Effects of Asprosin and Role of TLR4 as a Biomarker in Endometrial Cancer
Abstract
1. Introduction
2. Results
2.1. Expression of Asprosin Receptors in Endometrial Cancer (EC) Cell Lines
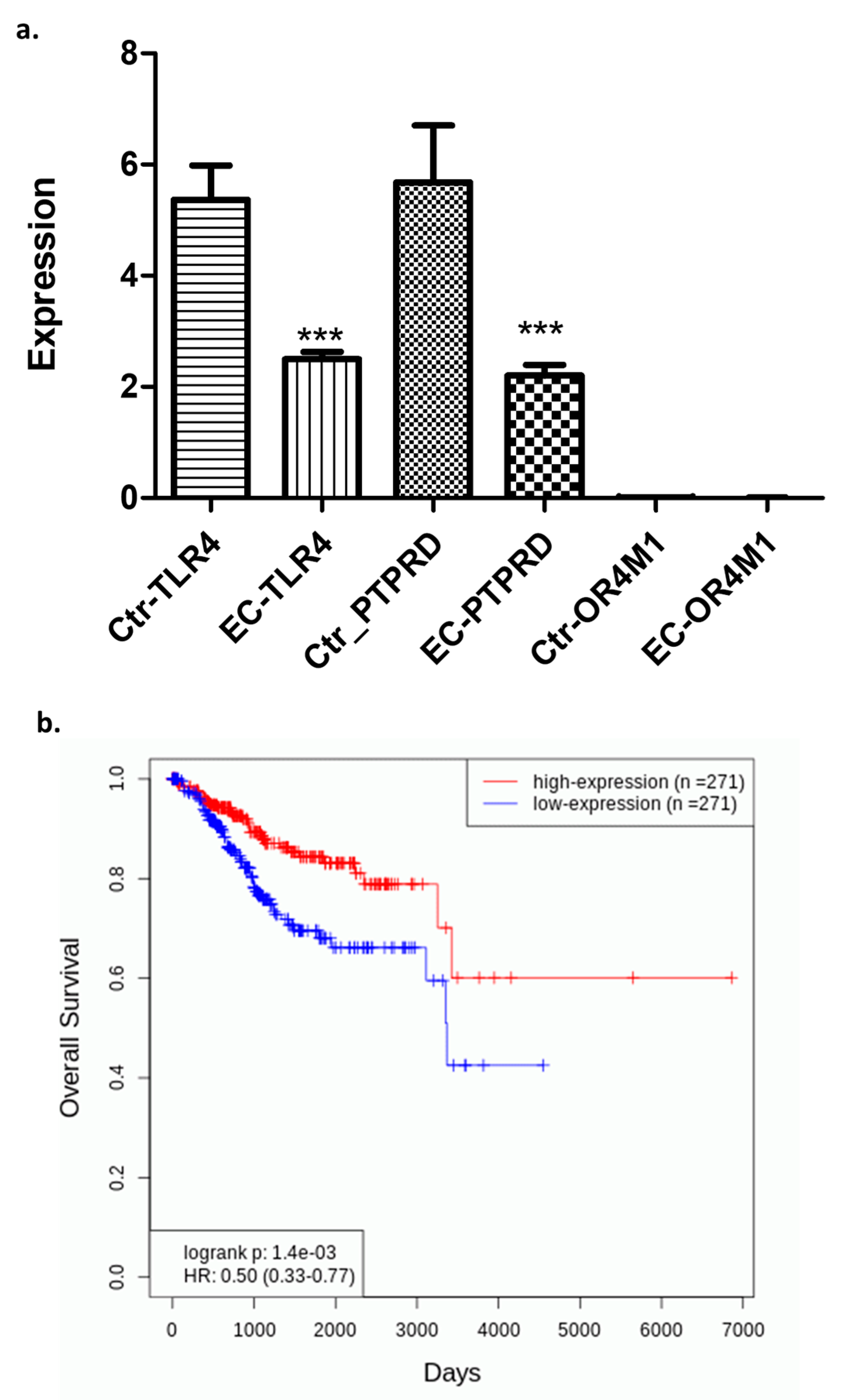
2.2. Differential Expression of TLR4, PTPRD, and OR4M1 in EC: Potential Role of TLR4 as a Biomarker
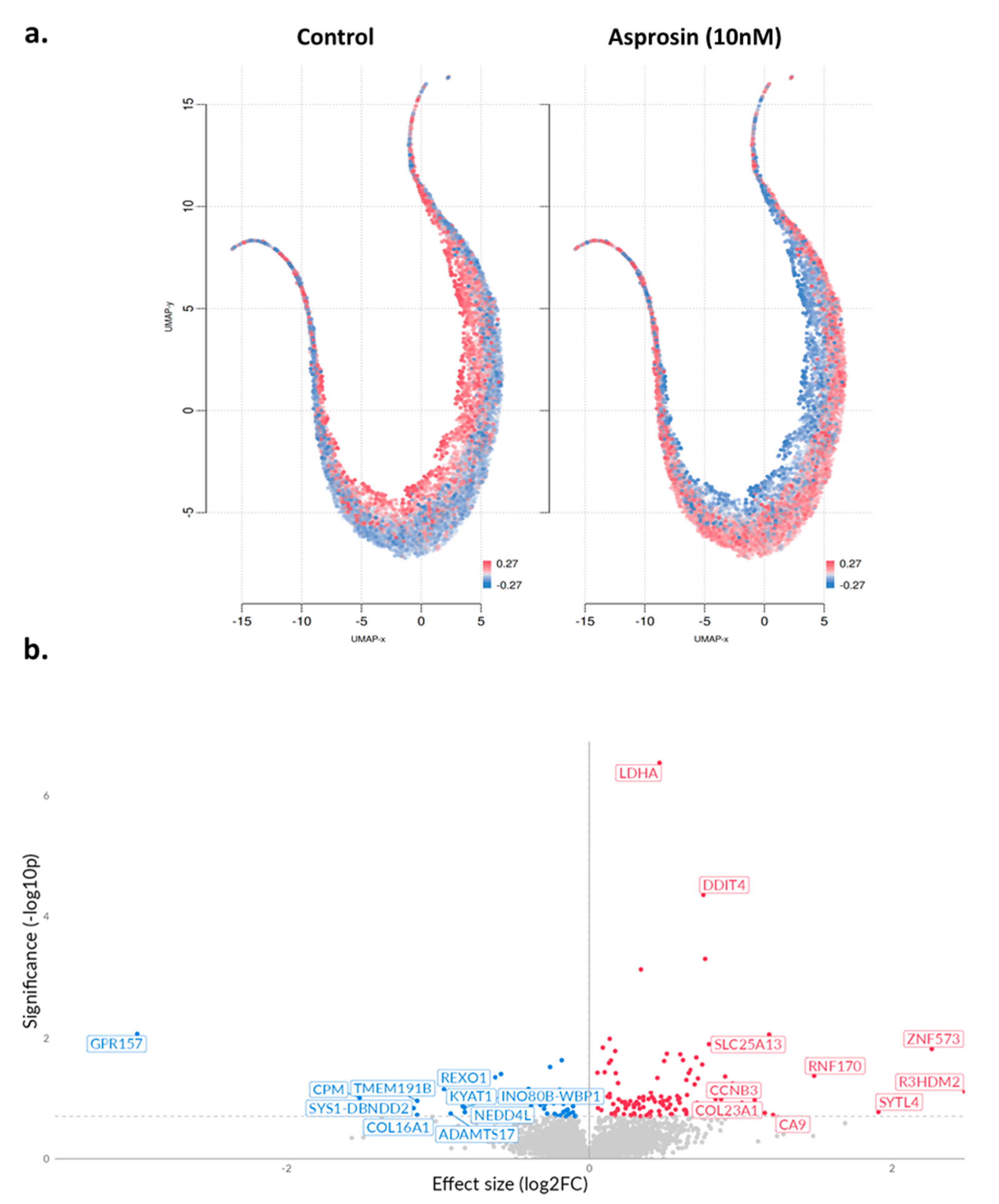
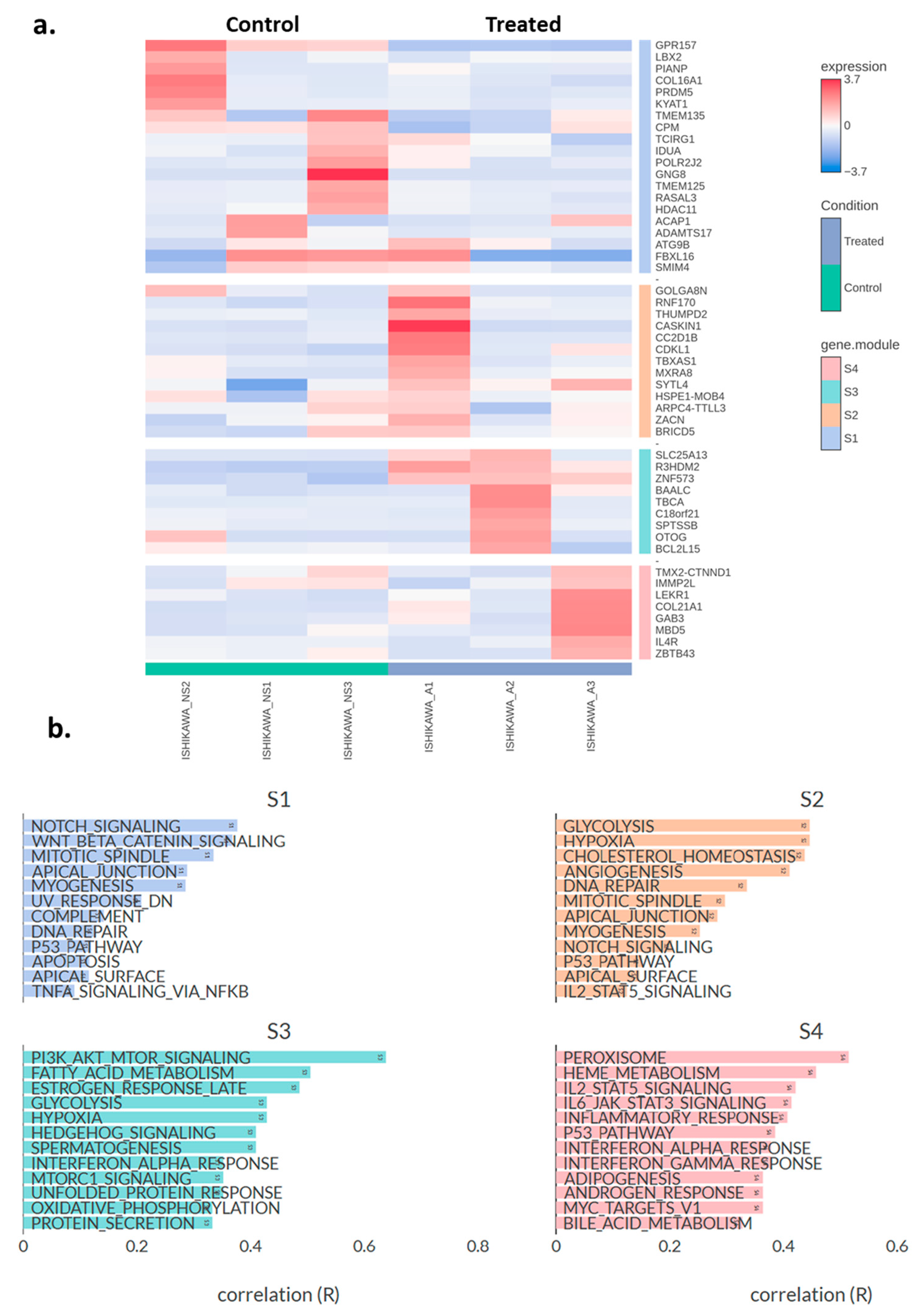
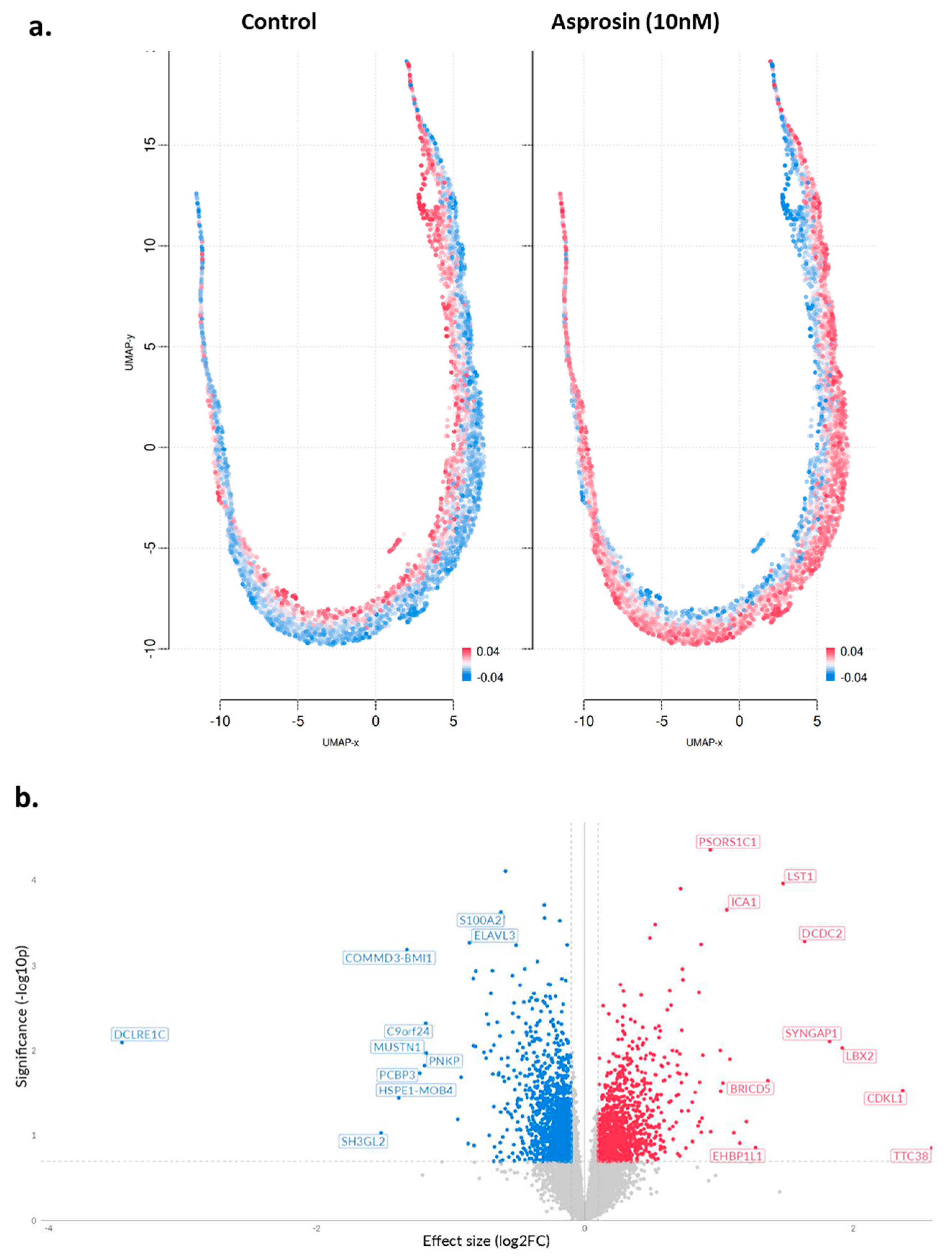
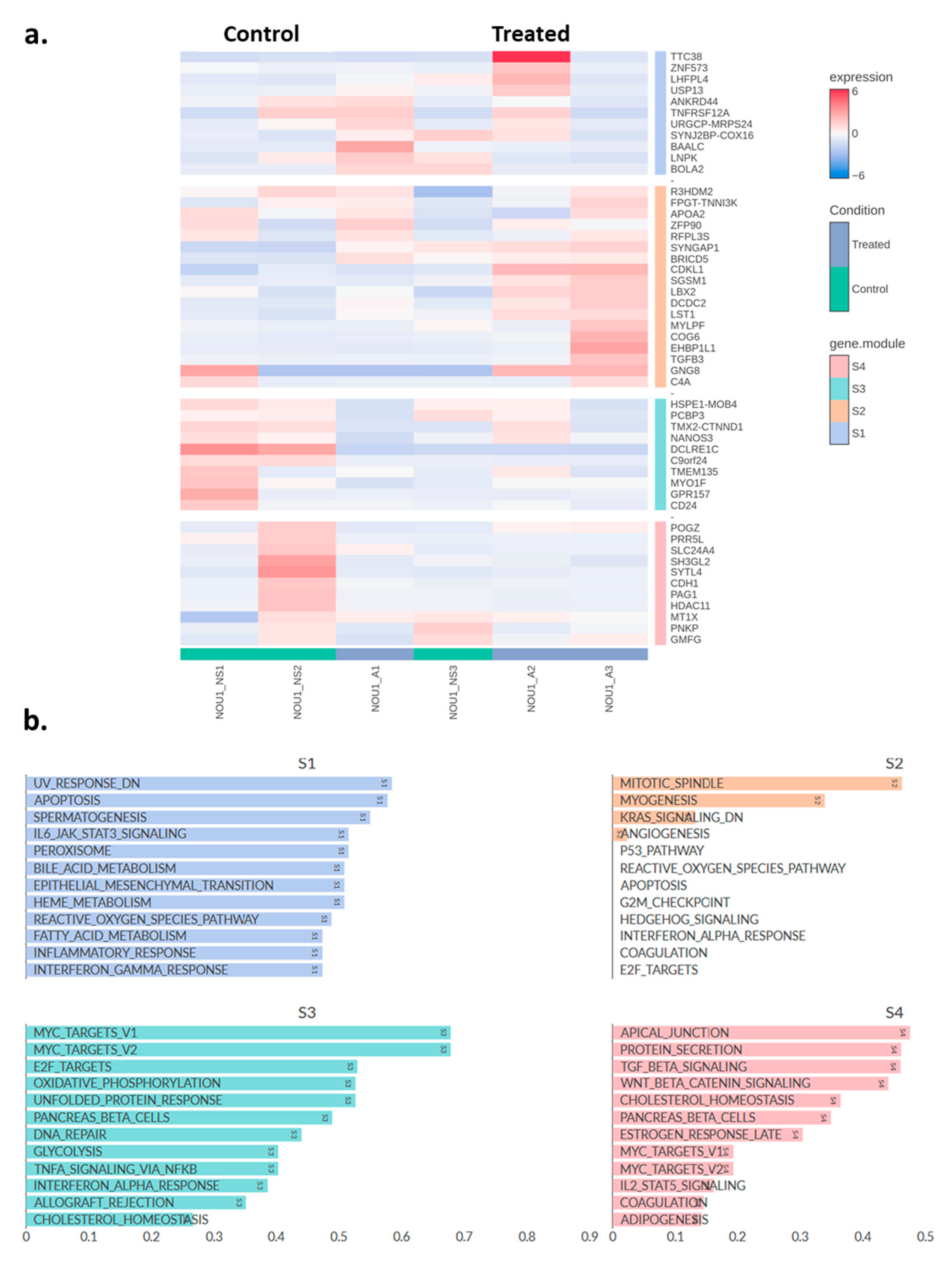
2.3. Transcriptomic Changes of Ishikawa and NOU-1 Cells Treated Wth Asprosin
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Immunofluorescence (IF)
4.3. In Silico Tools
4.4. RNA Sequencing
4.5. RT-qPCR
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Endometrial Cancer | EC |
| body mass index | BMI |
| polycystic ovary syndrome | PCOS |
| European Society of Gynaecological Oncology | ESGO |
| European Society for Radiotherapy and Oncology | ESTRO |
| European Society of Medical Oncology | ESMO |
| metabolic syndrome | MetS |
| Olfactory Receptor Family 4 Subfamily M Member 1 | OR4M1 |
| Toll Like Receptor 4 | TLR4 |
| Protein Tyrosine Phosphatase Receptor Delta | PTPRD |
| G protein-coupled receptor | GPCR |
| transcripts per million | TPM |
| differentially expressed genes | DEGs |
| no supplement | NS |
| epithelial-mesenchymal transition | EMT |
| reactive oxygen species | ROS |
| cAMP-Response Element Binding Proteins | CREB |
| mechanistic target of rapamycin complex 1 | mTORC1 |
| paraformaldehyde | PFA |
| bovine serum albumin | BSA |
Appendix A
| Gene_ID | Gene (Upregulated) | Gene_ID | Gene (Downregulated) |
|---|---|---|---|
| ENSG00000005961.19 | ITGA2B | ENSG00000028277.21 | POU2F2 |
| ENSG00000007350.17 | TKTL1 | ENSG00000079393.21 | DUSP13 |
| ENSG00000009950.16 | MLXIPL | ENSG00000087253.13 | LPCAT2 |
| ENSG00000050767.18 | COL23A1 | ENSG00000095587.9 | TLL2 |
| ENSG00000061656.11 | SPAG4 | ENSG00000099250.18 | NRP1 |
| ENSG00000066336.12 | SPI1 | ENSG00000100060.18 | MFNG |
| ENSG00000070526.15 | ST6GALNAC1 | ENSG00000105497.8 | ZNF175 |
| ENSG00000074047.22 | GLI2 | ENSG00000115590.14 | IL1R2 |
| ENSG00000074317.11 | SNCB | ENSG00000125804.14 | FAM182A |
| ENSG00000074410.14 | CA12 | ENSG00000132837.15 | DMGDH |
| ENSG00000081051.8 | AFP | ENSG00000139318.8 | DUSP6 |
| ENSG00000081237.20 | PTPRC | ENSG00000144063.4 | MALL |
| ENSG00000100336.18 | APOL4 | ENSG00000145934.16 | TENM2 |
| ENSG00000101441.5 | CST4 | ENSG00000151376.18 | ME3 |
| ENSG00000102287.19 | GABRE | ENSG00000151418.12 | ATP6V1G3 |
| ENSG00000102878.18 | HSF4 | ENSG00000159516.9 | SPRR2G |
| ENSG00000103269.14 | RHBDL1 | ENSG00000162267.12 | ITIH3 |
| ENSG00000104419.17 | NDRG1 | ENSG00000162571.14 | TTLL10 |
| ENSG00000104804.8 | TULP2 | ENSG00000163016.10 | ALMS1P1 |
| ENSG00000105650.22 | PDE4C | ENSG00000167588.13 | GPD1 |
| ENSG00000106631.8 | MYL7 | ENSG00000169715.15 | MT1E |
| ENSG00000108405.4 | P2RX1 | ENSG00000175874.10 | CREG2 |
| ENSG00000110777.12 | POU2AF1 | ENSG00000179362.14 | HMGN2P46 |
| ENSG00000112715.25 | VEGFA | ENSG00000182168.15 | UNC5C |
| ENSG00000114268.12 | PFKFB4 | ENSG00000183230.18 | CTNNA3 |
| ENSG00000114626.18 | ABTB1 | ENSG00000185271.9 | KLHL33 |
| ENSG00000115257.15 | PCSK4 | ENSG00000185864.17 | NPIPB4 |
| ENSG00000115548.17 | KDM3A | ENSG00000187243.16 | MAGED4B |
| ENSG00000116014.10 | KISS1R | ENSG00000196628.20 | TCF4 |
| ENSG00000117586.11 | TNFSF4 | ENSG00000197714.9 | ZNF460 |
| ENSG00000118407.15 | FILIP1 | ENSG00000204711.9 | C9orf135 |
| ENSG00000121966.7 | CXCR4 | ENSG00000211716.2 | TRBV9 |
| ENSG00000122756.15 | CNTFR | ENSG00000212724.3 | KRTAP2-3 |
| ENSG00000123977.10 | DAW1 | ENSG00000213309.3 | RPL9P18 |
| ENSG00000124116.19 | WFDC3 | ENSG00000214145.7 | LINC00887 |
| ENSG00000124749.17 | COL21A1 | ENSG00000218208.1 | RPS27AP11 |
| ENSG00000126233.2 | SLURP1 | ENSG00000224194.1 | AC008278.3 |
| ENSG00000127129.10 | EDN2 | ENSG00000227337.1 | RPL23AP43 |
| ENSG00000130540.14 | SULT4A1 | ENSG00000227777.1 | RP4-738P11.3 |
| ENSG00000130595.19 | TNNT3 | ENSG00000228624.7 | HDAC2-AS2 |
| ENSG00000132793.12 | LPIN3 | ENSG00000228793.2 | RP1-223B1.1 |
