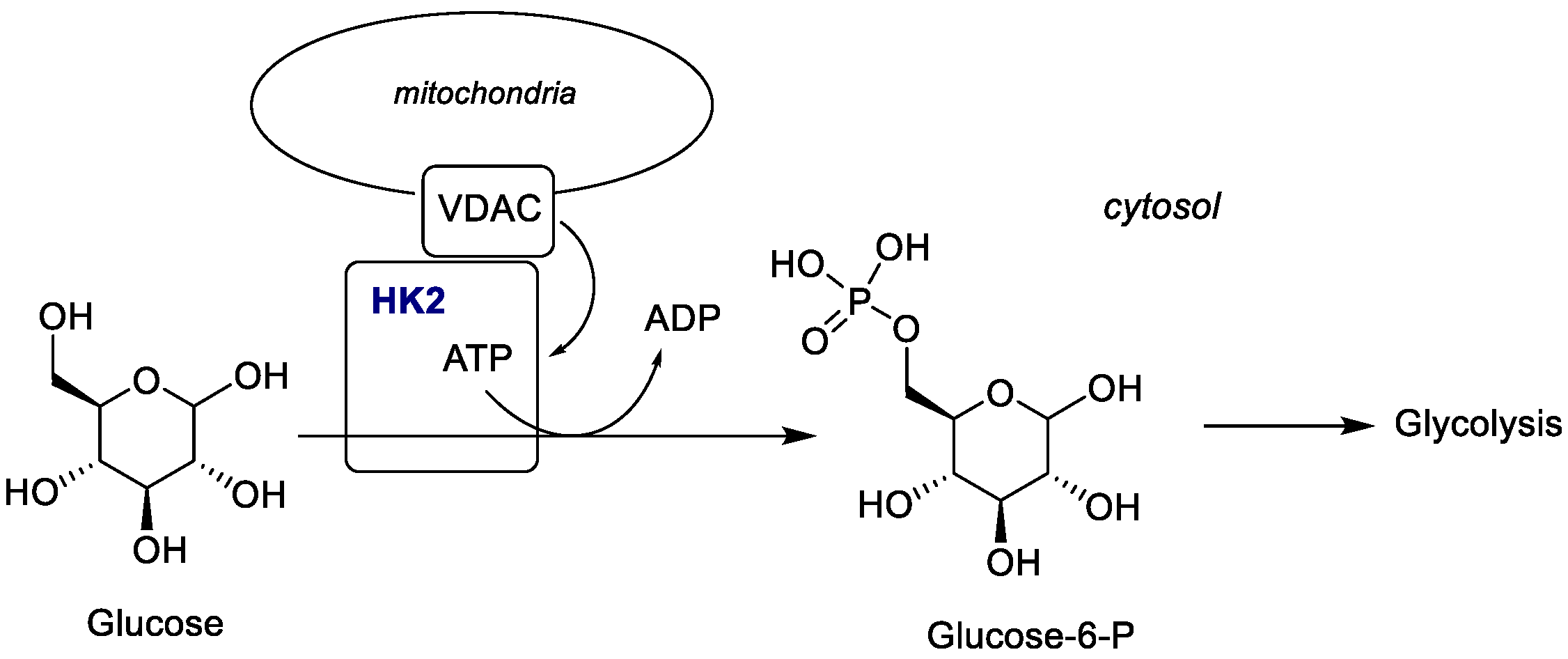
2.1.3. Hexokinase (HK)
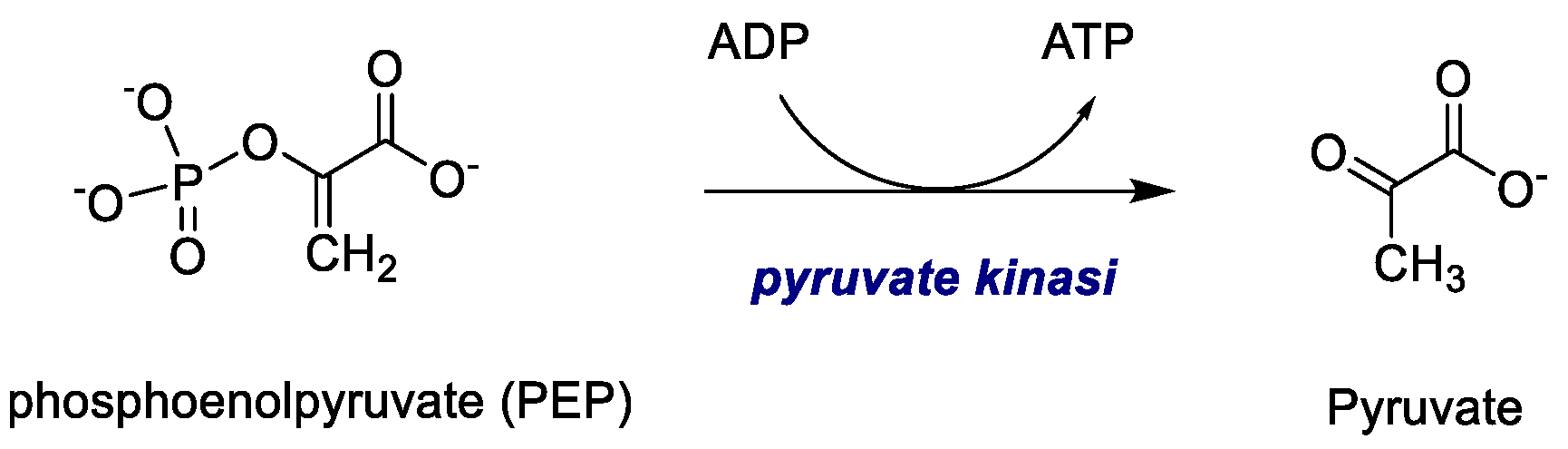
2.1.4. Pyruvate Kinase (PK)
| Name | Target | Year | Stage | Limitation | References |
|---|---|---|---|---|---|
| Sodium oxamate (1) | LDHA, AAT | 1965 | Preclinical studies | Not suitable for clinical utilization | [30,31,32] |
| GSK2837808A (2) | LDHA | 2013 | Preclinical studies | Poor oral bioavailability; very rapid clearance in vivo; very low plasma concentrations | [33,34,35,36] |
| GNE-140 (3) | LDHA | 2015 | Preclinical studies | Unfavorable pharmacokinetics; rapid clearance | [37,38,39,40,41] |
| TODP (5) | LDHA | 2013 | Preclinical studies | Unfavorable pharmacokinetics | [42] |
| NCI-006 (6) | LDHA | 2020 | Advanced preclinical candidate | Low cell permeability; high plasma binding; potential target saturation | [45] |
| (8), (9) | LDHA | 2020 | Preclinical Studies | Only enzymatic assay | [49] |
| (10) | Allosteric site LDH | 2020 | Preclinical studies | Only enzymatic assay; not selective for LDHA (also active on LDHB) | [49] |
| AZD3965 (11) | MCT1/2 | 2017 | Phase I (2020) | Reduced efficacy in glycolytic/resistant tumors; potential ocular and cardiac toxicities; limited to lymphomas; limited systemic pharmacology and long-term data | [52,53,54,55] |
| STF-31 (12) | GLUT1 | 2011 | Preclinical studies | No PK data available; necrotic cell death; complex mechanism | [62] |
| STF-62247 (13) | GLUT1 | 2011 | Preclinical studies | Undefined pharmacokinetics | [62,63] |
| Fasentin (14) | Fas pathway GLUT1 | 2006 | Preclinical studies | Undefined pharmacokinetics | [66] |
| WZB27 (15) | GLUT1 | 2010 | Preclinical studies | Moderate potency; no PK studies | [68,69] |
| WZB115 (16) | GLUT1 | 2010 | Preclinical studies | Moderate potency; no PK studies | [68,69] |
| WZB117 (17) | GLUT1 | 2012 | Preclinical studies | Limited selectivity; no PK studies; in vivo instability | [70,71] |
| BAY-876 (18) | GLUT1 | 2016 | Preclinical studies | Limited cellular activity; effects on normal cells; tumor metabolic adaptation | [72] |
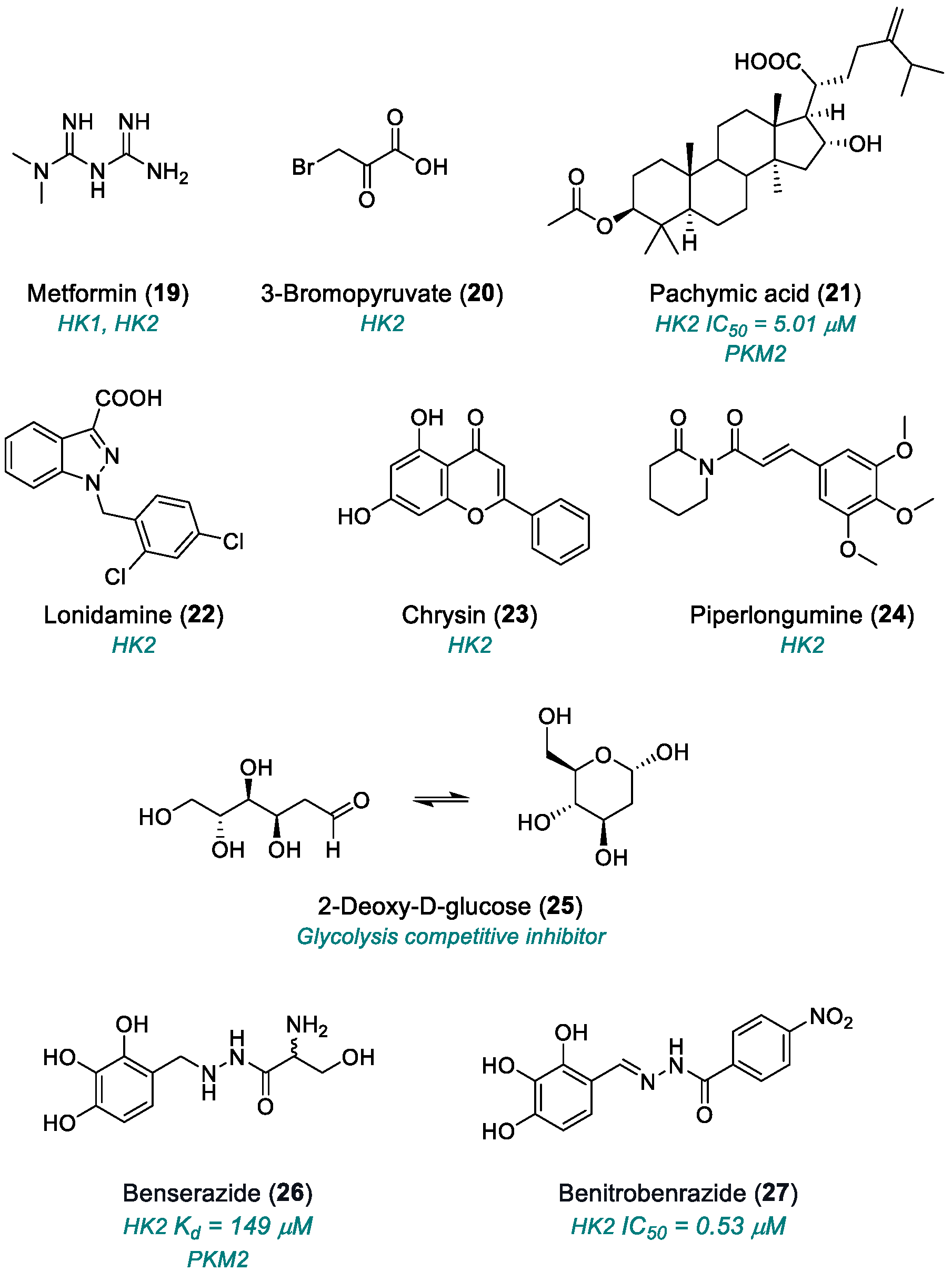
| Metformin (19) | HK1, HK2 | 2013 | Preclinical studies | Cytotoxic effect | [82,83] |
| ]3-BrPA (20) | HK2 | 2009 | Preclinical studies | Systemic toxicity; non-selective reactivity; acidic pH dependence; metabolic adaptation; | [86] |
| ]Pachymic acid (21) | HK2, PKM2 | 2015 | Preclinical studies | Poor in vivo bioavailability; narrow therapeutic window; moderate potency; multiple and partly selective mechanisms; lack of systematic toxicity data | [87,88] |
| Lonidamine (22) | HK2, MCT | 1981 | Fase II–III (only in combination) | Mitochondrial toxicity; low oral bioavailability | [89,90,91,92] |
| Chrysin (23) | HK2, PI3K/Akt, NF-κB | 2010 | Preclinical studies | Poor oral bioavailability | [93,94,95,96,97] |
| ]Piperlongumine (24) | PI3K/Akt/mTOR e HIF-1α | 2011 | Preclinical studies | Low bioavailability; poor metabolic stability | [98] |
| 2-DG (25) | Glycolysis competitive inhibitor | 2019 | Phase II completed | Systemic toxicity; metabolic compensation | [99,100,101,102,103,104] |
| Benserazide (26) | HK2, PKM2 | 2017 | Preclinical studies | Off-label repositioning; neurological effects | [113] |
| Benitrobenrazide (27) | HK2 | 2021 | Advanced preclinical studies | Aggregation tendency; poor ADME/Tox characterization; activity on minor HK isoenzymes | [115] |
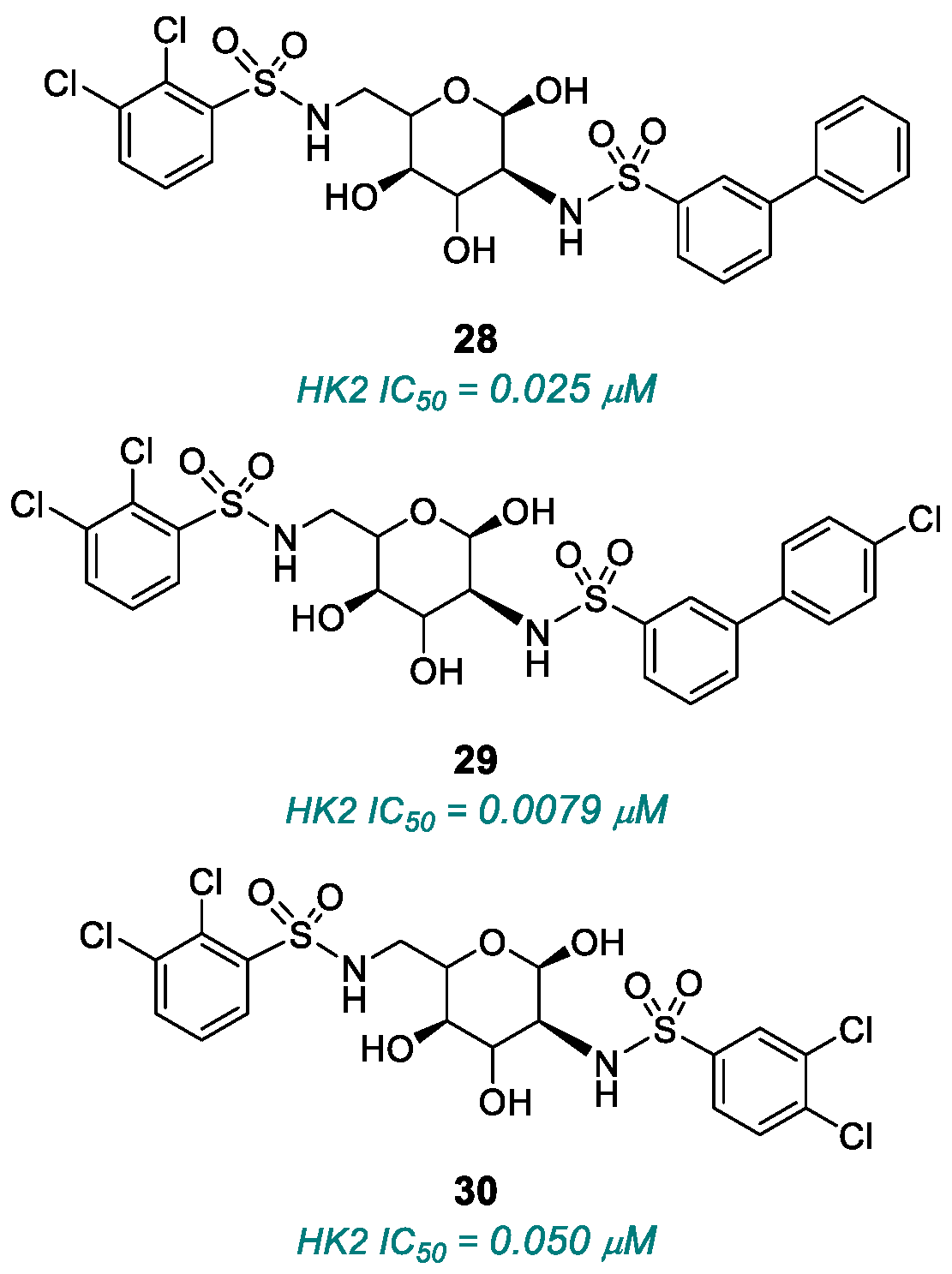
| (28), (29), (30) | HK2 | 2015 | Preclinical studies | Micromolar potency; incomplete selectivity | [117] |
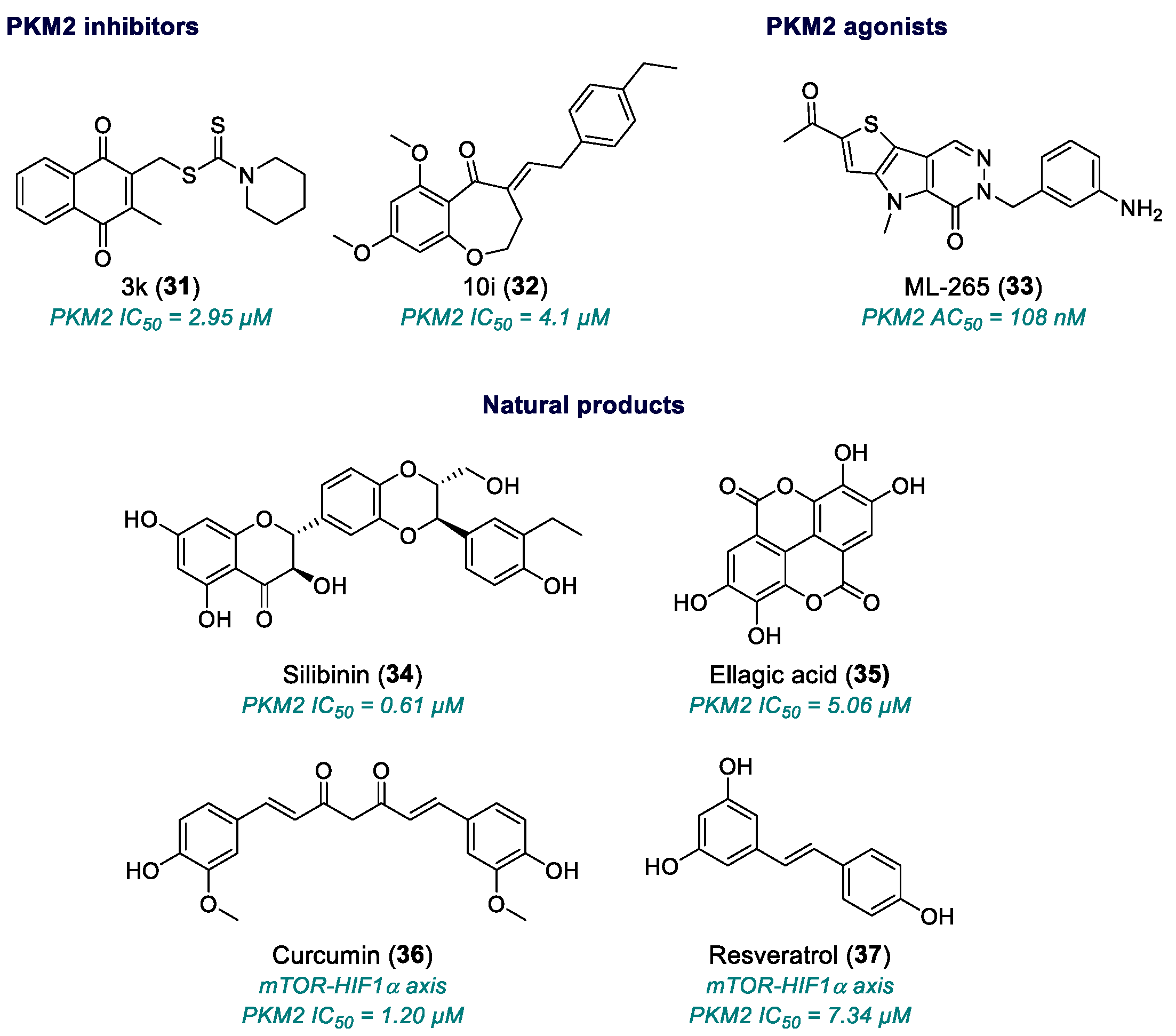
| 3k (31) | PKM2 | 2017 | Preclinical studies | Low potency; relative selectivity; no full pharmacological development | [126] |
| 10i (32) | PKM2 | 2019 | Preclinical studies | Neuroinflammatory/ischemic application only; no published antitumor validation | [128] |
| ML-265 (33) | PKM2 | 2012 | Advanced Preclinical studies | Variable cell-based potency; poor solubility; need for optimized formulation; incomplete toxicological data | [131,132,133] |
| Silibinin (34) | PKM2 | 2003 | Phase II | Effective only at micromolar range | [134,135,136,137,138,139] |
| Ellagic acid (35) | PKM2 | 1996 | Small phase I/II studies (in combination) | Effective only at micromolar range | [140,141,142,143,144,145] |