| ENSG00000134107.5 | BHLHE40 | ENSG00000229660.1 | RP5-1142J19.1 |
| ENSG00000134160.15 | TRPM1 | ENSG00000231704.6 | AC004895.4 |
| ENSG00000134365.13 | CFHR4 | ENSG00000231738.11 | TSPAN19 |
| ENSG00000134874.18 | DZIP1 | ENSG00000231971.6 | CT69 |
| ENSG00000137573.14 | SULF1 | ENSG00000233196.2 | GS1-304P7.1 |
| ENSG00000138696.11 | BMPR1B | ENSG00000234262.1 | RP1-20N18.4 |
| ENSG00000139445.18 | FOXN4 | ENSG00000234460.2 | XXyac-YM21GA2.4 |
| ENSG00000142609.19 | CFAP74 | ENSG00000236512.1 | RP3-336K20__B.2 |
| ENSG00000142973.14 | CYP4B1 | ENSG00000237664.1 | LINC00316 |
| ENSG00000143127.13 | ITGA10 | ENSG00000240179.1 | RPL26P3 |
| ENSG00000147256.12 | ARHGAP36 | ENSG00000241870.1 | CTB-14A14.1 |
| ENSG00000148926.10 | ADM | ENSG00000242288.9 | BMS1P4-AGAP5 |
| ENSG00000150625.16 | GPM6A | ENSG00000242439.1 | CTD-2349P21.1 |
| ENSG00000150722.11 | PPP1R1C | ENSG00000249464.6 | LINC01091 |
| ENSG00000151062.15 | CACNA2D4 | ENSG00000250337.7 | PURPL |
| ENSG00000151640.13 | DPYSL4 | ENSG00000250891.2 | LINC02208 |
| ENSG00000153237.18 | CCDC148 | ENSG00000251095.7 | RP11-115D19.1 |
| ENSG00000159167.12 | STC1 | ENSG00000253217.1 | KB-1991G8.1 |
| ENSG00000159871.15 | LYPD5 | ENSG00000254237.6 | RP11-115J16.2 |
| ENSG00000160219.12 | GAB3 | ENSG00000254508.5 | FBXO3-DT |
| ENSG00000162576.17 | MXRA8 | ENSG00000258599.2 | RP11-84C10.1 |
| ENSG00000162931.12 | TRIM17 | ENSG00000258754.8 | LINC01579 |
| ENSG00000163516.14 | ANKZF1 | ENSG00000259656.1 | RP11-325E5.1 |
| ENSG00000163631.17 | ALB | ENSG00000260352.1 | CTD-2288F12.1 |
| ENSG00000164362.21 | TERT | ENSG00000260475.1 | RP11-85A1.3 |
| ENSG00000164406.8 | LEAP2 | ENSG00000260615.1 | RPL23AP97 |
| ENSG00000164463.12 | CREBRF | ENSG00000260911.2 | RP11-196G11.2 |
| ENSG00000164877.19 | MICALL2 | ENSG00000262213.1 | AC144836.1 |
| ENSG00000165061.15 | ZMAT4 | ENSG00000264655.1 | PPIAP54 |
| ENSG00000165495.16 | PKNOX2 | ENSG00000267123.7 | SCAT1 |
| ENSG00000165935.9 | SMCO2 | ENSG00000267674.1 | RP11-813F20.2 |
| ENSG00000167178.16 | ISLR2 | ENSG00000267737.1 | AC061992.2 |
| ENSG00000167612.13 | ANKRD33 | ENSG00000268266.1 | AC003005.2 |
| ENSG00000167702.13 | KIFC2 | ENSG00000268350.8 | FAM156A |
| ENSG00000168060.16 | NAALADL1 | ENSG00000269489.1 | RP11-98D18.17 |
| ENSG00000168209.6 | DDIT4 | ENSG00000270049.3 | ATP6V0D1-DT |
| ENSG00000170074.19 | FAM153A | ENSG00000271021.1 | RP5-878I13.2 |
| ENSG00000170989.10 | S1PR1 | ENSG00000271185.1 | RP5-855F16.1 |
| ENSG00000171388.12 | APLN | ENSG00000271200.1 | PATJ-DT |
| ENSG00000172238.6 | ATOH1 | ENSG00000271369.1 | RP11-350D17.3 |
| ENSG00000172650.15 | AGAP5 | ENSG00000271803.1 | RP1-63M2.5 |
| ENSG00000175018.13 | TEX36 | ENSG00000272646.1 | RP11-188P17.2 |
| ENSG00000175265.18 | GOLGA8A | ENSG00000272989.1 | LINC02012 |
| ENSG00000175766.13 | EIF4E1B | ENSG00000273143.2 | DUSP5-DT |
| ENSG00000176171.11 | BNIP3 | ENSG00000273321.1 | RP11-621L6.3 |
| ENSG00000177453.7 | NIM1K | ENSG00000273384.1 | RP5-1098D14.1 |
| ENSG00000178015.5 | GPR150 | ENSG00000273542.2 | H4C12 |
| ENSG00000181856.15 | SLC2A4 | ENSG00000274012.1 | RN7SL2 |
| ENSG00000182310.15 | SPACA6 | ENSG00000274549.1 | RP11-676B18.2 |
| ENSG00000182511.12 | FES | ENSG00000274813.1 | RP11-21I4.3 |
| ENSG00000182985.18 | CADM1 | ENSG00000274818.1 | RP1-292L20.3 |
| ENSG00000184144.12 | CNTN2 | ENSG00000275409.1 | RP11-131L12.4 |
| ENSG00000184564.11 | SLITRK6 | ENSG00000276077.4 | CH507-254M2.2 |
| ENSG00000184786.6 | DYNLT2 | ENSG00000277027.1 | RMRP |
| ENSG00000184925.12 | LCN12 | ENSG00000277067.4 | CH507-254M2.1 |
| ENSG00000185101.13 | ANO9 | ENSG00000277991.4 | CH507-338C24.1 |
| ENSG00000185189.18 | NRBP2 | ENSG00000278558.5 | TMEM191B |
| ENSG00000187492.9 | CDHR4 | ENSG00000282787.1 | RP11-151A10.3 |
| ENSG00000197358.9 | BNIP3P1 | ENSG00000284696.2 | LINC02808 |
| ENSG00000197745.3 | SCGB1D4 | ENSG00000284735.1 | RP4-736L20.4 |
| ENSG00000197837.3 | H4-16 | ENSG00000285201.1 | RP4-552O12.3 |
| ENSG00000203685.10 | STUM | ENSG00000286064.1 | RP4-657E11.11 |
| ENSG00000203780.12 | FANK1 | ||
| ENSG00000204420.10 | MPIG6B | ||
| ENSG00000205702.11 | CYP2D7 | ||
| ENSG00000214140.11 | PRCD | ||
| ENSG00000215252.12 | GOLGA8B | ||
| ENSG00000223718.3 | AC093107.7 | ||
| ENSG00000223756.7 | TSSC2 | ||
| ENSG00000224635.2 | RP4-564F22.5 | ||
| ENSG00000225255.6 | PSLNR | ||
| ENSG00000225605.3 | RP11-550H2.1 | ||
| ENSG00000226043.2 | AP000705.7 | ||
| ENSG00000226250.2 | LINC00408 | ||
| ENSG00000226321.5 | CROCC2 | ||
| ENSG00000226956.1 | AP000432.2 | ||
| ENSG00000227877.7 | MRLN | ||
| ENSG00000228224.3 | NACA4P | ||
| ENSG00000228232.1 | GAPDHP1 | ||
| ENSG00000229474.8 | PATL2 | ||
| ENSG00000229609.1 | LINC01079 | ||
| ENSG00000230216.1 | HSPB1P2 | ||
| ENSG00000230323.6 | LINC00159 | ||
| ENSG00000231359.3 | AC072052.7 | ||
| ENSG00000231650.1 | RFESDP1 | ||
| ENSG00000231720.1 | RP11-568A7.3 | ||
| ENSG00000232065.1 | LINC01063 | ||
| ENSG00000232354.8 | VIPR1-AS1 | ||
| ENSG00000233090.1 | AC015922.7 | ||
| ENSG00000233198.4 | RNF224 | ||
| ENSG00000233928.6 | RP11-305F18.1 | ||
| ENSG00000234648.1 | AL162151.3 | ||
| ENSG00000235308.1 | RP11-82K18.2 | ||
| ENSG00000236581.9 | STARD13-AS | ||
| ENSG00000241529.3 | RN7SL767P | ||
| ENSG00000242101.3 | RN7SL416P | ||
| ENSG00000242338.6 | BMS1P4 | ||
| ENSG00000244573.3 | RPL30P11 | ||
| ENSG00000245750.10 | DRAIC | ||
| ENSG00000245870.4 | LINC00682 | ||
| ENSG00000247095.3 | MIR210HG | ||
| ENSG00000248099.4 | INSL3 | ||
| ENSG00000248459.1 | RP11-93K22.1 | ||
| ENSG00000249700.9 | SRD5A3-AS1 | ||
| ENSG00000249937.8 | LINC02223 | ||
| ENSG00000249965.2 | CDC42P4 | ||
| ENSG00000250122.1 | CTD-2013M15.1 | ||
| ENSG00000250241.6 | RP11-9G1.3 | ||
| ENSG00000250305.9 | TRMT9B | ||
| ENSG00000254599.1 | RP11-115E19.1 | ||
| ENSG00000254701.3 | RP11-1415C14.4 | ||
| ENSG00000254842.6 | LINC02551 | ||
| ENSG00000255647.3 | CTD-2373J6.2 | ||
| ENSG00000255966.1 | RP5-940J5.3 | ||
| ENSG00000256995.8 | RP11-114G22.1 | ||
| ENSG00000259039.3 | RP11-409I10.2 | ||
| ENSG00000259315.1 | ACTG1P17 | ||
| ENSG00000259704.2 | CTD-3094K11.1 | ||
| ENSG00000260025.1 | CRIM1-DT | ||
| ENSG00000261302.6 | NUP93-DT | ||
| ENSG00000261341.7 | CTD-2568A17.1 | ||
| ENSG00000261433.1 | CTC-297N7.1 | ||
| ENSG00000261786.1 | RP4-555D20.2 | ||
| ENSG00000262117.5 | BCAR4 | ||
| ENSG00000263154.1 | RP11-1055B8.2 | ||
| ENSG00000265542.5 | RP11-60A24.3 | ||
| ENSG00000265690.7 | RP11-5A19.5 | ||
| ENSG00000266714.9 | MYO15B | ||
| ENSG00000270069.1 | MIR222HG | ||
| ENSG00000270917.1 | RP11-27I1.6 | ||
| ENSG00000272128.1 | KB-1836B5.4 | ||
| ENSG00000272645.3 | GTF2IP20 | ||
| ENSG00000272784.1 | RP11-335L23.5 | ||
| ENSG00000272795.1 | GAR1-DT | ||
| ENSG00000273437.1 | RP11-434H6.7 | ||
| ENSG00000273712.1 | RP5-874C20.7 | ||
| ENSG00000274156.1 | RP11-407N8.6 | ||
| ENSG00000277453.1 | CTC-492K19.7 | ||
| ENSG00000278595.1 | RP5-1068H6.6 | ||
| ENSG00000279208.1 | bP-21264C1.1 | ||
| ENSG00000284523.2 | RP13-968A2.1 | ||
| ENSG00000285287.1 | RP11-643E14.2 | ||
| ENSG00000286794.1 | GHc-351F8.3 | ||
| ENSG00000287542.1 | RP11-2I7.1 | ||
| ENSG00000287737.1 | RP13-516M14.11 |
| Gene_ID | Gene (Upregulated) | Gene_ID | Gene (Downregulated) |
|---|---|---|---|
| ENSG00000170419.11 | VSTM2A | ENSG00000058335.16 | RASGRF1 |
| ENSG00000008517.19 | IL32 | ENSG00000135362.14 | PRR5L |
| ENSG00000102057.10 | KCND1 | ENSG00000158560.14 | DYNC1I1 |
| ENSG00000109339.24 | MAPK10 | ENSG00000189292.16 | ALKAL2 |
| ENSG00000132837.15 | DMGDH | ENSG00000244128.7 | LINC01322 |
| ENSG00000140853.16 | NLRC5 | ENSG00000287193.1 | RP13-20L14.11 |
| ENSG00000155511.18 | GRIA1 | ENSG00000105976.16 | MET |
| ENSG00000184908.19 | CLCNKB | ENSG00000125207.7 | PIWIL1 |
| ENSG00000215478.8 | CES5AP1 | ENSG00000148346.12 | LCN2 |
| ENSG00000229140.11 | CCDC26 | ENSG00000152670.19 | DDX4 |
| ENSG00000234722.5 | LINC01287 | ENSG00000163993.7 | S100P |
| ENSG00000271138.1 | IGLVIVOR22-1 | ENSG00000165495.16 | PKNOX2 |
| ENSG00000272255.1 | CTD-3224K15.3 | ENSG00000169862.19 | CTNND2 |
| ENSG00000274012.1 | RN7SL2 | ENSG00000173083.16 | HPSE |
| ENSG00000003147.19 | ICA1 | ENSG00000179520.11 | SLC17A8 |
| ENSG00000077092.19 | RARB | ENSG00000189129.14 | PLAC9 |
| ENSG00000100884.9 | CPNE6 | ENSG00000204140.10 | CLPSL1 |
| ENSG00000106128.19 | GHRHR | ENSG00000206172.8 | HBA1 |
| ENSG00000120555.13 | SEPTIN7P9 | ENSG00000224535.2 | LINC02771 |