| Curcumin (36) | mTOR-HIF1α axis | 1990 | Phase I/II completed, few phase III studies | Low bioavailability; high dosing; standardization issues | [146,147] |
| Resveratrol (37) | PI3K/Akt/mTOR, HIF1α | 1997 | Phase I/II | Very low bioavailability; rapid metabolism; high doses needed | [148,149,150,151,152] |
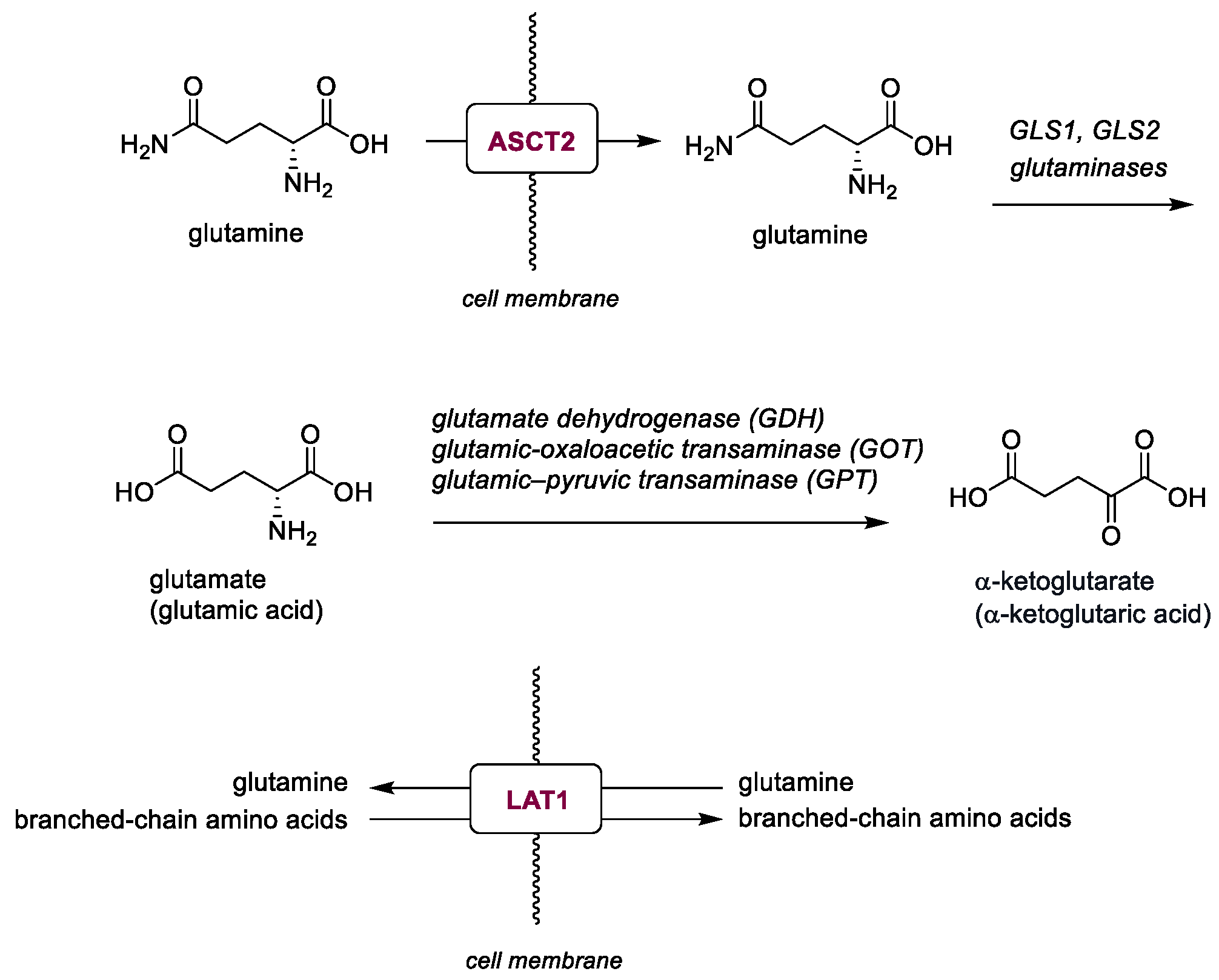
2.2. Glutamine Metabolisms
| Name | Target | Year | Stage | Limitation | References |
|---|---|---|---|---|---|
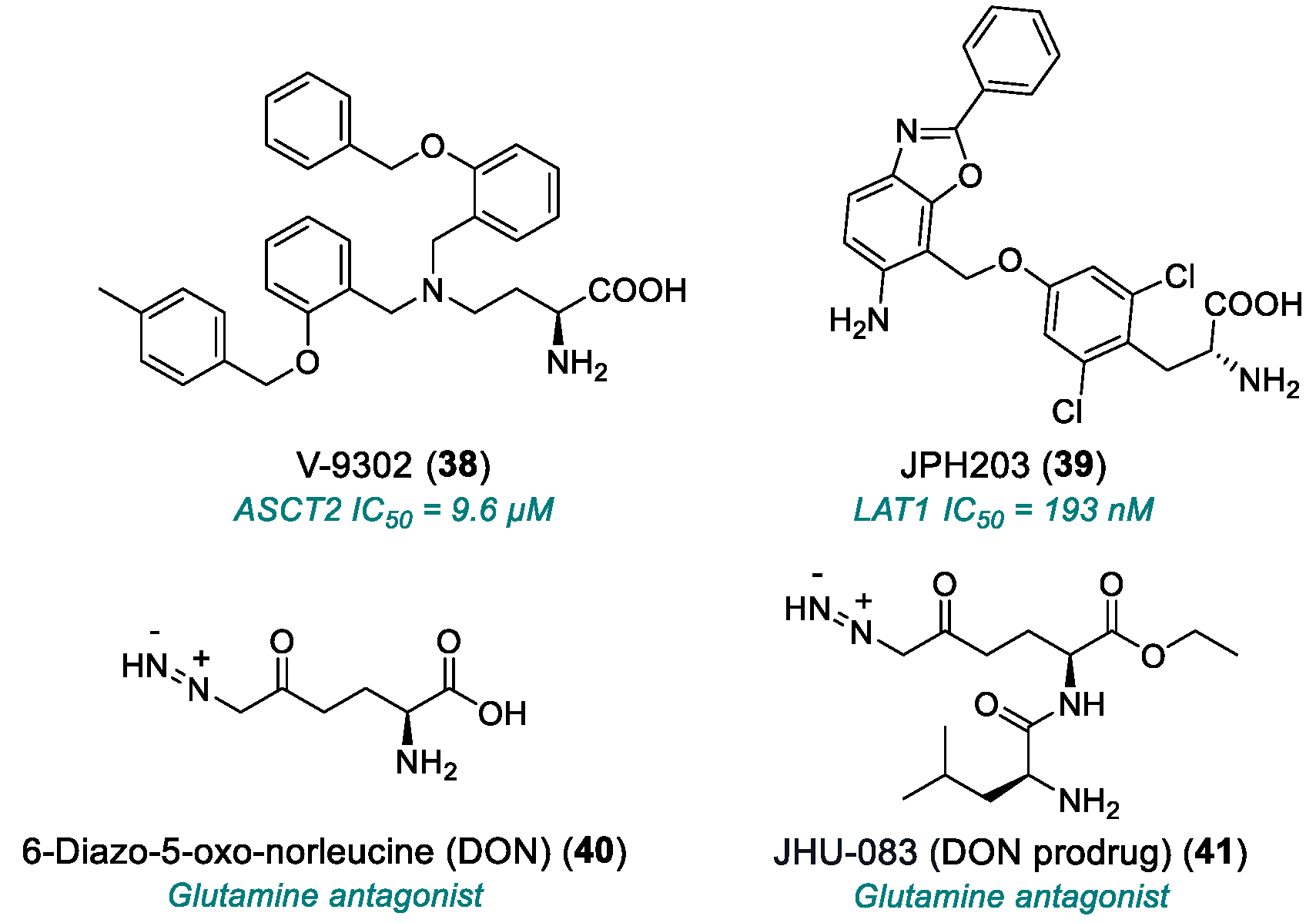
| V-9302 (38) | ASCT2 | 2016 | Preclinical studies | Effective only at micromolar range | [153,154,155,156,157] |
| JPH203 (39) | LAT1 | 2018 | Phase I completed | Hepatotoxicity; limited PK data | [158] |
| DON (40) | Glutamine antagonist | 1979 | Phase I/II completed | Weak in vitro efficacy; poo3,172–172r in vivo tolerability; severe GI toxicity | [159,160,161,162] |
| JHU-083 (41) | Glutamine antagonist (DON prodrug) | 2016 | Phase I/II | Toxicity at high doses | [163,164,165] |
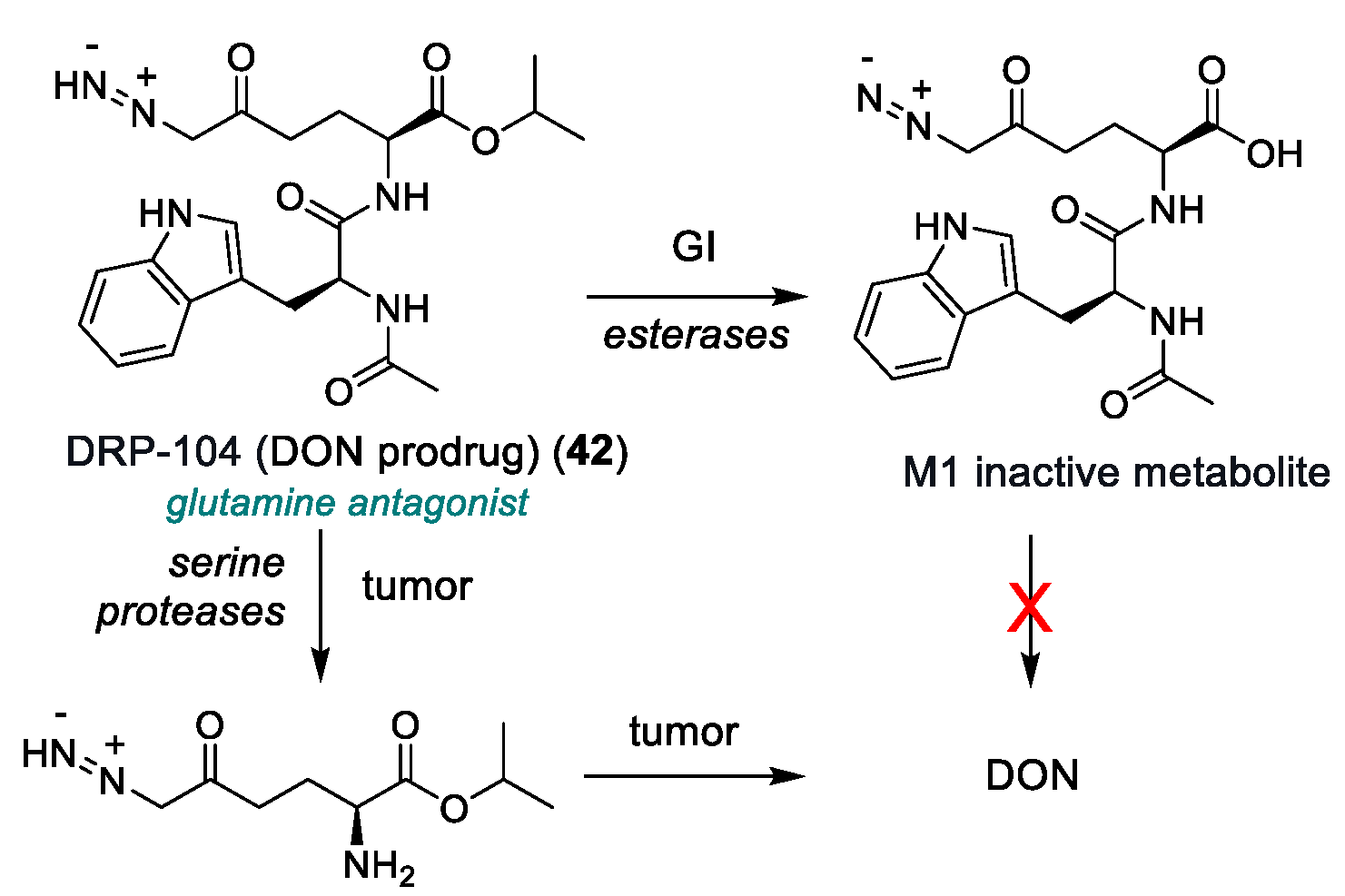
| DRP-104 (42) | Glutamine antagonist (DON prodrug) | 2022 | Phase I/II | Residual toxicity (GI and systemic); narrow therapeutic window; poor PK data | [166,167] |
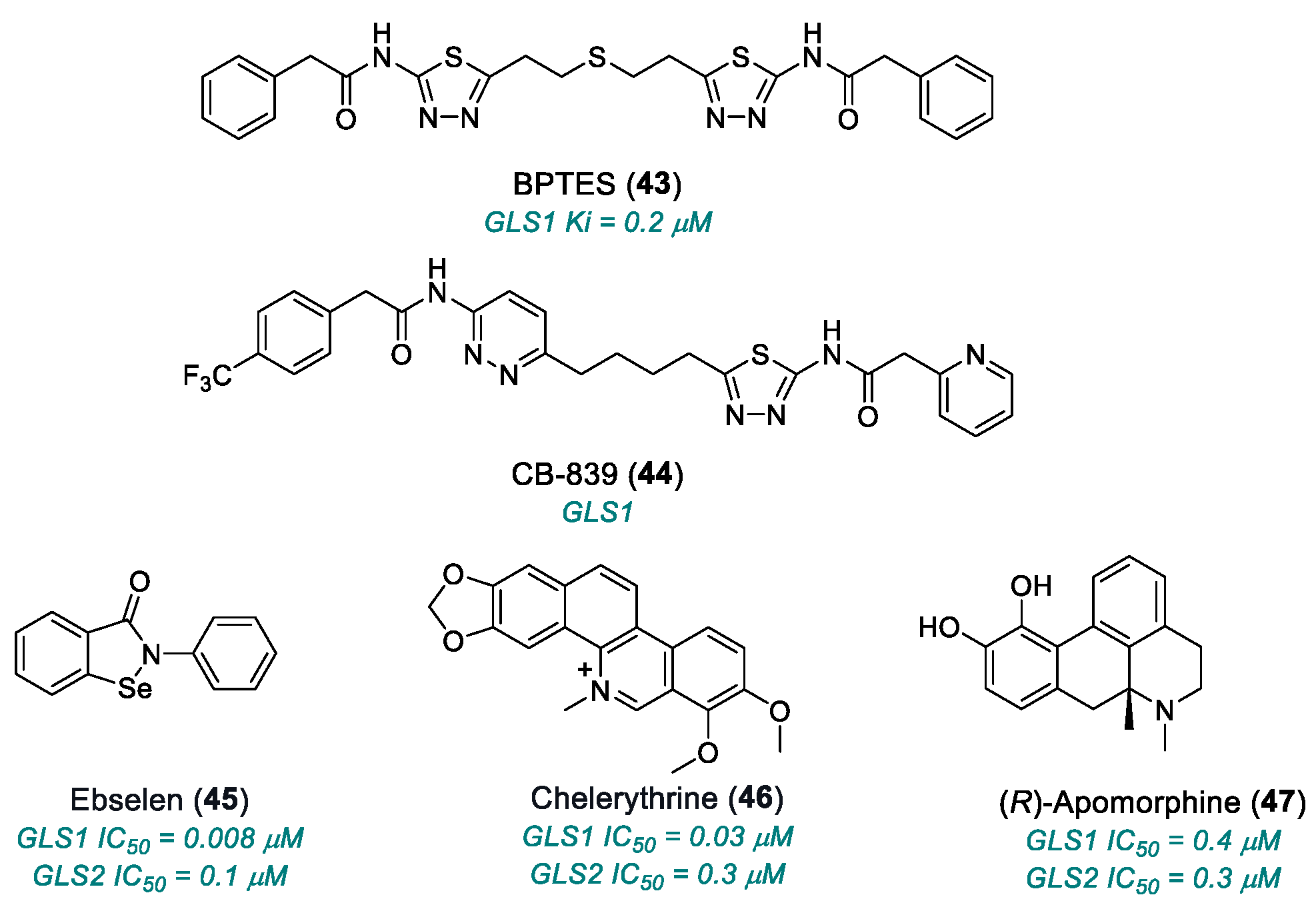
| BPTES (43) | GLS1 | 2010 | Preclinical studies | Limited solubility and stability; micromolar activity | [168,169,170,171] |
| CB-839 (telaglenastat) (44) | GLS1 | 2014 | Phase I completed | Non-universal efficacy; moderate toxicity | [170,171] |
| ebselen (45) | GLS1 | 2015 | Preclinical studies | Multi-target, redox reactive; off-target risk and mitochondrial toxicity | [162,172,173,174] |
| chelerythrine chloride (46) | GLS1 | 2015 | Preclinical studies | Significant inhibition but with off-target cytotoxic effects | [162,172,173,174] |
| (R)-apomorphine hydrochloride (47) | GLS1, GLS2 | 2015 | Preclinical studies | Low selectivity | [162,172,173,174] |
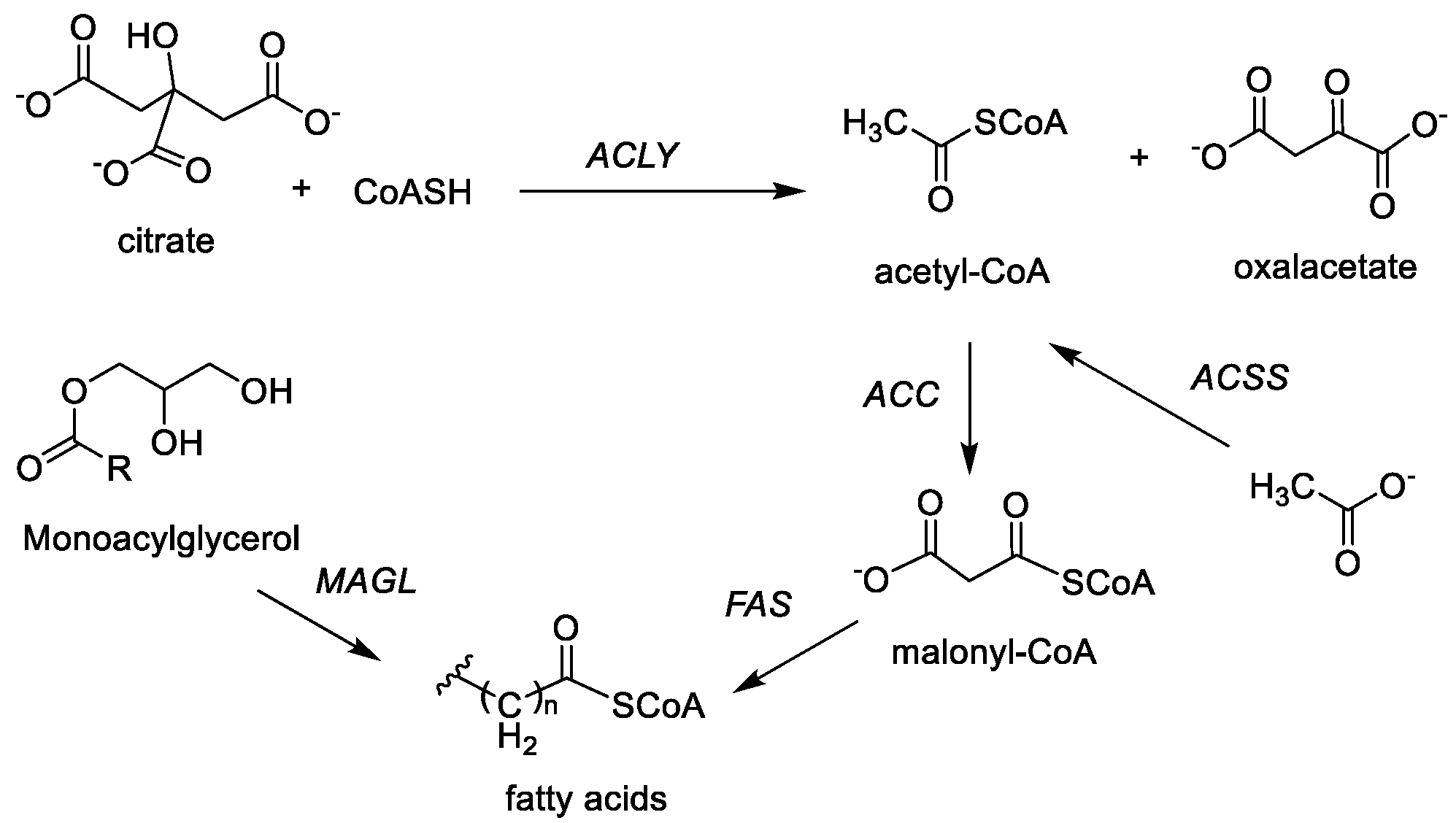
2.3. Fatty Acid Synthesis
| Name | Target | Year | Stage | Limitation | References |
|---|---|---|---|---|---|
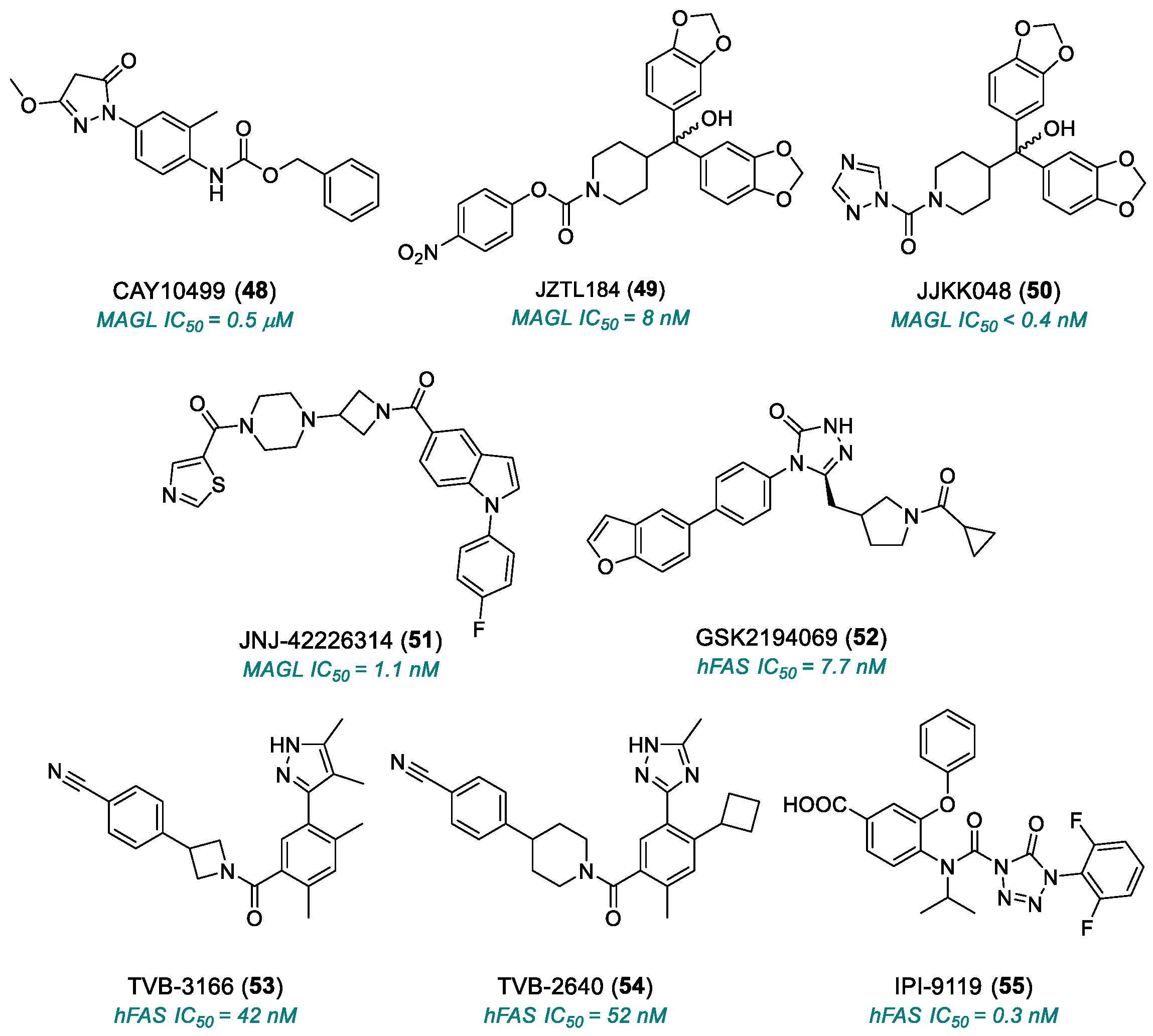
| CAY10499 (48) | MAGL | 2008 | Preclinical studies | Potential toxicity from normal lipid metabolism (liver, heart); poorly characterized bioavailability/toxicity; endocannabinoid accumulation | [180,181] |
| JZL184 (49) | MAGL | 2009 | Preclinical studies | Off-target effects at high doses; poorly characterized bioavailability/toxicity; endocannabinoid accumulation | [182,183,184] |
| JJKK-048 (50) | MAGL | 2013 | Preclinical studies | Off-target effects at high doses; poorly characterized bioavailability/toxicity; endocannabinoid accumulation | [186,187,188] |
| JNJ-42226314 (51) | MAGL | 2020 | Preclinical studies | Endocannabinoid accumulation | [189] |
| GSK2194069 (52) | FASN | 2011 | Preclinical studies | Limited oral bioavailability; metabolic and lipid side effects | [191,192] |
| TVB-3166 (53) | FASN | 2015 | Advanced preclinical studies | Metabolic and lipid side effects | [193] |
| TVB-2640 (54) | FASN | 2017 | Phase II | Risk of long-term resistance | [194,195] |
| IPI-9119 (55) | FASN | 2020 | Preclinical studies | Irreversible inhibitor | [196] |
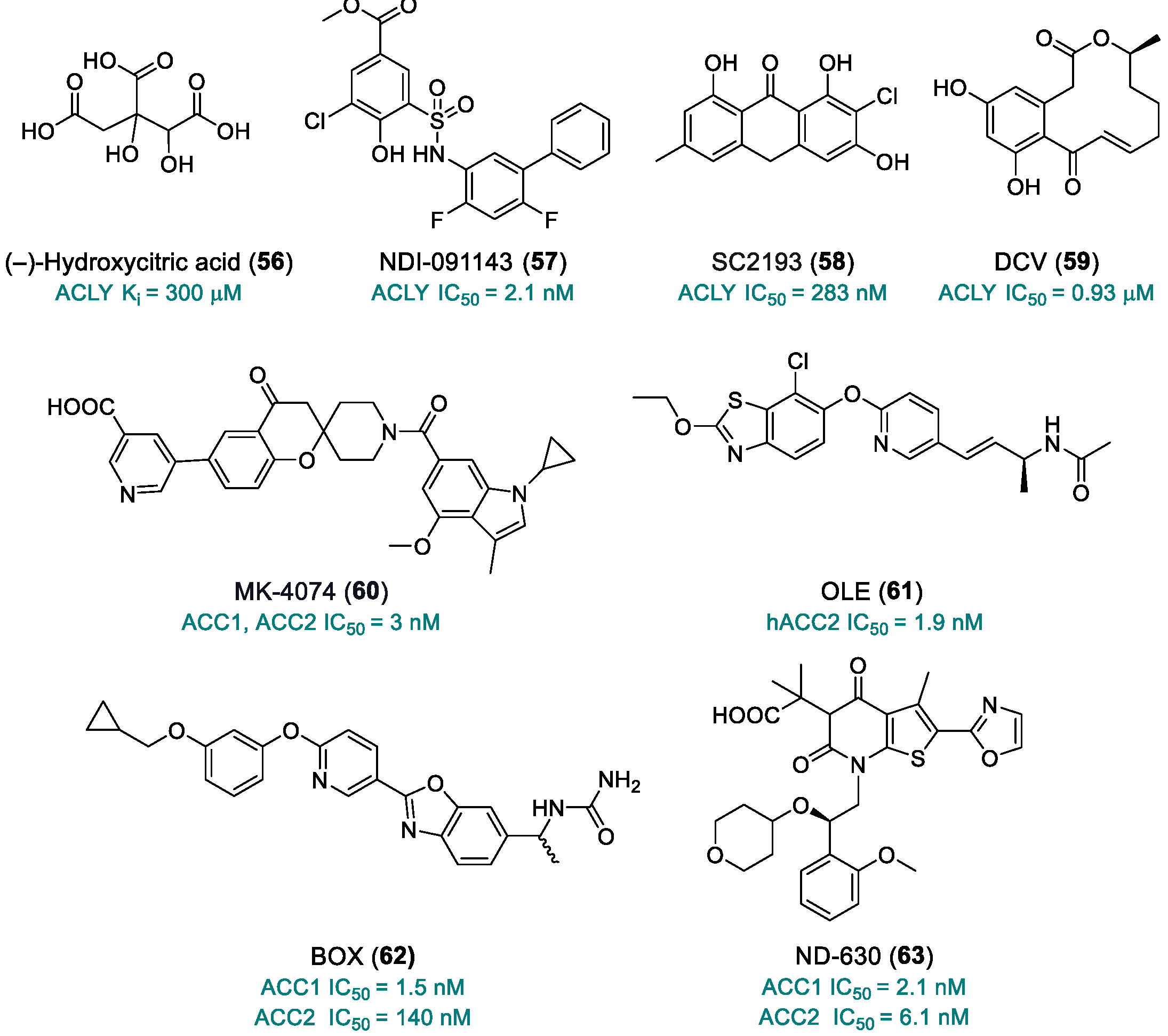
| (–)-Hydroxycitric acid (56) | ACLY | 2012 | Preclinical studies | Low bioavailability | [201] |
| NDI-091143 (57) | ACLY | 2019 | Preclinical studies | No PK studies; limited toxicity data | [202,203,204] |
| SC2193 (58) | ACLY | 2017 | Preclinical studies | No PK studies; limited toxicity data | [205] |
| 10,11-Dehydrocurvularin, DCV (59) | ACLY | 2015 | Preclinical studies | Poorly defined toxicity; instability; multi-target activity | [206,207] |
| MK-4074 (60) | ACC1, ACC2 | 2017 | Preclinical studies | No PK studies; limited toxicity data | [211] |
| OLE (61) | hACC2 | 2018 | Preclinical studies | No PK studies; limited toxicity data | [212] |
| BOX (62) | ACC1 | 2019 | Preclinical studies | No PK studies; limited toxicity data | [213] |
| ND-630 (63) | ACC1, ACC2 | 2016 | Preclinical studies | No PK studies; limited toxicity data | [214] |
2.4. Structural Considerations of Metabolic Targets
3. Perspectives and Emerging Strategies in Tumor Metabolism
3.1. Metabolic Crosstalk and Compensatory Mechanisms
3.2. Emerging Technologies: PROTACs and Targeted Degradation of Metabolic Proteins
3.3. Precision Metabolic Oncology: New Therapeutic Frontiers
3.4. Concluding Remark