| ENSG00000120907.18 | ADRA1A | ENSG00000230216.1 | HSPB1P2 |
| ENSG00000124440.16 | HIF3A | ENSG00000232732.11 | AC073043.1 |
| ENSG00000130377.14 | ACSBG2 | ENSG00000237126.8 | AC073254.1 |
| ENSG00000130592.17 | LSP1 | ENSG00000239705.2 | RP11-65N13.8 |
| ENSG00000147443.13 | DOK2 | ENSG00000248538.8 | RP11-115J16.1 |
| ENSG00000156113.24 | KCNMA1 | ENSG00000253816.3 | GUSBP14 |
| ENSG00000158482.10 | SNX29P1 | ENSG00000261314.1 | RP11-359E8.5 |
| ENSG00000169064.13 | ZBBX | ENSG00000261341.7 | CTD-2568A17.1 |
| ENSG00000169877.10 | AHSP | ENSG00000266950.1 | CTD-2623N2.5 |
| ENSG00000170374.6 | SP7 | ENSG00000271734.1 | RP1-111B22.3 |
| ENSG00000175265.18 | GOLGA8A | ENSG00000272361.2 | GS1-166A23.2 |
| ENSG00000179954.16 | SSC5D | ENSG00000273375.1 | RP11-803P9.1 |
| ENSG00000182621.18 | PLCB1 | ENSG00000276832.2 | CDRT15P6 |
| ENSG00000186642.16 | PDE2A | ENSG00000277764.1 | ENSG10010137820.1 |
| ENSG00000188257.12 | PLA2G2A | ENSG00000286102.1 | FAM246A |
| ENSG00000188517.16 | COL25A1 | ENSG00000006606.9 | CCL26 |
| ENSG00000189068.11 | VSTM1 | ENSG00000018236.15 | CNTN1 |
| ENSG00000189149.12 | CRYM-AS1 | ENSG00000107105.15 | ELAVL2 |
| ENSG00000214193.11 | SH3D21 | ENSG00000107954.10 | NEURL1 |
| ENSG00000223855.2 | HRAT92 | ENSG00000110203.9 | FOLR3 |
| ENSG00000224389.9 | C4B | ENSG00000111319.13 | SCNN1A |
| ENSG00000224637.1 | PDSS1P2 | ENSG00000116016.14 | EPAS1 |
| ENSG00000225783.8 | MIAT | ENSG00000124507.11 | PACSIN1 |
| ENSG00000226889.3 | RP11-474I16.8 | ENSG00000124788.19 | ATXN1 |
| ENSG00000227438.1 | AP001471.1 | ENSG00000131620.17 | ANO1 |
| ENSG00000228526.7 | MIR34AHG | ENSG00000134121.10 | CHL1 |
| ENSG00000230096.1 | RP11-34C15.2 | ENSG00000134531.10 | EMP1 |
| ENSG00000232801.1 | SDCBPP3 | ENSG00000140682.19 | TGFB1I1 |
| ENSG00000242866.10 | STRC | ENSG00000141668.10 | CBLN2 |
| ENSG00000245532.9 | NEAT1 | ENSG00000144668.12 | ITGA9 |
| ENSG00000247982.6 | LINC00926 | ENSG00000151490.15 | PTPRO |
| ENSG00000249476.2 | CTD-2587M2.1 | ENSG00000158517.15 | NCF1 |
| ENSG00000253848.1 | RP11-10N23.5 | ENSG00000164398.15 | ACSL6 |
| ENSG00000254833.1 | RP11-50B3.2 | ENSG00000172349.18 | IL16 |
| ENSG00000256091.1 | MTRF1LP1 | ENSG00000173926.6 | MARCHF3 |
| ENSG00000260628.5 | RP11-1166P10.1 | ENSG00000175564.13 | UCP3 |
| ENSG00000262877.5 | RP11-1055B8.4 | ENSG00000176358.15 | TAC4 |
| ENSG00000263946.1 | RP11-434D2.11 | ENSG00000182676.5 | PPP1R27 |
| ENSG00000267505.1 | CTC-296K1.3 | ENSG00000185078.3 | RPL9P14 |
| ENSG00000269993.1 | AF003625.3 | ENSG00000185847.8 | LINC01405 |
| ENSG00000271265.1 | RP11-230C9.4 | ENSG00000198838.14 | RYR3 |
| ENSG00000272192.1 | PDE7A-DT | ENSG00000204625.10 | HCG9 |
| ENSG00000280325.1 | AC074183.3 | ENSG00000205277.10 | MUC12 |
| ENSG00000286379.1 | RP11-268J15.6 | ENSG00000214243.3 | AC004980.10 |
| ENSG00000287263.1 | CTD-2201E18.8 | ENSG00000224382.2 | LINC00703 |
| ENSG00000287331.1 | RP11-29H20.2 | ENSG00000224566.2 | FAM96AP2 |
| ENSG00000011590.14 | ZBTB32 | ENSG00000225099.1 | ATP6V1E1P1 |
| ENSG00000055732.13 | MCOLN3 | ENSG00000226856.7 | THORLNC |
| ENSG00000056558.11 | TRAF1 | ENSG00000228232.1 | GAPDHP1 |
| ENSG00000066032.19 | CTNNA2 | ENSG00000228541.1 | AC093159.1 |
| ENSG00000066405.13 | CLDN18 | ENSG00000229203.1 | AC103564.7 |
| ENSG00000072422.17 | RHOBTB1 | ENSG00000229914.1 | RP11-404O13.4 |
| ENSG00000073605.19 | GSDMB | ENSG00000230268.3 | SSU72P8 |
| ENSG00000090238.12 | YPEL3 | ENSG00000231327.1 | LINC01816 |
| ENSG00000099338.23 | CATSPERG | ENSG00000232028.1 | AC007391.2 |
| ENSG00000102271.14 | KLHL4 | ENSG00000232065.1 | LINC01063 |
| ENSG00000105339.11 | DENND3 | ENSG00000232599.1 | RP1-161N10.1 |
| ENSG00000108602.18 | ALDH3A1 | ENSG00000233017.3 | RP5-908M14.5 |
| ENSG00000108849.8 | PPY | ENSG00000233084.2 | RPL23AP25 |
| ENSG00000111644.8 | ACRBP | ENSG00000234175.1 | RP11-730A19.9 |
| ENSG00000120645.12 | IQSEC3 | ENSG00000234318.1 | RP4-771M4.3 |
| ENSG00000126583.12 | PRKCG | ENSG00000235126.1 | AC128709.3 |
| ENSG00000128655.18 | PDE11A | ENSG00000235214.1 | FAM83C-AS1 |
| ENSG00000130518.17 | IQCN | ENSG00000235828.5 | RPL36AP26 |
| ENSG00000130653.16 | PNPLA7 | ENSG00000243069.8 | ARHGEF26-AS1 |
| ENSG00000131480.9 | AOC2 | ENSG00000249148.2 | AC006445.7 |
| ENSG00000132205.11 | EMILIN2 | ENSG00000250662.1 | HNRNPKP5 |
| ENSG00000132259.13 | CNGA4 | ENSG00000254731.1 | CTD-2005H7.1 |
| ENSG00000137098.14 | SPAG8 | ENSG00000255362.1 | LINC02761 |
| ENSG00000138795.10 | LEF1 | ENSG00000256312.1 | RP13-977J11.8 |
| ENSG00000139890.10 | REM2 | ENSG00000258754.8 | LINC01579 |
| ENSG00000144868.14 | TMEM108 | ENSG00000261564.1 | RP11-616M22.10 |
| ENSG00000146038.12 | DCDC2 | ENSG00000263711.6 | LINC02864 |
| ENSG00000151790.9 | TDO2 | ENSG00000271002.1 | RP11-599B13.8 |
| ENSG00000153071.15 | DAB2 | ENSG00000272057.1 | ANKH-DT |
| ENSG00000154529.15 | CNTNAP3B | ENSG00000275773.1 | RP11-343D24.2 |
| ENSG00000159958.7 | TNFRSF13C | ENSG00000277602.1 | AC005363.11 |
| ENSG00000160201.12 | U2AF1 | ENSG00000278077.1 | RP11-157L3.10 |
| ENSG00000161509.14 | GRIN2C | ENSG00000278276.1 | RP3-324O17.8 |
| ENSG00000162777.17 | DENND2D | ENSG00000278463.2 | H2AC4 |
| ENSG00000163017.14 | ACTG2 | ENSG00000280331.1 | RP11-545D22.1 |
| ENSG00000163083.6 | INHBB | ENSG00000283698.1 | VINAC1P |
| ENSG00000164037.17 | SLC9B1 | ENSG00000286064.1 | RP4-657E11.11 |
| ENSG00000164330.17 | EBF1 | ENSG00000286508.1 | AC000050.1 |
| ENSG00000166833.22 | NAV2 | ENSG00000286850.1 | RP11-533K9.5 |
| ENSG00000167037.19 | SGSM1 | ENSG00000287961.1 | CTD-2562J15.8 |
| ENSG00000167588.13 | GPD1 | ||
| ENSG00000168785.8 | TSPAN5 | ||
| ENSG00000169885.10 | CALML6 | ||
| ENSG00000172005.11 | MAL | ||
| ENSG00000172014.12 | ANKRD20A4P | ||
| ENSG00000172232.10 | AZU1 | ||
| ENSG00000174469.23 | CNTNAP2 | ||
| ENSG00000175097.8 | RAG2 | ||
| ENSG00000180712.4 | LINC02363 | ||
| ENSG00000183929.7 | DUSP5P1 | ||
| ENSG00000185215.11 | TNFAIP2 | ||
| ENSG00000186115.13 | CYP4F2 | ||
| ENSG00000187045.19 | TMPRSS6 | ||
| ENSG00000188710.3 | QRFP | ||
| ENSG00000188981.11 | MSANTD1 | ||
| ENSG00000196338.14 | NLGN3 | ||
| ENSG00000196421.9 | C20orf204 | ||
| ENSG00000196604.13 | POTEF | ||
| ENSG00000197558.13 | SSPOP | ||
| ENSG00000197714.9 | ZNF460 | ||
| ENSG00000197826.12 | CFAP299 | ||
| ENSG00000197971.16 | MBP | ||
| ENSG00000204540.11 | PSORS1C1 | ||
| ENSG00000213199.8 | ASIC3 | ||
| ENSG00000214279.13 | SCART1 | ||
| ENSG00000214402.7 | LCNL1 | ||
| ENSG00000214784.4 | RPS3AP21 | ||
| ENSG00000214900.11 | LINC01588 | ||
| ENSG00000218208.1 | RPS27AP11 | ||
| ENSG00000220161.4 | LINC02076 | ||
| ENSG00000223749.11 | MIR503HG | ||
| ENSG00000224019.1 | RPL21P32 | ||
| ENSG00000224473.1 | CCND3P1 | ||
| ENSG00000224934.5 | RP11-441O15.3 | ||
| ENSG00000225156.2 | AC012354.6 | ||
| ENSG00000225828.1 | FAM229A | ||
| ENSG00000226229.2 | RPLP0P1 | ||
| ENSG00000226314.8 | ZNF192P1 | ||
| ENSG00000226985.7 | LINC01203 | ||
| ENSG00000227036.8 | LINC00511 | ||
| ENSG00000227232.5 | WASH7P | ||
| ENSG00000228649.9 | SNHG26 | ||
| ENSG00000229807.13 | XIST | ||
| ENSG00000229869.1 | RP11-363N22.2 | ||
| ENSG00000230914.3 | KIF19BP | ||
| ENSG00000230994.1 | FGFR3P1 | ||
| ENSG00000231057.4 | RP11-122M14.1 | ||
| ENSG00000231120.3 | BTF3P10 | ||
| ENSG00000233178.7 | RP11-88I18.2 | ||
| ENSG00000234296.1 | RP13-16H11.7 | ||
| ENSG00000234367.1 | PFN1P3 | ||
| ENSG00000234498.3 | RPL13AP20 | ||
| ENSG00000237094.12 | RP4-669L17.4 | ||
| ENSG00000242715.8 | CCDC169 | ||
| ENSG00000243802.2 | RP11-390K5.1 | ||
| ENSG00000245017.2 | LINC02453 | ||
| ENSG00000245750.10 | DRAIC | ||
| ENSG00000246922.9 | UBAP1L | ||
| ENSG00000248124.8 | RRN3P1 | ||
| ENSG00000250420.8 | AACSP1 | ||
| ENSG00000251221.1 | LINC01337 | ||
| ENSG00000253641.6 | LINCR-0001 | ||
| ENSG00000254632.2 | RP11-21L23.4 | ||
| ENSG00000255452.1 | RP11-107P7.1 | ||
| ENSG00000255741.1 | RP11-757G1.5 | ||
| ENSG00000256732.1 | RP11-407A16.3 | ||
| ENSG00000259030.8 | FPGT-TNNI3K | ||
| ENSG00000259156.7 | CHEK2P2 | ||
| ENSG00000259215.1 | RP11-253M7.4 | ||
| ENSG00000260236.1 | RP11-708J19.1 | ||
| ENSG00000260911.2 | RP11-196G11.2 | ||
| ENSG00000261408.7 | TEN1-CDK3 | ||
| ENSG00000266274.2 | RN7SL138P | ||
| ENSG00000267224.1 | AC005498.4 | ||
| ENSG00000268307.1 | LINC02560 | ||
| ENSG00000270019.1 | RP11-141B14.1 | ||
| ENSG00000270248.1 | CTC-471J1.10 | ||
| ENSG00000271327.1 | RP11-1109F11.3 | ||