4. Summary and Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AAT | aspartate aminotransferase |
| ACC | acetyl-coa carboxylase |
| ACLY | human atp citrate lyase |
| AIF | apoptosis-inducing factor |
| ASCT2 | alanine, serine, cysteine transporter 2 |
| ATP | adenosine triphosphate |
| BBB | blood–brain barrier |
| CDKs | cyclin-dependent kinases |
| CRPC | castration-resistant prostate cancer |
| DLBCL | diffuse large b cell lymphoma |
| DNMT | DNA methyltransferase |
| ECM | extracellular matrix |
| FADH | flavin adenine dinucleotide |
| FAS | fatty acid synthase |
| FASN | fatty acid synthase |
| G6P | glucose-6-phosphate |
| GLS1 | glutaminase 1 |
| GLUT | glucose transporter |
| GLUT1 | glucose transporter type 1 |
| GLUT1DS | glucose transporter type 1 deficiency syndrome |
| GOT | glutamic-oxaloacetic transaminase |
| GPT | glutamic–pyruvic transaminase |
| HCC | hepatocellular carcinoma |
| HK | hexokinase |
| HKla | histone lysine lactylation |
| HNSCC | head and neck squamous cell carcinoma |
| LAT1 | l-type amino acid transporter 1 |
| LDH | lactate dehydrogenase |
| LDHA | lactate dehydrogenase type a |
| LDHB | lactate dehydrogenase type b |
| MAGL | monoacylglycerol lipase |
| MCT1 | monocarboxylate transporter 1 |
| NADH | nicotinamide adenine dinucleotide |
| NHL | non-Hodgkin lymphomas |
| NSCLC | non-small-cell lung cancer |
| OMM | outer mitochondrial membrane |
| OXPHOS | oxidative phosphorylation |
| PK | pyruvate kinase |
| PKM1 | pyruvate kinase m1 |
| PKM2 | pyruvate kinase m2 |
| RCCs | renal cell carcinomas |
| ROS | reactive oxygen species |
| SAR | structure-activity relationship |
| TCA | tricarboxylic acid |
| TMJOA | temporomandibular joint osteoarthritis |
| TNBC | in triple-negative breast cancer |
| VDAC | voltage-dependent anion channels |
| VHL | von Hippel–Lindau |
References
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Martínez-Reyes, I.; Chandel, N.S. Cancer Metabolism: Looking Forward. Nat. Rev. Cancer 2021, 21, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Ngo, D.C.; Ververis, K.; Tortorella, S.M.; Karagiannis, T.C. Introduction to the Molecular Basis of Cancer Metabolism and the Warburg Effect. Mol. Biol. Rep. 2015, 42, 819–823. [Google Scholar] [CrossRef]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic Glycolysis: Meeting the Metabolic Requirements of Cell Proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef]
- Kim, J.W.; Dang, C.V. Cancer’s Molecular Sweet Tooth and the Warburg Effect. Cancer Res. 2006, 66, 8927–8930. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The Biology of Cancer: Metabolic Reprogramming Fuels Cell Growth and Proliferation. Cell Metab. 2008, 7, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Liberti, V.M.; Locasale, J.W. The Warburg effect: How does it benefit cancer cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, L.; Supuran, T.C.; Alfarouk, O.K. The Warburg effect and the hallmarks of cancer. Anti-Cancer Agents Med. Chem. 2017, 17, 164–170. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Shaw, R.J. Glucose metabolism and cancer. Curr. Opin. Cell Biol. 2006, 18, 598–608. [Google Scholar] [CrossRef]
- Gillies, R.J.; Robey, I.; Gatenby, R.A. Causes and consequences of increased glucose metabolism of cancers. J. Nucl. Med. 2008, 49, 24S–42S. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of cancer: New dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R. V Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef]
- Beloribi-Djefaflia, S.; Vasseur, S.; Guillaumond, F. Lipid metabolic reprogramming in cancer cells. Oncogenesis 2016, 5, e189. [Google Scholar] [CrossRef]
- Montesdeoca, N.; López, M.; Ariza, X.; Herrero, L.; Makowski, K. Inhibitors of lipogenic enzymes as a potential therapy against cancer. FASEB J. 2020, 34, 11355–11381. [Google Scholar] [CrossRef]
- Stine, Z.E.; Schug, Z.T.; Salvino, J.M.; Dang, C.V. Targeting cancer metabolism in the era of precision oncology. Nat. Rev. Drug Discov. 2022, 21, 141–162. [Google Scholar] [CrossRef]
- Masci, D.; Puxeddu, M.; Silvestri, R.; La Regina, G. Metabolic rewiring in cancer: Small molecule inhibitors in colorectal cancer therapy. Molecules 2024, 29, 2110. [Google Scholar] [CrossRef] [PubMed]
- Rattigan, Y.; Patel, B.; Ackerstaff, E.; Sukenick, G.; Koutcher, J.; Glod, J.; Banerjee, D. Lactate is a mediator of metabolic cooperation between stromal carcinoma associated fibroblasts and, glycolytic tumor cells in the tumor microenvironment. Exp. Cell Res. 2012, 318, 326–335. [Google Scholar] [CrossRef]
- Ristow, M. Oxidative metabolism in cancer growth. Curr. Opin. Clin. Nutr. Metab. Care 2006, 9, 339–345. [Google Scholar] [CrossRef]
- Fantin, V.R.; St-Pierre, J.; Philip, L. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 2006, 9, 425–434. [Google Scholar] [CrossRef]
- Di Magno, L.; Coluccia, A.; Bufano, M.; Ripa, S.; La Regina, G.; Nalli, M.; Di Pastena, F.; Canettieri, G.; Silvestri, R.; Frati, L. Discovery of novel human lactate dehydrogenase inhibitors: Structure-based virtual screening studies and biological assessment. Eur. J. Med. Chem. 2022, 240, 114605. [Google Scholar] [CrossRef]
- Zhou, Y.; Tao, P.; Wang, M.; Xu, P.; Lu, W.; Lei, P.; You, Q. Development of novel human lactate dehydrogenase A inhibitors: High-throughput screening, synthesis, and biological evaluations. Eur. J. Med. Chem. 2019, 177, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Bononi, G.; Di Bussolo, V.; Tuccinardi, T.; Minutolo, F.; Granchi, C. A patent review of lactate dehydrogenase inhibitors (2014–present). Expert Opin. Ther. Pat. 2024, 34, 1121–1135. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.L.; He, Y.; Tam, K.Y. Targeting cancer metabolism to develop human lactate dehydrogenase (hLDH)5 inhibitors. Drug Discov. Today 2018, 23, 1407–1415. [Google Scholar] [CrossRef] [PubMed]
- Farhana, A.; Lappin, S.L. Biochemistry, Lactate Dehydrogenase; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Dang, C.V.; Semenza, G.L. Oncogenic alterations of metabolism. Trends Biochem. Sci. 1999, 24, 68–72. [Google Scholar] [CrossRef]
- Han, J.H.; Lee, E.J.; Park, W.; Ha, K.T.; Chung, H.S. Natural compounds as lactate dehydrogenase inhibitors: Potential therapeutics for lactate dehydrogenase inhibitors-related diseases. Front. Pharmacol. 2023, 14, 1275000. [Google Scholar] [CrossRef]
- Granchi, C.; Bertini, S.; Macchia, M.; Minutolo, F. Inhibitors of lactate dehydrogenase isoforms and their therapeutic potentials. Curr. Med. Chem. 2010, 17, 672–697. [Google Scholar] [CrossRef]
- Thornburg, J.M.; Nelson, K.K.; Clem, B.F.; Lane, A.N.; Arumugam, S.; Simmons, A.; Eaton, J.W.; Telang, S.; Chesney, S.J. Targeting aspartate aminotransferase in breast cancer. Breast Cancer Res. 2008, 10, R84. [Google Scholar] [CrossRef]
- Fiume, L.; Manerba, M.; Vettraino, M.; Di Stefano, G. Impairment of aerobic glycolysis by inhibitors of lactic dehydrogenase hinders the growth of human hepatocellular carcinoma cell lines. Pharmacology 2010, 86, 157–162. [Google Scholar] [CrossRef]
- Goldberg, E.; Nitowsky, H.; Colowick, S. The role of glycolysis in the growth of tumor cells The basis of glucose toxicity in oxamate-treated cultured cells. J. Biol. Chem. 1965, 240, 2791–2796. [Google Scholar] [CrossRef]
- Billiard, J.; Dennison, J.B.; Briand, J.; Annan, R.S.; Chai, D.; Colon, M.; Dodson, C.S.; Gilbert, S.A.; Greshock, J.; Jing, J.; et al. Quinoline 3-sulfonamides inhibit lactate dehydrogenase A and reverse aerobic glycolysis in cancer cells. Cancer Metab. 2013, 1, 19. [Google Scholar] [CrossRef]
- Cisewski, S.E.; Zhang, L.; Kuo, J.; Wright, G.J.; Wu, Y.; Kern, M.J.; Yao, H. Thee ffects of oxygen level and glucose concentration on the metabolism of porcine TMJ disc cells. Osteoarthr. Cartil. 2015, 23, 1790–1796. [Google Scholar] [CrossRef]
- Li, H.M.; Guo, H.L.; Xu, C.; Liu, L.; Hu, S.Y.; Hu, Z.H.; Jiang, H.H.; He, Y.M.; Li, Y.J.; Ke, J.; et al. Inhibition of glycolysis by targeting lactate dehydrogenase A facilitates hyaluronan synthase 2 synthesis in synovial fibroblasts of temporomandibular joint osteoarthritis. Bone 2020, 141, 115584. [Google Scholar] [CrossRef]
- Wong, N.; De Melo, J.; Tang, D. PKM2, a central point of regulation in cancer metabolism. Int. J. Cell Biol. 2013, 2013, 242513. [Google Scholar] [CrossRef] [PubMed]
- Boudreau, A.; Purkey, H.E.; Hitz, A.; Robarge, K.; Peterson, D.; Labadie, S.; Kwong, M.; Hong, R.; Gao, M.; Del Nagro, C.; et al. Metabolic plasticity underpins innate and acquired resistance to LDHA inhibition. Nat. Chem. Biol. 2016, 12, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Abcam. Oxidative Phosphorylation Pathway. Available online: https://www.abcam.com/en-us/technical-resources/pathways/oxidative-phosphorylation-pathway (accessed on 1 November 2024).