| ENSG00000272631.1 | RP11-359E3.4 | ||
| ENSG00000272645.3 | GTF2IP20 | ||
| ENSG00000272662.1 | TIMMDC1-DT | ||
| ENSG00000272942.1 | CTA-246H3.12 | ||
| ENSG00000273230.1 | RP11-1246C19.1 | ||
| ENSG00000275265.1 | RP11-15J22.8 | ||
| ENSG00000277112.3 | ANKRD20A21P | ||
| ENSG00000277157.2 | H4C4 | ||
| ENSG00000278546.1 | RP1-41C23.4 | ||
| ENSG00000278683.1 | RP11-132A1.6 | ||
| ENSG00000279161.1 | CTB-12A17.2 | ||
| ENSG00000279617.1 | AC005796.2 | ||
| ENSG00000280047.1 | CTC-463A16.1 | ||
| ENSG00000283189.2 | RP11-949J7.8 | ||
| ENSG00000286058.1 | RP11-32C20.1 | ||
| ENSG00000286562.1 | RP1-128O3.7 | ||
| ENSG00000286656.1 | CTD-2201E18.7 | ||
| ENSG00000287506.1 | RP11-443G13.4 | ||
| ENSG00000287766.1 | AC006130.5 |
References
- SHutt, D.; Mihaies, E.; Karteris, A.; Michael, A.; Payne, M.; Chatterjee, J. Statistical Meta-Analysis of Risk Factors for Endometrial Cancer and Development of a Risk Prediction Model Using an Artificial Neural Network Algorithm. Cancers 2021, 13, 3689. [Google Scholar] [CrossRef]
- Johnson, J.-E.; Daley, D.; Tarta, C.; Stanciu, P.I. Risk of endometrial cancer in patients with polycystic ovarian syndrome: A meta-analysis. Oncol. Lett. 2023, 25, 168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, H.; Yang, S.; Zhang, J.; Qian, L.; Chen, X. Overweight, Obesity and Endometrial Cancer Risk: Results from a Systematic Review and Meta-Analysis. Int. J. Biol. Markers 2014, 29, e21–e29. [Google Scholar] [CrossRef]
- Höhn, A.K.; Brambs, C.E.; Hiller, G.G.R.; May, D.; Schmoeckel, E.; Horn, L.C. 2020 WHO Classification of Female Genital Tumors. Geburtshilfe Frauenheilkd. 2021, 81, 1145–1153. [Google Scholar] [CrossRef] [PubMed]
- Murali, R.; Soslow, R.A.; Weigelt, B. Classification of endometrial carcinoma: More than two types. Lancet Oncol. 2014, 15, e268–e278. [Google Scholar] [CrossRef]
- Colombo, N.; Creutzberg, C.; Amant, F.; Bosse, T.; González-Martín, A.; Ledermann, J.; Marth, C.; Nout, R.; Querleu, D.; Mirza, M.R.; et al. ESMO-ESGO-ESTRO Consensus Conference on Endometrial Cancer: Diagnosis, treatment and follow-up. Ann. Oncol. 2016, 27, 16–41. [Google Scholar] [CrossRef]
- Concin, N.; Matias-Guiu, X.; Vergote, I.; Cibula, D.; Mirza, M.R.; Marnitz, S.; Ledermann, J.; Bosse, T.; Chargari, C.; Fagotti, A.; et al. ESGO/ESTRO/ESP guidelines for the management of patients with endometrial carcinoma. Int. J. Gynecol. Cancer 2021, 31, 12–39. [Google Scholar] [CrossRef]
- Karkia, R.; Maccarthy, G.; Payne, A.; Karteris, E.; Pazoki, R.; Chatterjee, J. The Association Between Metabolic Syndrome and the Risk of Endometrial Cancer in Pre- and Post-Menopausal Women: A UK Biobank Study. J. Clin. Med. 2025, 14, 751. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Lin, T.-A.; Liu, K.-H.; Liao, C.-H.; Liu, Y.-Y.; Wu, V.C.-C.; Wen, M.-S.; Yeh, T.-S. Serum asprosin levels and bariatric surgery outcomes in obese adults. Int. J. Obes. 2019, 43, 1019–1025. [Google Scholar] [CrossRef]
- Ugur, K.; Erman, F.; Turkoglu, S.; Aydin, Y.; Aksoy, A.; Lale, A.; Karagöz, Z.K.; Ugur, I.; Akkoc, R.F.; Yalniz, M. Asprosin, visfatin and subfatin as new biomarkers of obesity and metabolic syndrome. Eur. Rev. Med. Pharmacol.Sci. 2022, 26, 2124–2133. [Google Scholar] [CrossRef] [PubMed]
- Mishra, I.; Xie, W.R.; Bournat, J.C.; He, Y.; Wang, C.; Silva, E.S.; Liu, H.; Ku, Z.; Chen, Y.; Erokwu, B.O.; et al. Protein tyrosine phosphatase receptor δ serves as the orexigenic asprosin receptor. Cell Metab. 2022, 34, 549–563.e8. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Yun, S.; Jeong, J.H.; Jung, T.W. Asprosin impairs insulin secretion in response to glucose and viability through TLR4/JNK-mediated inflammation. Mol. Cell. Endocrinol. 2019, 486, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Shabir, K.; Gharanei, S.; Orton, S.; Patel, V.; Chauhan, P.; Karteris, E.; Randeva, H.S.; Brown, J.E.; Kyrou, I. Asprosin Exerts Pro-Inflammatory Effects in THP-1 Macrophages Mediated via the Toll-like Receptor 4 (TLR4) Pathway. Int. J. Mol. Sci. 2022, 24, 227. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Shan, H.; Chen, L.; Long, A.; Zhang, Y.; Liu, Y.; Jia, L.; Wei, F.; Han, J.; Li, T.; et al. OLFR734 Mediates Glucose Metabolism as a Receptor of Asprosin. Cell Metab. 2019, 30, 319–328.e8. [Google Scholar] [CrossRef]