- Zhao, S.S.; Liu, J.; Wu, Q.C.; Zhou, X.L. Lactate regulates pathological cardiac hypertrophy via histone lactylation modification. J. Cell Mol. Med. 2024, 28, e70022. [Google Scholar] [CrossRef] [PubMed]
- Zhai, M.; Cui, S.; Li, L.; Cheng, C.; Zhang, Z.; Liu, J.; Wei, F. Mechanical force modulates alveolar bone marrow mesenchymal cells characteristics for bone remodeling during orthodontic tooth movement through lactate production. Cells 2022, 11, 3724. [Google Scholar] [CrossRef]
- Ma, W.; Ao, S.; Zhou, J.; Li, J.; Liang, X.; Yang, X.; Zhang, H.; Liu, B.; Tang, W.; Liu, H.; et al. Methylsulfonylmethane protects against lethal dose MRSA-induced sepsis through promoting M2 macrophage polarization. Mol. Immunol. 2022, 146, 69–77. [Google Scholar] [CrossRef]
- Dragovich, P.S.; Fauber, B.P.; Corson, L.B.; Ding, C.Z.; Eigenbrot, C.; Ge, H.; Giannetti, A.M.; Hunsaker, T.; Labadie, S.; Liu, Y.; et al. Identification of substituted 2-thio-6-oxo-1,6-dihydropyrimidines as inhibitors of human lactate dehydrogenase. Bioorg. Med. Chem. Lett. 2013, 23, 3186–3194. [Google Scholar] [CrossRef]
- Fauber, B.P.; Dragovich, P.S.; Chen, J.; Corson, L.B.; Ding, C.Z.; Eigenbrot, C.; Giannetti, A.M.; Hunsaker, T.; Labadie, S.; Liu, Y.; et al. Identification of 2-amino-5-aryl-pyrazines as inhibitors of human lactate dehydrogenase. Bioorg. Med. Chem. Lett. 2013, 23, 5533–5539. [Google Scholar] [CrossRef]
- Rai, G.; Brimacombe, K.R.; Mott, B.T.; Urban, D.J.; Hu, X.; Yang, S.M.; Lee, T.D.; Cheff, D.M.; Kouznetsova, J.; Benavides, G.A.; et al. Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH). J. Med. Chem. 2017, 60, 9184–9204. [Google Scholar] [CrossRef]
- Rai, G.; Urban, D.J.; Mott, B.T.; Hu, X.; Yang, S.M.; Benavides, G.A.; Johnson, M.S.; Squadrito, G.L.; Brimacombe, K.R.; Lee, T.D.; et al. Pyrazole-Based Lactate Dehydrogenase Inhibitors with Optimized Cell Activity and Pharmacokinetic Properties. J. Med. Chem. 2020, 63, 10984–11011. [Google Scholar] [CrossRef]
- Ludwig, J.A. Ewing sarcoma: Historical perspectives, current state-of-the-art, and opportunities for targeted therapy in the future. Curr. Opin. Oncol. 2008, 20, 412–418. [Google Scholar] [CrossRef]
- Durer, S.; Gasalberti, D.P.; Shaikh, H. Ewing Sarcoma. In StatPearls [Internet]; NIH, National Library of Medicine: Bethesda, MD, USA, 2024. Available online: https://www.ncbi.nlm.nih.gov/books/NBK559183/ (accessed on 15 November 2024).
- Yeung, C.; Gibson, A.E.; Issaq, S.H.; Oshima, N.; Baumgart, J.T.; Edessa, L.D.; Rai, G.; Urban, D.J.; Johnson, M.S.; Benavides, G.A.; et al. Targeting Glycolysis through inhibition of lactate dehydrogenase impairs tumor growth in preclinical models of Ewing sarcoma. Cancer Res. 2019, 79, 5060–5073. [Google Scholar] [CrossRef]
- Friberg, A.; Rehwinkel, H.; Nguyen, D.; Pütter, V.; Quanz, M.; Weiske, J.; Eberspächer, U.; Heisler, I.; Langer, G. Structural Evidence for Isoform-Selective Allosteric Inhibition of Lactate Dehydrogenase A. ACS Omega 2020, 5, 13034–13041. [Google Scholar] [CrossRef] [PubMed]
- Saito, M. Subunit cooperativity in the action of lactate dehydrogenase. Biochim. Biophys. Acta 1972, 258, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Masterson, J.E.; Schwartz, S.D. Changes in protein architecture and subpicosecond protein dynamics impact the reaction catalyzed by lactate dehydrogenase. J. Phys. Chem. A 2013, 17, 7107–7113. [Google Scholar] [CrossRef]
- Curtis, N.J.; Mooney, L.; Hopcroft, L.; Michopoulos, F.; Whalley, N.; Zhong, H.; Murray, C.; Logie, A.; Revill, M.; Byth, K.F.; et al. Pre-clinical pharmacology of AZD3965, a selective inhibitor of MCT1: DLBCL, NHL and Burkitt’s lymphoma anti-tumor activity. Oncotarget 2017, 8, 69219–69236. [Google Scholar] [CrossRef]
- Doherty, J.R.; Yang, C.; Scott, K.E.; Cameron, M.D.; Fallahi, M.; Li, W.; Hall, M.A.; Amelio, A.L.; Mishra, J.K.; Li, F.; et al. Blocking lactate export by inhibiting the Myc target MCT1 Disables glycolysis and glutathione synthesis. Cancer Res. 2014, 74, 908–920. [Google Scholar] [CrossRef] [PubMed]
- Beloueche-Babari, M.; Casals Galobart, T.; Delgado-Goni, T.; Wantuch, S.; Parkes, H.G.; Tandy, D.; Harker, J.A.; Leach, M.O. Monocarboxylate transporter 1 blockade with AZD3965 inhibits lipid biosynthesis and increases tumour immune cell infiltration. Br. J. Cancer 2020, 122, 895–903. [Google Scholar] [CrossRef]
- Halford, S.; Veal, G.J.; Wedge, S.R.; Payne, G.S.; Bacon, C.M.; Sloan, P.; Dragoni, I.; Heinzmann, K.; Potter, S.; Salisbury, B.M.; et al. A phase I dose-escalation dtudy of AZD3965, an oral Monocarboxylate Transporter 1 inhibitor, in patients with advanced cancer. Clin. Cancer Res. 2023, 29, 1429–1439. [Google Scholar] [CrossRef] [PubMed]
- Landowski, C.P.; Suzuki, Y.; Hediger, M.A. The Mammalian Transporter Families. In Seldin and Giebisch’s The Kidney: Physiology and Pathophysiology, 4th ed.; Alpern, R.J., Hebert, S.C., Eds.; Elsevier: Oxford, UK, 2008; pp. 91–146. [Google Scholar]
- Mueckler, M.; Caruso, C.; Baldwin, S.A.; Panico, M.; Blench, I.; Morris, H.R.; Allard, W.J.; Lienhard, G.E.; Lodish, H.F. Sequence and structure of a human glucose transporter. Science 1985, 229, 941–945. [Google Scholar] [CrossRef]
- Tang, M.; Park, S.H.; De Vivo, D.C.; Monani, U.R. Therapeutic strategies for glucose transporter 1 deficiency syndrome. Ann. Clin. Transl. Neurol. 2019, 6, 1923–1932. [Google Scholar] [CrossRef]
- De Vivo, D.C.; Trifiletti, R.R.; Jacobson, R.I.; Ronen, G.M.; Behmand, R.A.; Harik, S.I. Defective glucose transport across the blood-brain barrier as a cause of persistent hypoglycorrhachia, seizures, and developmental delay. N. Engl. J. Med. 1991, 325, 703–709. [Google Scholar] [CrossRef]
- Vulturar, R.; Chiș, A.; Pintilie, S.; Farcaș, I.M.; Botezatu, A.; Login, C.C.; Sitar-Taut, A.V.; Orasan, O.H.; Stan, A.; Lazea, C.L.; et al. One molecule for mental nourishment and more: Glucose transporter type 1-biology and deficiency syndrome. Biomedicines 2022, 10, 1249. [Google Scholar] [CrossRef] [PubMed]
- Santer, R.; Klepper, J. Disorders of Glucose Transport in Inherited Metabolic Diseases, Diagnostic and Treatment, 6th ed.; Saudubray, J.M., Baumgartner, M., Waler, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 175–184. [Google Scholar]
- Chan, D.A.; Sutphin, P.D.; Nguyen, P.; Turcotte, S.; Lai, E.W.; Banh, A.; Reynolds, G.E.; Chi, J.T.; Wu, J.; Solow-Cordero, D.E.; et al. Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci. Transl. Med. 2011, 3, 94ra70. [Google Scholar] [CrossRef]
- Turcotte, S.; Chan, D.A.; Sutphin, P.D.; Hay, M.P.; Denny, W.A.; Giaccia, A.J. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell 2008, 14, 90–102. [Google Scholar] [CrossRef]
- Hay, M.P.; Turcotte, S.; Flanagan, J.U.; Bonnet, M.; Chan, D.A.; Sutphin, P.D.; Nguyen, P.; Giaccia, A.J.; Denny, W.A. 4-Pyridylanilinothiazoles that selectively target von Hippel–Lindau deficient renal cell carcinoma cells by inducing autophagic cell death. J. Med. Chem. 2010, 53, 787–797. [Google Scholar] [CrossRef]
- Wood, T.E.; Dalili, S.; Simpson, C.D.; Hurren, R.; Mao, X.; Saiz, F.S.; Gronda, M.; Eberhard, Y.; Minden, M.D.; Bilan, P.J.; et al. A novel inhibitor of glucose uptake sensitizes cells to FAS-induced cell death. Mol. Cancer Ther. 2008, 7, 3546–3555. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Montgomery, B.K.; Chatain, G.P.; Bugarini, A.; Zhang, Q.; Wang, X.; Edwards, N.A.; Ray-Chaudhury, A.; Merrill, M.J.; Lonser, R.R.; et al. Corticotropin releasing hormone can selectively stimulate glucose uptake in corticotropinoma via glucose transporter 1. Mol. Cell Endocrinol. 2018, 470, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Ocaña, M.C.; Martínez-Poveda, B.; Marí-Beffa, M.; Quesada, A.R.; Medina, M.Á. Fasentin diminishes endothelial cell proliferation, differentiation and invasion in a glucose metabolism-independent manner. Sci. Rep. 2020, 10, 6132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, Y.; Chen, X.; Bergmeier, S.C. Novel inhibitors of basal glucose transport as potential anticancer agents. Bioorg. Med. Chem. Lett. 2010, 20, 2191–2194. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, W.; Cao, Y.; Bergmeier, S.; Chen, X. Small compound inhibitors of basal glucose transport inhibit cell proliferation and induce apoptosis in cancer cells via glucose-deprivation-like mechanisms. Cancer Lett. 2010, 298, 176–185. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, Y.; Zhang, W.; Bergmeier, S.; Qian, Y.; Akbar, H.; Colvin, R.; Ding, J.; Tong, L.; Wu, S.; et al. A small-molecule inhibitor of glucose transporter 1 downregulates glycolysis, induces cell-cycle arrest, and inhibits cancer cell growth in vitro and in vivo. Mol. Cancer Ther. 2012, 11, 1672–1682. [Google Scholar] [CrossRef]
- Ojelabi, O.A.; Lloyd, K.P.; Simon, A.H.; De Zutter, J.K.; Carruthers, A. WZB117 (2-Fluoro-6-(m-hydroxybenzoyloxy) Phenyl m-Hydroxybenzoate) Inhibits GLUT1-mediated Sugar Transport by Binding Reversibly at the Exofacial Sugar Binding Site. J. Biol. Chem. 2016, 291, 26762–26772. [Google Scholar] [CrossRef]
- Siebeneicher, H.; Cleve, A.; Rehwinkel, H.; Neuhaus, R.; Heisler, I.; Müller, T.; Bauser, M.; Buchmann, B. Identification and Optimization of the First Highly Selective GLUT1 Inhibitor BAY-876. ChemMedChem 2016, 11, 2261–2271. [Google Scholar] [CrossRef]
- Miller, Z.A.; Muthuswami, S.; Mueller, A.; Ma, R.Z.; Sywanycz, S.M.; Naik, A.; Huang, L.; Brody, R.M.; Diab, A.; Carey, R.M.; et al. GLUT1 inhibitor BAY-876 induces apoptosis and enhances anti-cancer effects of bitter receptor agonists in head and neck squamous carcinoma cells. Cell Death Discov. 2024, 10, 339. [Google Scholar] [CrossRef]
- Pedersen, P.L. Warburg, me and Hexokinase 2: Multiple discoveries of key molecular events underlying one of cancers’ most common phenotypes, the “Warburg Effect”, i.e., elevated glycolysis in the presence of oxygen. J. Bioenerg. Biomembr. 2007, 39, 211–222. [Google Scholar] [CrossRef]
- Irwin, D.M.; Tan, H. Molecular evolution of the vertebrate hexokinase gene family: Identification of a conserved fifth vertebrate hexokinase gene. Comp. Biochem. Physiol. Part D Genom. Proteom. 2008, 3, 96–107. [Google Scholar] [CrossRef]
- Patra, K.C.; Wang, Q.; Bhaskar, P.T.; Miller, L.; Wang, Z.; Wheaton, W.; Chandel, N.; Laakso, M.; Muller, W.J.; Allen, E.L.; et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell 2013, 24, 213–228. [Google Scholar] [CrossRef]
- Anderson, M.; Marayati, R.; Moffitt, R.; Yeh, J.J. Hexokinase 2 promotes tumor growth and metastasis by regulating lactate production in pancreatic cancer. Oncotarget 2016, 8, 56081–56094. [Google Scholar] [CrossRef] [PubMed]
- Shan, W.; Zhou, Y.; Tam, K.Y. The development of small-molecule inhibitors targeting hexokinase 2. Drug Discov. Today 2022, 27, 2574–2585. [Google Scholar] [CrossRef]
- Benz, R. Permeation of hydrophilic solutes through mitochondrial outer membranes-review on mitochondrial porins. Biophys. Acta 1994, 1197, 167–196. [Google Scholar] [CrossRef] [PubMed]
- De Pinto, V.; Guarino, F.; Guarnera, A.; Messina, A.; Reina, S.; Tomasello, F.M.; Palermo, V.; Mazzoni, C. Characterization of human VDAC isoforms: A peculiar function for VDAC3? Biochim. Biophys. Acta 2010, 1797, 1268–1275. [Google Scholar] [CrossRef] [PubMed]
- Pastorino, J.G.; Hoek, J.B. Regulation of hexokinase binding to VDAC. J. Bioenerg. Biomembr. 2008, 40, 171–182. [Google Scholar] [CrossRef]
- Salani, B.; Marini, C.; Rio, A.D.; Ravera, S.; Massollo, M.; Orengo, A.M.; Amaro, A.; Passalacqua, M.; Maffioli, S.; Pfeffer, U.; et al. Metformin impairs glucose con- sumption and survival in Calu-1 cells by direct inhibition of hexokinase-II. Sci. Rep. 2013, 3, 2070. [Google Scholar] [CrossRef]
- Marini, C.; Salani, B.; Massollo, M.; Amaro, A.; Esposito, A.I.; Orengo, A.M.; Capitanio, S.; Emionite, L.; Riondato, M.; Bottoni, G.; et al. Direct inhibition of hexokinase activity by metformin at least partially impairs glucose metabolism and tumor growth in experimental breast cancer. Cell Cycle 2013, 12, 3490–3499. [Google Scholar] [CrossRef]
- John, S.; Weiss, J.N.; Ribalet, B. Subcellular localization of hexokinases I and II directs the metabolic fate of glucose. PLoS ONE 2011, 6, e17674. [Google Scholar] [CrossRef]
- Bustamante, E.; Pedersen, P.L. High aerobic glycolysis of rat hepatoma cells in culture: Role of mitochondrial hexokinase. Proc. Natl. Acad. Sci. USA 1977, 74, 3735–3739. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, H.; Lu, W.; Huang, P. Role of mitochondria-associated hexokinase II in cancer cell death induced by 3-bromopyruvate. Biochim. Biophys. Acta 2009, 1787, 553–560. [Google Scholar] [CrossRef]
- Ríos, J.L. Chemical constituents and pharmacological properties of Poria cocos. Planta Med. 2011, 77, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Miao, G.; Han, J.; Zhang, J.; Wu, Y.; Tong, G. Targeting Pyruvate Kinase M2 and Hexokinase II, Pachymic acid impairs glucose metabolism and induces mitochondrial apoptosis. Biol. Pharm. Bull. 2019, 42, 123–129. [Google Scholar] [CrossRef]
- Floridi, A.; Paggi, M.G.; Marcante, M.L.; Silvestrini, B.; Caputo, A.; De Martino, C. Lonidamine, a selective inhibitor of aerobic glycolysis of murine tumor cells. J. Natl. Cancer Inst. 1981, 66, 497–499. [Google Scholar]
- Ben-Yoseph, O.; Lyons, J.C.; Song, C.W.; Ross, B.D. Mechanism of action of lonidamine in the 9L brain tumor model involves inhibition of lactate efflux and intracellular acidification. J. Neurooncol. 1998, 36, 149–157. [Google Scholar] [CrossRef]
- Ravagnan, L.; Marzo, I.; Costantini, P.; Susin, S.A.; Zamzami, N.; Petit, P.X.; Hirsch, F.; Goulbern, M.; Poupon, M.F.; Miccoli, L.; et al. Lonidamine triggers apoptosis via a direct, Bcl-2-inhibited effect on the mitochondrial permeability transition pore. Oncogene 1999, 18, 2537–2546. [Google Scholar] [CrossRef]
- Tidmarsh, G. Therapies for the Treatment of Cancer. U.S. Patent US20060276527A1, 7 December 2006. [Google Scholar]
- Tang, Q.; Ji, F.; Guo, J.; Wang, J.; Li, Y.; Bao, Y. Directional modification of chrysin for exerting apoptosis and enhancing significantly anti-cancer effects of 10- hydroxy camptothecin. Biomed. Pharmacother. 2016, 82, 693–703. [Google Scholar] [CrossRef]
- Pichichero, E.; Cicconi, R.; Mattei, M.; Muzi, M.G.; Canini, A. Acacia honey and chrysin reduce proliferation of melanoma cells through alterations in cell cycle progression. Int. J. Oncol. 2010, 37, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Lian, S.; Khoi, P.N.; Yoon, H.J.; Han, J.Y.; Chay, K.O.; Kim, K.K.; Jung, Y.D. Chrysin inhibits cell invasion by inhibition of Recepteur d’origine Nantais via suppressing early growth response-1 and NF-kappaB transcription factor activities in gastric cancer cells. Int. J. Oncol. 2015, 46, 1835–1843. [Google Scholar] [CrossRef] [PubMed]
- Gao, A.M.; Ke, Z.P.; Shi, F.; Sun, G.C.; Chen, H. Chrysin enhances sensitivity of BEL- 7402/ADM cells to doxorubicin by suppressing PI3K/Akt/Nrf2 and ERK/Nrf2 pathway. Chem. Biol. Interact. 2013, 206, 100–108. [Google Scholar] [CrossRef]
- Xu, D.; Jin, J.; Yu, H.; Zhao, Z.; Ma, D.; Zhang, C.; Jiang, H. Chrysin inhibited tumor glycolysis and induced apoptosis in hepatocellular carcinoma by targeting hexokinase-2. J. Exp. Clin. Cancer Res. 2017, 36, 44. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Li, M.; Yu, X.; Gao, F.; Li, W. Repression of hexokinases II-mediated glycolysis contributes to piperlongumine-induced tumor suppression in non- small cell lung cancer cells. Int. J. Biol. Sci. 2019, 15, 826–837. [Google Scholar] [CrossRef]
- Pelicano, H.; Martin, D.S.; Xu, R.A.; Huang, P. Glycolysis inhibition for anticancer treatment. Oncogene 2006, 25, 4633–4646. [Google Scholar] [CrossRef]
- Zhu, G.; Guo, N.; Yong, Y.; Xiong, Y.; Tong, Q. Effect of 2-deoxy-D-glucose on gellan gum biosynthesis by Sphingomonas paucimobilis. Bioprocess. Biosyst. Eng. 2019, 42, 897–900. [Google Scholar] [CrossRef]
- Singh, R.; Gupta, V.; Kumar, A.; Singh, K. 2-Deoxy-D-glucose: A novel pharmacological agent for killing hypoxic tumor cells, oxygen dependence-lowering in COVID-19, and other pharmacological activities. Adv. Pharmacol. Pharm. Sci. 2023, 2023, 9993386. [Google Scholar] [CrossRef]
- IFokt, S.; Skora, C.; Conrad, T.; Madden, M.; Emmett, W. Priebe- D-glucose and D-mannose-based metabolic probes. Part 3: Synthesis of specifcally deuterated D-glucose, -mannose, and 2-deoxy-D-glucose. Carbohydr. Res. 2013, 368, 111–119. [Google Scholar]
- Xi, H.; Kurtoglu, M.; Lampidis, T.J. The wonders of 2- deoxy-D-glucose. IUBMB Life 2014, 66, 110–121. [Google Scholar] [CrossRef]
- Cunha, A.; Rocha, A.C.; Barbosa, F.; Baião, A.; Silva, P.; Sarmento, B.; Queirós, O. Glycolytic inhibitors potentiated the activity of paclitaxel and their nanoencapsulation increased their delivery in a lung cancer model. Pharmaceutics 2022, 14, 2021. [Google Scholar] [CrossRef] [PubMed]
- Francis, R.; Singh, P.K.; Singh, S.; Giri, S.; Kumar, A. Glycolytic inhibitor 2-deoxyglucose suppresses inflammatory response in innate immune cells and experimental staphylococcal endophthalmitis. Exp. Eye Res. 2020, 97, 108079. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, J.G.; Rosenthal, K.S. 2-deoxy-D-glucose inhibition of herpes simplex virus type-1 receptor expression. Antivir. Res. 1986, 6, 137–149. [Google Scholar] [CrossRef]
- Dey, S.; Murmu, N.; Mondal, T.; Saha, I.; Chatterjee, S.; Manna, R.; Haldar, S.; Dash, S.K.; Sarkar, T.R.; Giri, B. Multifaceted entrancing role of glucose and its analogue, 2-deoxy-D-glucose in cancer cell proliferation, inflammation, and virus infection. Biomed. Pharmacother. 2022, 156, 113801. [Google Scholar] [CrossRef]
- Yamaguchi, R.; Janssen, E.; Perkins, G.; Ellisman, M.; Kitada, S.; Reed, J.C. Efficient elimination of cancer cells by deoxyglucose-ABT-263/737 combination therapy. PLoS ONE 2011, 6, e24102. [Google Scholar] [CrossRef]
- Zhang, D.; Li, J.; Wang, F.; Hu, J.; Wang, S.; Sun, Y. 2-Deoxy-D-glucose targeting of glucose metabolism in cancer cells as a potential therapy. Cancer Lett. 2014, 355, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Lampidis, T.J.; Kurtoglu, M.; Maher, J.C.; Liu, H.; Krishan, A.; Sheft, V.; Szymanski, S.; Fokt, I.; Rudnicki, W.R.; Ginalski, K.; et al. Efficacy of 2-halogen substituted D-glucose analogs in blocking glycolysis and killing “hypoxic tumor cells”. Cancer Chemother. Pharmacol. 2006, 58, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Ma, Y.; Zhang, Y.; Fu, B.; Wu, X.; Li, Q.; Cai, G.; Chen, X.; Bai, X.Y. Low-dose 2-deoxyglucose and metformin synergically inhibit proliferation of human polycystic kidney cells by modulating glucose metabolism. Cell Death Discov. 2019, 5, 76. [Google Scholar] [CrossRef]
- Cheong, J.H.; Park, E.S.; Liang, J.; Dennison, J.B.; Tsavachidou, D.; Nguyen-Charles, C.; Wa Cheng, K.; Hall, H.; Zhang, D.; Lu, Y.; et al. Dual inhibition of tumor energy pathway by 2-deoxyglucose and metformin is effective against a broad spectrum of preclinical cancer models. Mol. Cancer Ther. 2011, 10, 2350–2362. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zheng, M.; Wu, S.; Gao, S.; Yang, M.; Li, Z.; Min, Q.; Sun, W.; Chen, L.; Xiang, G.; et al. Benserazide, a dopadecarboxylase inhibitor, suppresses tumor growth by targeting hexokinase 2. J. Exp. Clin. Cancer Res. 2017, 36, 58. [Google Scholar] [CrossRef]
- Chiara, F.; Castellaro, D.; Marin, O.; Petronilli, V.; Brusilow, W.S.; Juhaszova, M.; Sollott, S.J.; Forte, M.; Bernardi, P.; Rasola, A. Hexokinase II detachment from mitochondria triggers apoptosis through the permeability transition pore independent of voltage-dependent anion channels. PLoS ONE 2008, 3, e1852. [Google Scholar] [CrossRef]
- Zheng, M.; Wu, C.; Yang, K.; Yang, Y.; Liu, Y.; Gao, S.; Wang, Q.; Li, C.; Chen, L.; Li, H. Novel selective hexokinase 2 inhibitor Benitrobenrazide blocks cancer cells growth by targeting glycolysis. Pharmacol. Res. 2021, 164, 105367. [Google Scholar] [CrossRef] [PubMed]
- Mulichak, A.M.; Wilson, J.E.; Padmanabhan, K.; Garavito, R.M. The mammalian hexokinase-1. Nat. Struct. Biol. 1998, 5, 555–560. [Google Scholar] [CrossRef]
- Lin, H.; Zeng, J.; Xie, R.; Schulz, M.J.; Tedesco, R.; Qu, J.; Erhard, K.F.; Mack, J.F.; Raha, K.; Rendina, A.R.; et al. Discovery of a novel 2,6-disubstituted glucosamine series of potent and selective Hexokinase 2 inhibitors. ACS Med. Chem. Lett. 2015, 7, 217–222. [Google Scholar] [CrossRef]
- Deeb, S.S.; Malkki, M.; Laakso, M. human hexokinase II: Sequence and homology to other hexokinases. Biochem. Biophys. Res. Commun. 1993, 197, 68–74. [Google Scholar] [CrossRef]
- Yang, W.; Lu, Z. Pyruvate kinase M2 at a glance. J. Cell Sci. 2015, 128, 1655–1660. [Google Scholar] [CrossRef]
- Heng, T.S.; Painter, M.W.; The Immunological Genome Project Consortium; Elpek, K.; Lukacs-Kornek, V.; Mauermann, N.; Turley, S.J.; Koller, D.; Kim, F.S.; Wagers, A.J.; et al. The Immunological Genome Project: Networks of gene expression in immune cells. Nat. Immunol. 2008, 9, 1091–1094. [Google Scholar] [CrossRef]
- Alves-Filho, J.C.; Pålsson-McDermott, E.M. Pyruvate kinase M2: A potential target for regulating inflammation. Front. Immunol. 2016, 7, 145. [Google Scholar] [CrossRef]
- Toller-Kawahisa, J.E.; Hiroki, C.H.; de Souza Silva, C.M.; Nascimento, D.C.; Pùblio, G.A.; Varela Martins, T.; Alves Damascenco, L.E.; Protàsio Veras, F.; Ramos Viacava, P.; Yuji Sukesada, F.; et al. The metabolic function of pyruvate kinase M2 regulates reactive oxygen species production and microbial killing by neutrophils. Nat. Commun. 2023, 14, 4280. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Puri, S.; Leland, P.; Puri, A.; Moudgil, T.; Fox, B.A.; Puri, R.K.; Joshi, B.H. Subcellular compartmentalization of PKM2 identifies anti-PKM2 therapy response in vitro and in vivo mouse model of human non-small-cell lung cancer. PLoS ONE 2019, 14, e0217131. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Huang, Z.; Su, J.; Li, J.; Zhao, S.; Wu, L.; Zhang, J.; He, Y.; Zhang, G.; Tao, J.; et al. Benserazide is a novel inhibitor targeting PKM2 for melanoma treatment. Int. J. Cancer 2020, 147, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Druzhyna, N.; Szczesny, B.; Olah, G.; Módis, K.; Asimakopoulou, A.; Pavlidou, A.; Szoleczky, P.; Gerö, D.; Yanagi, K.; Törö, G.; et al. Screening of a composite library of clinically used drugs and well-characterized pharmacological compounds for cystathionine β-synthase inhibition identifies benserazide as a drug potentially suitable for repurposing for the experimental therapy of colon cancer. Pharmacol. Res. 2016, 113, 18–37. [Google Scholar]
- Ning, X.; Qi, H.; Li, R.; Li, Y.; Jin, Y.; McNutt, M.A.; Liu, J.; Yin, Y. Discovery of novel naphthoquinone derivatives as inhibitors of the tumor cell specific M2 isoform of pyruvate kinase. Eur. J. Med. Chem. 2017, 138, 343–352. [Google Scholar] [CrossRef]
- Ning, X.; Qi, H.; Li, R.; Jin, Y.; McNutt, M.A.; Yin, Y. Synthesis and antitumor activity of novel 2, 3-didithiocarbamate substituted naphthoquinones as inhibitors of pyruvate kinase M2 isoform. J. Enzym. Inhib. Med. Chem. 2018, 33, 126–129. [Google Scholar] [CrossRef]
- Gao, C.L.; Hou, G.G.; Liu, J.; Ru, T.; Xu, Y.Z.; Zhao, S.Y.; Ye, H.; Zhang, L.Y.; Chen, K.X.; Guo, Y.W.; et al. Synthesis and target identification of benzoxepane derivatives as potential anti-neuroinflammatory agents for ischemic stroke. Angew. Chem. Int. Ed. Engl. 2020, 59, 2429–2439. [Google Scholar] [CrossRef]
- Zhong, W.J.; Yang, H.H.; Guan, X.X.; Xiong, J.B.; Sun, C.C.; Zhang, C.Y.; Luo, X.Q.; Zhang, Y.F.; Zhang, J.; Duan, J.X.; et al. Inhibition of glycolysis alleviates lipopolysaccharide-induced acute lung injury in a mouse model. J. Cell Physiol. 2019, 234, 4641–4654. [Google Scholar] [CrossRef]
- Xie, M.; Yu, Y.; Kang, R.; Zhu, S.; Yang, L.; Zeng, L.; Sun, X.; Yang, M.; Billiar, T.R.; Wang, H.; et al. PKM2-dependent glycolysis promotes NLRP3 and AIM2 inflammasome activation. Nat. Commun. 2016, 7, 13280. [Google Scholar] [CrossRef]
- Wubben, T.J.; Pawar, M.; Weh, E.; Smith, A.; Sajjakulnukit, P.; Zhang, L.; Dai, L.; Hager, H.; Pai, M.P.; Lyssiotis, C.A.; et al. Small molecule activation of metabolic enzyme pyruvate kinase muscle isozyme 2, PKM2, circumvents photoreceptor apoptosis. Sci. Rep. 2020, 10, 2990. [Google Scholar] [CrossRef]
- Sayyad, Z.; Sirohi, K.; Radha, V.; Swarp, G. 661W is a retinal ganglion precursor-like cell line in which glaucoma-associated optineurin mutants induce cell death selectively. Sci. Rep. 2017, 7, 16855. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.K.; Walsh, M.J.; Brimacombe, K.R.; Anastasiou, D.; Yu, Y.; Israelsen, W.J.; Hong, B.S.; Tempel, W.; Dimov, S.; Veith, H.; et al. ML265: A potent PKM2 activator induces tetramerization and reduces tumor formation and size in a mouse xenograft model. In Probe Reports from the NIH Molecular Libraries Program [Internet]; National Center for Biotechnology Information: Bethesda, MD, USA, 2012. Available online: https://www.ncbi.nlm.nih.gov/books/NBK153222/ (accessed on 9 July 2025).