- Kerslake, R.; Sisu, C.; Panfilov, S.; Hall, M.; Khan, N.; Jeyaneethi, J.; Randeva, H.; Kyrou, I.; Karteris, E. Differential Regulation of Genes by the Glucogenic Hormone Asprosin in Ovarian Cancer. J. Clin. Med. 2022, 11, 5942. [Google Scholar] [CrossRef]
- Maggi, L.B.; Weber, J.D. Forget Transcription: Translation Is Where the Action Is. Mol. Cell. Biol. 2013, 33, 1884–1885. [Google Scholar] [CrossRef]
- Orton, S.; Karkia, R.; Mustafov, D.; Gharanei, S.; Braoudaki, M.; Filipe, A.; Panfilov, S.; Saravi, S.; Khan, N.; Kyrou, I.; et al. In Silico and In Vitro Mapping of Receptor-Type Protein Tyrosine Phosphatase Receptor Type D in Health and Disease: Implications for Asprosin Signalling in Endometrial Cancer and Neuroblastoma. Cancers 2024, 16, 582. [Google Scholar] [CrossRef]
- Bae, W.J.; Ahn, J.M.; Byeon, H.E.; Kim, S.; Lee, D. PTPRD-inactivation-induced CXCL8 promotes angiogenesis and metastasis in gastric cancer and is inhibited by metformin. J. Exp. Clin. Cancer Res. 2019, 38, 484. [Google Scholar] [CrossRef]
- Kohno, T.; Otsuka, A.; Girard, L.; Sato, M.; Iwakawa, R.; Ogiwara, H.; Sanchez-Cespedes, M.; Minna, J.D.; Yokota, J. A catalog of genes homozygously deleted in human lung cancer and the candidacy of PTPRD as a tumor suppressor gene. Genes Chromosomes Cancer 2010, 49, 342–352. [Google Scholar] [CrossRef]
- Funato, K.; Yamazumi, Y.; Oda, T.; Akiyama, T. Tyrosine phosphatase PTPRD suppresses colon cancer cell migration in coordination with CD44′. Exp. Ther. Med. 2011, 2, 457–463. [Google Scholar] [CrossRef]
- Painter, J.N.; O’mara, T.A.; Morris, A.P.; Cheng, T.H.; Gorman, M.; Martin, L.; Hodson, S.; Jones, A.; Martin, N.G.; Gordon, S.; et al. Genetic overlap between endometriosis and endometrial cancer: Evidence from cross-disease genetic correlation and GWAS meta-analyses. Cancer Med. 2018, 7, 1978–1987. [Google Scholar] [CrossRef]
- Sameer, A.S.; Nissar, S. Toll-Like Receptors (TLRs): Structure, Functions, Signaling, and Role of Their Polymorphisms in Colorectal Cancer Susceptibility. BioMed Res. Int. 2021, 2021, 1157023. [Google Scholar] [CrossRef]
- CKüper, F.-X.B.; Neuhofer, W. Toll-like receptor 4 activates NF-κB and MAP kinase pathways to regulate expression of proinflammatory COX-2 in renal medullary collecting duct cells. Am. J. Physiol.-Ren. Physiol. 2012, 302, F38–F46. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, Y.X.; Lu, J.J.; Hou, D.Y.; Abudukeyoumu, A.; Zhang, H.W.; Li, M.Q.; Xie, F. Active Heme Metabolism Suppresses Macrophage Phagocytosis via the TLR4/Type I IFN Signaling/CD36 in Uterine Endometrial Cancer. Am. J. Reprod. Immunol. 2024, 92, e13916. [Google Scholar] [CrossRef]
- Gulbay, G.; Secme, M.; Mutlu, D. Fusaric acid inhibits cell proliferation and downregulates expressions of toll-like receptors pathway genes in Ishikawa endometrial cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2023, 27, 7431–7436. [Google Scholar] [CrossRef]
- Ashton, K.A.; Proietto, A.; Otton, G.; Symonds, I.; McEvoy, M.; Attia, J.; Scott, R.J. Toll-Like Receptor (TLR) and Nucleosome-binding Oligomerization Domain (NOD) gene polymorphisms and endometrial cancer risk. BMC Cancer 2010, 10, 382. [Google Scholar] [CrossRef]
- Allhorn, S.; Böing, C.; Koch, A.A.; Kimmig, R.; Gashaw, I. TLR3 and TLR4 expression in healthy and diseased human endometrium. Reprod. Biol. Endocrinol. 2008, 6, 40. [Google Scholar] [CrossRef]
- Hossain, M.A.; Islam, S.M.S.; Quinn, J.M.W.; Huq, F.; Moni, M.A. Machine learning and bioinformatics models to identify gene expression patterns of ovarian cancer associated with disease progression and mortality. J. Biomed. Inform. 2019, 100, 103313. [Google Scholar] [CrossRef] [PubMed]
- Hao, B.; Chen, Z.; Bi, B.; Yu, M.; Yao, S.; Feng, Y.; Yu, Y.; Pan, L.; Di, D.; Luo, G.; et al. Role of TLR4 as a prognostic factor for survival in various cancers: A meta-analysis. Oncotarget 2018, 9, 13088–13099. [Google Scholar] [CrossRef] [PubMed]