- Sarfraz, I.; Rasul, A.; Jabeen, F.; Sultana, T.; Adem, Ş. Identification of natural compounds as inhibitors of pyruvate kinase M2 for cancer treatment. Molecules 2022, 27, 7113. [Google Scholar] [CrossRef]
- Tuli, H.S.; Mittal, S.; Aggarwal, D.; Parashar, G.; Parashar, N.C.; Upadhyay, S.K.; Barwal, T.S.; Jain, A.; Kaur, G.; Savla, R.; et al. Path of Silibinin from diet to medicine: A dietary polyphenolic flavonoid having potential anti-cancer therapeutic significance. Semin. Cancer Biol. 2021, 73, 196–218. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.X.; Ding, Y.H.; Wu, Y.; Qian, L.Y.; Zou, H.; He, Q. Silibinin: A potential old drug for cancer therapy. Expert. Rev. Clin. Pharmacol. 2016, 9, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Mateen, S.; Raina, K.; Agarwal, C.; Chan, D.; Agarwal, R. Silibinin synergizes with histone deacetylase and DNA methyltransferase inhibitors in upregulating E-cadherin expression together with inhibition of migration and invasion of human non-small cell lung cancer cells. J. Pharmacol. Exp. Ther. 2013, 345, 206–214. [Google Scholar] [CrossRef]
- Kauntz, H.; Bousserouel, S.; Gossé, F.; Raul, F. Epigenetic effects of the natural flavonolignan silibinin on colon adenocarcinoma cells and their derived metastatic cells. Oncol. Lett. 2013, 5, 1273–1277. [Google Scholar] [CrossRef]
- Iqbal, M.A.; Chattopadhyay, S.; Siddiqui, F.A.; Ur Rehman, A.; Siddiqui, S.; Prakasam, G.; Khan, A.; Sultana, S.; Bamezai, R.N. Silibinin induces metabolic crisis in triple-negative breast cancer cells by modulating EGFR-MYC-TXNIP axis: Potential therapeutic implications. FEBS J. 2021, 288, 471–485. [Google Scholar] [CrossRef]
- Existing Food Additives, Ministry of Health, Labor and Welfare of Japan, Notification n.120. 1996. Available online: https://www.ffcr.or.jp/en/tenka/list-of-existing-food-additives/list-of-existing-food-additives.html (accessed on 26 February 2020).
- Tasaki, M.; Umemura, T.; Maeda, M.; Ishii, Y.; Okamura, T.; Inoue, T.; Kuroiwa, Y.; Hirose, M.; Nishikawa, A. Safety assessment of ellagic acid, a food additive, in a subchronic toxicity study using F344 rats. Food Chem. Toxicol. 2008, 46, 1119–1124. [Google Scholar] [CrossRef]
- Devipriya, N.; Srinivasan, M.; Sudheer, A.R.; Menon, V.P. Effect of ellagic acid, a natural polyphenol, on alcohol-induced prooxidant and antioxidant imbalance: A drug dose dependent study. Singap. Med. J. 2007, 48, 311–318. [Google Scholar]
- Ceci, C.; Lacal, P.M.; Tentori, L.; De Martino, M.G.; Miano, R.; Graziani, G. Experimental evidence of the antitumor, antimetastatic and antiangiogenic activity of ellagic acid. Nutrients 2018, 10, 1756. [Google Scholar] [CrossRef]
- Yousuf, M.; Shamsi, A.; Khan, P.; Shahbaaz, M.; AlAjmi, M.F.; Hussain, A.; Hassan, G.M.; Islam, A.; Rizwanul Haque, Q.M.; Hassan, M.I. Ellagic acid controls cell proliferation and induces apoptosis in breast cancer cells via inhibition of cyclin-dependent kinase 6. Int. J. Mol. Sci. 2020, 21, 3526. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Mohammad, T.; Khan, P.; Alajmi, M.F.; Hussain, A.; Rehman, M.T.; Hassan, M.I. Evaluation of ellagic acid as an inhibitor of sphingosine kinase 1: A targeted approach towards anticancer therapy. Biomed. Pharmacother. 2019, 118, 109245. [Google Scholar] [CrossRef]
- Stanić, Z. Curcumin, a compound from natural sources, a true scientific challenge—A review. Plant Foods Hum. Nutr. 2017, 72, 1–12. [Google Scholar] [CrossRef]
- Siddiqui, F.A.; Prakasam, G.; Chattopadhyay, S.; Rehman, A.U.; Padder, R.A.; Ansari, M.A.; Irshad, R.; Mangalhara, K.; Bamezai, R.N.K.; Husain, M.; et al. Curcumin decreases Warburg effect in cancer cells by down-regulating pyruvate kinase M2 via mTOR-HIF1α inhibition. Sci. Rep. 2018, 8, 8323. [Google Scholar] [CrossRef] [PubMed]
- Baur, J.A.; Sinclair, D.A. Therapeutic potential of resveratrol: The in vivo evidence. Nat. Rev. Drug Discov. 2006, 5, 493–506. [Google Scholar] [CrossRef]
- Jang, M.; Cai, L.; Udeani, G.O.; Slowing, K.V.; Thomas, C.F.; Beecher, C.W.; Fong, H.H.; Farnsworth, N.R.; Kinghorn, A.D.; Mehta, R.G.; et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science 1997, 275, 218–220. [Google Scholar] [CrossRef]
- Iqbal, M.A.; Bamezai, R.N. Resveratrol inhibits cancer cell metabolism by down regulating pyruvate kinase M2 via inhibition of mammalian target of rapamycin. PLoS ONE 2012, 7, e36764. [Google Scholar] [CrossRef]
- Sun, Q.; Chen, X.; Ma, J.; Peng, H.; Wang, F.; Zha, X.; Wang, Y.; Jing, Y.; Yang, H.; Chen, R.; et al. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proc. Natl. Acad. Sci. USA 2011, 108, 4129–4134. [Google Scholar] [CrossRef]
- Varoni, E.M.; Lo Faro, A.F.; Sharifi-Rad, J.; Iriti, M. Anticancer molecular mechanisms of resveratrol. Front. Nutr. 2016, 3, 8. [Google Scholar] [CrossRef]
- Schulte, M.L.; Khodadadi, A.B.; Cuthbertson, M.L.; Smith, J.A.; Manning, H.C. 2-Amino-4-bis(aryloxybenzyl)aminobutanoic acids: A novel scaffold for inhibition of ASCT2-mediated glutamine transport. Bioorg. Med. Chem. Lett. 2016, 26, 1044–1047. [Google Scholar] [CrossRef]
- Shimizu, K.; Kaira, K.; Tomizawa, Y.; Sunaga, N.; Kawashima, O.; Oriuchi, N.; Tominaga, H.; Nagamori, S.; Kanai, Y.; Yamada, M.; et al. ASC amino-acid transporter 2 (ASCT2) as a novel prognostic marker in non-small cell lung cancer. Br. J. Cancer 2014, 110, 2030. [Google Scholar] [CrossRef]
- Kaira, K.; Sunose, Y.; Arakawa, K.; Sunaga, N.; Shimizu, K.; Tominaga, H.; Oriuchi, N.; Nagamori, S.; Kanai, Y.; Oyama, T.; et al. Clinicopathological significance of ASC amino acid transporter-2 expression in pancreatic ductal carcinoma. Histopathology. 2015, 66, 234–243. [Google Scholar] [CrossRef]
- Bröer, A.; Fairweather, S.; Bröer, S. Disruption of amino acid homeostasis by novel ASCT2 inhibitors involves multiple targets. Front. Pharmacol. 2018, 9, 785. [Google Scholar] [CrossRef]
- Najumudeen, A.K.; Ceteci, F.; Fey, S.K.; Hamm, G.; Steven, R.T.; Hall, H.; Nikula, C.J.; Dexter, A.; Murta, T.; Race, A.M.; et al. The amino acid transporter SLC7A5 is required for efficient growth of KRAS-mutant colorectal cancer. Nat. Genet. 2021, 53, 16–26. [Google Scholar] [CrossRef]
- Okunushi, K.; Furihata, T.; Morio, H.; Muto, Y.; Higuchi, K.; Kaneko, M.; Otsuka, Y.; Ohno, Y.; Watanabe, Y.; Reien, Y.; et al. JPH203, a newly developed anti-cancer drug, shows a preincubation inhibitory effect on L-type amino acid transporter 1 function. J. Pharmacol. Sci. 2020, 144, 16–22. [Google Scholar] [CrossRef]
- Investigational Drug Branch CTTP. Clinical Brochure: DON—NSC 7365; Investigational Drug Branch CTTP: Bethesda, MD, USA, 1979; pp. 1–53. [Google Scholar]
- Lemberg, K.M.; Vornov, J.J.; Rais, R.; Slusher, B.S. We’re Not “DON” Yet: Optimal Dosing and Prodrug Delivery of 6-Diazo-5-oxo-L-norleucine. Mol. Cancer Ther. 2018, 17, 1824–1832. [Google Scholar] [CrossRef]
- Pinkus, L.M. Glutamine binding sites. Methods Enzymol. 1977, 46, 414–427. [Google Scholar]
- Thomas, A.G.; Rojas, C.; Tanega, C.; Shen, M.; Simeonov, A.; Boxer, M.B.; Auld, D.S.; Ferraris, D.V.; Tsukamoto, T.; Slusher, B.S. Kinetic characterization of ebselen, chelerythrine and apomorphine as glutaminase inhibitors. Biochem. Biophys. Res. Commun. 2013, 438, 243–248. [Google Scholar] [CrossRef]
- Rais, R.; Jančařík, A.; Tenora, L.; Nedelcovych, M.; Alt, J.; Englert, J.; Rojas, C.; Le, A.; Elgogary, A.; Tan, J.; et al. Discovery of 6-diazo-5-oxo-l-norleucine (DON) prodrugs with enhanced csf delivery in monkeys: A potential treatment for glioblastoma. J. Med. Chem. 2016, 59, 8621–8633. [Google Scholar] [CrossRef]
- Tenora, L.; Alt, J.; Dash, R.P.; Gadiano, A.J.; Novotná, K.; Veeravalli, V.; Lam, J.; Kirkpatrick, Q.R.; Lemberg, K.M.; Majer, P.; et al. Tumor-Targeted Delivery of 6-Diazo-5-oxo-l-norleucine (DON) Using Substituted Acetylated Lysine Prodrugs. J. Med. Chem. 2019, 62, 3524–3538. [Google Scholar] [CrossRef]
- Hanaford, A.R.; Alt, J.; Rais, R.; Wang, S.Z.; Kaur, H.; Thorek, D.L.J.; Eberhart, C.G.; Slusher, B.S.; Martin, A.M.; Raabe, E.H. Orally bioavailable glutamine antagonist prodrug JHU-083 penetrates mouse brain and suppresses the growth of MYC-driven medulloblastoma. Transl. Oncol. 2019, 12, 1314–1322. [Google Scholar] [CrossRef]
- Rais, R.; Lemberg, K.M.; Tenora, L.; Arwood, M.L.; Pal, A.; Alt, J.; Wu, Y.; Lam, J.; Aguilar, J.M.H.; Zhao, L.; et al. Discovery of DRP-104, a tumor-targeted metabolic inhibitor prodrug. Sci. Adv. 2022, 8, eabq5925. [Google Scholar] [CrossRef]
- Pillai, R.; LeBoeuf, S.E.; Hao, Y.; New, C.; Blum, J.L.E.; Rashidfarrokhi, A.; Huang, S.M.; Bahamon, C.; Wu, W.L.; Karadal-Ferrena, B.; et al. Glutamine antagonist DRP-104 suppresses tumor growth and enhances response to checkpoint blockade in KEAP1 mutant lung cancer. Sci. Adv. 2024, 10, eadm9859. [Google Scholar] [CrossRef]
- Seltzer, M.J.; Bennett, B.D.; Joshi, A.D.; Gao, P.; Thomas, A.G.; Ferraris, D.V.; Tsukamoto, T.; Rojas, C.J.; Slusher, B.S.; Rabinowitz, J.D.; et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010, 70, 8981–8987. [Google Scholar] [CrossRef]
- Elgadi, K.M.; Meguid, R.A.; Qian, M.; Souba, W.W.; Abcouwer, S.F. Cloning and analysis of unique human glutaminase isoforms generated by tissue-specific alternative splicing. Physiol. Genom. 1999, 1, 51–62. [Google Scholar] [CrossRef]
- Hartwick, E.W.; Curthoys, N.P. BPTES inhibition of hGA(124–551), a truncated form of human kidney-type glutaminase. J. Enzym. Inhib. Med. Chem. 2012, 27, 861–867. [Google Scholar] [CrossRef]
- Shukla, K.; Ferraris, D.V.; Thomas, A.G.; Stathis, M.; Duvall, B.; Delahanty, G.; Alt, J.; Rais, R.; Rojas, C.; Gao, P.; et al. Design, synthesis, and pharmacological evaluation of bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide 3 (BPTES) analogs as glutaminase inhibitors. J. Med. Chem. 2012, 5, 10551–10563. [Google Scholar] [CrossRef]
- Sakurai, T.; Kanayama, M.; Shibata, T.; Itoh, K.; Kobayashi, A.; Yamamoto, M.; Uchida, K. Ebselen, a seleno-organic antioxidant, as an electrophile. Chem. Res. Toxicol. 2006, 19, 1196–1204. [Google Scholar] [CrossRef]
- De La Barre, B.; Gross, S.; Fang, C.; Gao, Y.; Jha, A.; Jiang, F.; Song, J.J.; Wei, W.; Hurov, J.B. Full-length human glutaminase in complex with an allosteric inhibitor. Biochemistry 2011, 50, 10764–10770. [Google Scholar] [CrossRef]
- Santos, C.R.; Schulze, A. Lipid metabolism in cancer. FEBS J. 2012, 279, 2610–2623. [Google Scholar] [CrossRef]
- Baenke, F.; Peck, B.; Miess, H.; Schulze, A. Hooked on fat: The role of lipid synthesis in cancer metabolism and tumour development. Dis. Models Mech. 2013, 6, 1353–1363. [Google Scholar] [CrossRef]
- Nomura, D.K.; Long, J.Z.; Niessen, S.; Hoover, H.S.; Ng, S.W.; Cravatt, B.F. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell 2010, 140, 49–61. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Z.; Lian, Z.; Liao, R.; Chen, Y.; Qin, Y.; Wang, J.; Jiang, Q.; Wang, X.; Gong, J. Monoacylglycerol Lipase: A Novel Potential Therapeutic Target and Prognostic Indicator for Hepatocellular Carcinoma. Sci. Rep. 2016, 6, 35784. [Google Scholar] [CrossRef]
- Deng, H.; Li, W. Monoacylglycerol lipase inhibitors: Modulators for lipid metabolism in cancer malignancy, neurological and met- abolic disorders. Acta Pharm. Sin. B 2020, 10, 582–602. [Google Scholar] [CrossRef]
- Muccioli, G.G.; Labar, G.; Lambert, D.M. CAY10499, a novel monoglyceride lipase inhibitor evidenced by an expeditious MGL assay. ChemBioChem 2008, 9, 2704–2710. [Google Scholar] [CrossRef]
- Iglesias, J.; Lamontagne, J.; Erb, H.; Gezzar, S.; Zhao, S.; Joly, E.; Truong, V.L.; Skorey, K.; Crane, S.; Madiraju, S.R.; et al. Simplified assays of lipolysis enzymes for drug discovery and specificity assessment of known inhibitors. J. Lipid Res. 2016, 57, 131–141. [Google Scholar] [CrossRef]
- Cravatt, B.F.; Wright, A.T.; Kozarich, J.W. Activity-based protein profiling: From enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 2008, 77, 383–414. [Google Scholar] [CrossRef]
- Long, J.Z.; Li, W.; Booker, L.; Burston, J.J.; Kinsey, S.G.; Schlosburg, J.E.; Pavón, F.J.; Serrano, A.M.; Selley, D.E.; Parsons, L.H.; et al. Selective blockade of -arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat. Chem. Biol. 2009, 5, 37–44. [Google Scholar] [CrossRef]
- Marino, S.; de Ridder, D.; Bishop, R.T.; Renema, N.; Ponzetti, M.; Sophocleous, A.; Capulli, M.; Aljeffery, A.; Carrasco, G.; Gens, M.D.; et al. Paradoxical effects of JZL184, an inhibitor of monoacylglycerol lipase, on bone remodelling in healthy and cancer-bearing mice. EBioMedicine 2019, 44, 452–466. [Google Scholar] [CrossRef]
- Marino, S.; Carrasco, G.; Li, B.; Shah, K.M.; Lath, D.L.; Sophocleous, A.; Lawson, M.A.; Idris, A.I. JZL184, A Monoacylglycerol Lipase Inhibitor, Induces Bone Loss in a Multiple Myeloma Model of Immunocompetent Mice. Calcif. Tissue Int. 2020, 107, 72–85. [Google Scholar] [CrossRef]
- Aaltonen, N.; Savinainen, J.R.; Ribas, C.R.; Rönkkö, J.; Kuusisto, A.; Korhonen, J.; Navia-Paldanius, D.; Häyrinen, J.; Takabe, P.; Käsnänen, H.; et al. Piperazine and piperidine triazole ureas as ultrapotent and highly selective inhibitors of monoacylglycerol lipase. Chem. Biol. 2013, 20, 379–390. [Google Scholar] [CrossRef]
- Zhu, W.; Zhao, Y.; Zhou, J.; Wang, X.; Pan, Q.; Zhang, N.; Wang, L.; Wang, M.; Zhan, D.; Liu, Z.; et al. Monoacylglycerol lipase promotes progression of hepatocellular carcinoma via NF-κB-mediated epithelial-mesenchymal transition. J. Hematol. Oncol. 2016, 9, 127. [Google Scholar] [CrossRef]
- Puris, E.; Petralla, S.; Auriola, S.; Kidron, H.; Fricker, G.; Gynther, M. Monoacylglycerol Lipase Inhibitor JJKK048 Ameliorates ABCG2 Transporter-Mediated Regorafenib Resistance Induced by Hypoxia in Triple Negative Breast Cancer Cells. J. Pharm. Sci. 2023, 112, 2581–2590. [Google Scholar] [CrossRef]
- Wyatt, R.M.; Fraser, I.; Welty, N.; Lord, B.; Wennerholm, M.; Sutton, S.; Ameriks, M.K.; Dugovic, C.; Yun, S.; White, A.; et al. Pharmacologic Characterization of JNJ-42226314, [1-(4-fluorophenyl)indol-5-yl]-[3-[4-(thiazole-2-carbonyl)piperazin-1-yl]azetidin-1-yl]methanone, a reversible, selective, and potent monoacylglycerol lipase inhibitor. J. Pharmacol. Exp. Ther. 2020, 372, 339–353. [Google Scholar] [CrossRef]
- Jones, S.F.; Infante, J.R. Molecular pathways: Fatty acid synthase. Clin. Cancer Res. 2015, 21, 5434–5438. [Google Scholar] [CrossRef]
- Adams, N.A.; Aquino, C.J.; Chaudhari, A.M.; Ghergurovich, J.M.; Kiesow, T.J.; Parrish, C.A.; Reif, A.J.; Wiggall, K. Triazolones as Fatty Acid Synthase inhibitors. World Intellectual Property. Organization. Patent No. WO2011103546, 25 August 2011. [Google Scholar]
- Hardwicke, M.A.; Rendina, A.R.; Williams, S.P.; Moore, M.L.; Wang, L.; Krueger, J.A.; Plant, R.N.; Totoritis, R.D.; Zhang, G.; Briand, J.; et al. A human fatty acid synthase inhibitor binds β-ketoacyl reductase in the keto-substrate site. Nat. Chem. Biol. 2014, 10, 774–779. [Google Scholar] [CrossRef]
- Aquino, I.G.; Bastos, D.C.; Cuadra-Zelaya, F.J.M.; Teixeira, I.F.; Salo, T.; Coletta, R.D.; Graner, E. Anticancer properties of the fatty acid synthase inhibitor TVB-3166 on oral squamous cell carcinoma cell lines. Arch. Oral Biol. 2020, 113, 104707. [Google Scholar] [CrossRef]
- Syed-Abdul, M.M.; Parks, E.J.; Gaballah, A.H.; Bingham, K.; Hammoud, G.M.; Kemble, G.; Buckley, D.; McCulloch, W.; Manrique-Acevedo, C. Fatty Acid Synthase Inhibitor TVB-2640 Reduces Hepatic de Novo Lipogenesis in Males With Metabolic Abnormalities. Hepatology 2020, 72, 103–118. [Google Scholar] [CrossRef]
- Loomba, R.; Mohseni, R.; Lucas, K.J.; Gutierrez, J.A.; Perry, R.G.; Trotter, J.F.; Rahimi, R.S.; Harrison, S.A.; Ajmera, V.; Wayne, J.D.; et al. TVB-2640 (FASN Inhibitor) for the Treatment of Nonalcoholic Steatohepatitis: FASCINATE-1, a Randomized, Placebo-Controlled Phase 2a Trial. Gastroenterology. 2021, 161, 1475–1486. [Google Scholar] [CrossRef]
- Zadra, G.; Ribeiro, C.F.; Chetta, P.; Ho, Y.; Cacciatore, S.; Gao, X.; Syamala, S.; Bango, C.; Photopoulos, C.; Huang, Y.; et al. Inhibition of de novo lipogenesis targets androgen receptor signaling in castration-resistant prostate cancer. Proc. Natl. Acad. Sci. USA 2020, 116, 631–640, Erratum in Proc. Natl. Acad. Sci. USA 2020, 117, 18893. [Google Scholar] [CrossRef]
- Chypre, M.; Zaidi, N.; Smans, K. ATP-citrate lyase: A mini-review. Biochem. Biophys. Res. Commun. 2012, 422, 1–4. [Google Scholar] [CrossRef]
- Granchi, C. ATP citrate lyase (ACLY) inhibitors: An anti-cancer strategy at the crossroads of glucose and lipid metabolism. Eur. J. Med. Chem. 2018, 157, 1276–1291. [Google Scholar] [CrossRef]
- Shi, L.; Tu, B.P. Acetyl-CoA and the regulation of metabolism: Mechanisms and consequences. Curr. Opin. Cell Biol. 2015, 33, 125–131. [Google Scholar] [CrossRef]
- Lemus, H.N.; Mendivil, C.O. Adenosine triphosphate citrate lyase: Emerging target in the treatment of dyslipidemia. J. Clin. Lipidol. 2015, 9, 384–389. [Google Scholar] [CrossRef]
- Guais, A.; Baronzio, G.; Sanders, E.; Campion, F.; Mainini, C.; Fiorentini, G.; Montagnani, F.; Behzadi, M.; Schwartz, L.; Abolhassani, M. Adding a combination of hydroxycitrate and lipoic acid (METABLOC™) to chemotherapy improves effectiveness against tumor development: Experimental results and case report. Investig. New Drugs 2012, 30, 200–211. [Google Scholar] [CrossRef]
- Wei, J.; Leit, S.; Kuai, J.; Therrien, E.; Rafi, S.; Harwood, H.J.; De La Barre, B.; Tong, L. An allosteric mechanism for potent inhibition of human ATP-citrate lyase. Nature 2019, 568, 566–570. [Google Scholar] [CrossRef]
- Granchi, C. Discovery of Allosteric Inhibition of Human ATP-Citrate Lyase. Trends Pharmacol. Sci. 2019, 40, 364–366. [Google Scholar] [CrossRef]
- Li, J.J.; Wang, H.; Tino, J.A.; Robl, J.A.; Herpin, T.F.; Lawrence, R.M.; Biller, S.; Jamil, H.; Ponticiello, R.; Chen, L.; et al. 2-hydroxy-N-arylbenzenesulfonamides as ATP-citrate lyase inhibitors. Bioorg. Med. Chem. Lett. 2007, 17, 3208–3211. [Google Scholar] [CrossRef]
- Oleynek, J.J.; Barrow, C.J.; Burns, M.P.; Sedlock, D.M.; Murphy, D.J.; Kaplita, P.V.; Sun, H.H.; Copper, R.; Gillum, M.G.; Chadwick, C.C. Anthrones, naturally occurring competitive inhibitors of adenosine-triphosphate-citrate lyase. Drug Dev. Res. 1995, 36, 35–42. [Google Scholar] [CrossRef]
- Denga, Z.; Denga, A.; Luoa, D.; Gongb, D.; Zoua, K.; Pengc, Y.; Guoa, Z. Biotransformation of (-)-(10E,15S)-10,11-Dehydrocurvularin. Nat. Prod. Commun. 2015, 10, 1277–1278. [Google Scholar] [CrossRef]
- Deng, Z.; Wong, N.K.; Guo, Z.; Zou, K.; Xiao, Y.; Zhou, Y. Dehydrocurvularin is a potent antineoplastic agent irreversibly blocking ATP-citrate lyase: Evidence from chemoproteomics. Chem. Commun. 2019, 55, 4194–4197. [Google Scholar] [CrossRef]
- Wang, C.; Ma, J.; Zhang, N.; Yang, Q.; Jin, Y.; Wang, Y. The acetyl-CoA carboxylase enzyme: A target for cancer therapy? Expert Rev. Anticancer Ther. 2015, 15, 667–676. [Google Scholar] [CrossRef]
- Yahagi, N.; Shimano, H.; Hasegawa, K.; Ohashi, K.; Matsuzaka, T.; Najima, Y.; Sekiya, M.; Tomita, S.; Okazaki, H.; Tamura, Y.; et al. Co-ordinate activation of lipogenic enzymes in hepatocellular carcinoma. Eur. J. Cancer 2005, 41, 1316–1322. [Google Scholar] [CrossRef]
- Brownsey, R.W.; Boone, A.N.; Elliott, J.E.; Kulpa, J.E.; Lee, W.M. Regulation of acetyl-CoA carboxylase. Biochem. Soc. Trans. 2006, 34, 223–227. [Google Scholar] [CrossRef]
- Kim, C.W.; Addy, C.; Kusunoki, J.; Anderson, N.N.; Deja, S.; Fu, X.; Burgess, S.C.; Li, C.; Ruddy, M.; Chakravarthy, M.; et al. Acetyl CoA Carboxylase Inhibition Reduces Hepatic Steatosis but Elevates Plasma Triglycerides in Mice and Humans: A Bedside to Bench Investigation. Cell Metab. 2017, 23, 94–406, Erratum in Cell Metab. 2017, 26, 576. [Google Scholar]
- Nishiura, Y.; Matsumura, A.; Kobayashi, N.; Shimazaki, A.; Sakamoto, S.; Kitade, N.; Tonomura, Y.; Ino, A.; Okuno, T. Discovery of a novel olefin derivative as a highly potent and selective acetyl-CoA carboxylase 2 inhibitor with in vivo efficacy. Bioorg. Med. Chem. Lett. 2018, 28, 2498–2503. [Google Scholar] [CrossRef] [PubMed]
- Mizojiri, R.; Asano, M.; Sasaki, M.; Satoh, Y.; Yamamoto, Y.; Sumi, H.; Maezaki, H. The identification and pharmacological evaluation of potent, selective and orally available ACC1 inhibitor. Bioorg. Med. Chem. Lett. 2019, 29, 126749. [Google Scholar] [CrossRef]
- Harriman, G.; Greenwood, J.; Bhat, S.; Huang, X.; Wang, R.; Paul, D.; Tong, L.; Saha, A.K.; Westlin, W.F.; Kapeller, R.; et al. Acetyl-CoA carboxylase inhibition by ND-630 reduces hepatic steatosis, improves insulin sensitivity, and modulates dyslipidemia in rats. Proc. Natl. Acad. Sci. USA 2016, 113, E1796–E1805. [Google Scholar] [CrossRef]
- Almahmoud, S.; Wang, X.; Vennerstrom, J.L.; Zhong, H.A. Conformational Studies of Glucose Transporter 1 (GLUT1) as an Anticancer Drug Target. Molecules 2019, 24, 2159. [Google Scholar] [CrossRef]
- Yang, W. Structural basis of PKM2 regulation. Protein Cell 2015, 6, 238–240. [Google Scholar] [CrossRef]
- Vander Steen, T.; Espinoza, I.; Duran, C.; Casadevall, G.; Serrano-Hervás, E.; Cuyàs, E.; Verdura, S.; Kemble, G.; Kaufmann, S.; McWilliams, R.; et al. Fatty acid synthase (FASN) inhibition cooperates with BH3 mimetic drugs to overcome resistance to mitochondrial apoptosis in pancreatic cancer. Neoplasia 2025, 62, 101143. [Google Scholar] [CrossRef]
- Leone, R.D.; Zhao, L.; Englert, J.M.; Sun, I.M.; Sun, I.H.; Wen, J.; Tam, A.J.; Blosser, R.L.; Siska, P.J.; Rathmell, J.C. Crosstalk between glutaminolysis and mitochondrial respiration in cancer resistance. Trends Cancer 2023, 9, 345–358. [Google Scholar]
- Faubert, B.; Li, L.; Cai, L.; Hensley, C.T.; Kim, J.; Zacharias, L.G.; Yang, C.; Do, Q.N.; Doucette, S.; Burguete, D.; et al. Targeting Lactate Transport Augments Glutaminase Inhibition in Lymphoma. Nat. Cancer 2023, 4, 1193–1205. [Google Scholar]
- Vàzquez, A.; Chandel, N.S.; DeBerardinis, R.J.; Mehta, M.M.; Venneti, S. Phenformin Enhances LDH Inhibitor Sensitivity via AMPK Reprogramming. Cell Metab. 2024, 36, 235–248. [Google Scholar]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Lactylation-Dependent Epigenetic Regulation Links Metabolism to Inflammation. Nat. Rev. Mol. Cell Biol. 2022, 23, 587–604. [Google Scholar]
- Vetma, V.; Yang, X.; Liu, X.; Novakova, M.; Xiang, W.; Garza, C.; Hirsch, M.; Cao, Y. Development of PROTAC Degrader Drugs for Cancer. Annu. Rev. Cancer Biol. 2025, 9, 119–140. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Zhang, H.; Liu, X.; Zhang, Y.; Sun, C.; Wang, C.; Zhang, W. Development of Tumor-Specific PROTAC Prodrugs for LDHA Degradation. Mol. Cancer 2024, 23, 75. [Google Scholar]
- Li, W.; Zhang, H.; Ma, Y.; Tian, W.; Wang, C.; Lin, W.; Han, Y.; Wang, Y. Emerging Targeted Protein Degradation Platforms: AUTACs, ATTECs and LYTACs. Signal Transduct. Target. Ther. 2023, 8, 111. [Google Scholar]
- Rajagopalan, K.; Liu, Y.; Wu, H.; Zhang, J.; Tan, H.; Sun, J.; Liu, J.; Zhao, Y.; Peterson, T.E.; Kaufman, D.R.; et al. DRP-104 Induces Durable Immune-Mediated Tumor Regression. Cancer Immunol. Res. 2024, 12, 420–434. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puxeddu, M.; Silvestri, R.; Regina, G.L. Metabolism, a Blossoming Target for Small-Molecule Anticancer Drugs. Molecules 2025, 30, 3457. https://doi.org/10.3390/molecules30173457
Puxeddu M, Silvestri R, Regina GL. Metabolism, a Blossoming Target for Small-Molecule Anticancer Drugs. Molecules. 2025; 30(17):3457. https://doi.org/10.3390/molecules30173457
Chicago/Turabian StylePuxeddu, Michela, Romano Silvestri, and Giuseppe La Regina. 2025. "Metabolism, a Blossoming Target for Small-Molecule Anticancer Drugs" Molecules 30, no. 17: 3457. https://doi.org/10.3390/molecules30173457
APA StylePuxeddu, M., Silvestri, R., & Regina, G. L. (2025). Metabolism, a Blossoming Target for Small-Molecule Anticancer Drugs. Molecules, 30(17), 3457. https://doi.org/10.3390/molecules30173457