- Lupi, L.A.; Cucielo, M.S.; Silveira, H.S.; Gaiotte, L.B.; Cesário, R.C.; Seiva, F.R.F.; de Almeida Chuffa, L.G. The role of Toll-like receptor 4 signaling pathway in ovarian, cervical, and endometrial cancers. Life Sci. 2020, 247, 117435. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.-Q.; Peng, X.-C.; Liu, L.; Yang, F.-Y.; Qian, F. Olfactory receptors and human diseases. Cell Tissue Res. 2025, 401, 1–14. [Google Scholar] [CrossRef]
- Kerslake, R.; Hall, M.; Randeva, H.S.; Spandidos, D.A.; Chatha, K.; Kyrou, I.; Karteris, E. Co-expression of peripheral olfactory receptors with SARS-CoV-2 infection mediators: Potential implications beyond loss of smell as a COVID-19 symptom. Int. J. Mol. Med. 2020, 46, 949–956. [Google Scholar] [CrossRef]
- Kerslake, R.; Hall, M.; Vagnarelli, P.; Jeyaneethi, J.; Randeva, H.S.; Pados, G.; Kyrou, I.; Karteris, E. A pancancer overview of FBN1, asprosin and its cognate receptor OR4M1 with detailed expression profiling in ovarian cancer. Oncol. Lett. 2021, 22, 650. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.; Tyson-Capper, A.J.; Gilmore, K.; Robson, S.C.; Europe-Finner, G.N. Identification of human myometrial target genes of the cAMP pathway: The role of cAMP-response element binding (CREB) and modulator (CREMα and CREMτ2α) proteins. J. Mol. Endocrinol. 2005, 34, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Tang, Y.; Deng, M.J.; Huang, C.T.; Lan, D.; Nong, W.Z.; Li, L.; Wang, Q. A combined biomarker panel shows improved sensitivity and specificity for detection of ovarian cancer. J. Clin. Lab. Anal. 2022, 36, e24232. [Google Scholar] [CrossRef]
- Pastorekova, S.; Gillies, R.J. The role of carbonic anhydrase IX in cancer development: Links to hypoxia, acidosis, and beyond. Cancer Metastasis Rev. 2019, 38, 65–77. [Google Scholar] [CrossRef]
- Ronca, R.; Supuran, C.T. Carbonic anhydrase IX: An atypical target for innovative therapies in cancer. Biochim. Biophys. Acta (BBA)—Rev. Cancer 2024, 1879, 189120. [Google Scholar] [CrossRef]
- Xie, Q.; Huang, J.; Xie, Y.; Hu, J.; Jin, L. Identification of prognostic biomarkers for endometrioid endometrial carcinoma based on the miRNA and mRNA co-expression network regulated by estradiol. Clinics 2025, 80, 100672. [Google Scholar] [CrossRef]
- Jin, M.; Yin, C.; Yang, J.; Yang, X.; Wang, J.; Zhu, J.; Yuan, J. Identification and validation of calcium extrusion-related genes prognostic signature in colon adenocarcinoma. PeerJ 2024, 12, e17582. [Google Scholar] [CrossRef]
- Sharma, A.; Kumar, R.; Aier, I.; Semwal, R.; Tyagi, P.; Varadwaj, P. Sense of Smell: Structural, Functional, Mechanistic Advancements and Challenges in Human Olfactory Research. Curr. Neuropharmacol. 2019, 17, 891–911. [Google Scholar] [CrossRef] [PubMed]
- Boekhoff, I.; Inglese, J.; Schleicher, S.; Koch, W.J.; Lefkowitz, R.J.; Breer, H. Olfactory desensitization requires membrane targeting of receptor kinase mediated by beta gamma-subunits of heterotrimeric G. proteins. J. Biol. Chem. 1994, 269, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Peppel, K.; Boekhoff, I.; McDonald, P.; Breer, H.; Caron, M.G.; Lefkowitz, R.J. G protein-coupled receptor kinase 3 (GRK3) gene disruption leads to loss of odorant receptor desensitization. J. Biol. Chem. 1997, 272, 25425–25428. [Google Scholar] [CrossRef]
- Mashukova, A.; Spehr, M.; Hatt, H.; Neuhaus, E.M. β-Arrestin2-Mediated Internalization of Mammalian Odorant Receptors. J. Neurosci. 2006, 26, 9902–9912. [Google Scholar] [CrossRef]
- Fattahi, F.; Saeednejad Zanjani, L.; Habibi Shams, Z.; Kiani, J.; Mehrazma, M.; Najafi, M.; Madjd, Z. High expression of DNA damage-inducible transcript 4 (DDIT4) is associated with advanced pathological features in the patients with colorectal cancer. Sci. Rep. 2021, 11, 13626. [Google Scholar] [CrossRef]
- Yoshikawa, N.; Yoshida, K.; Liu, W.; Matsukawa, T.; Hattori, S.; Yoshihara, M.; Tamauchi, S.; Ikeda, Y.; Yokoi, A.; Shimizu, Y.; et al. The prognostic significance of DDIT4 in endometrial cancer. Cancer Biomark. 2023, 37, 217–225. [Google Scholar] [CrossRef]
- Chang, B.; Liu, G.; Yang, G.; Mercado-Uribe, I.; Huang, M.; Liu, J. REDD1 is required for RAS-mediated transformation of human ovarian epithelial cells. Cell Cycle 2009, 8, 780–786. [Google Scholar] [CrossRef]
- Ji, Y.; Xie, M.; Lan, H.; Zhang, Y.; Long, Y.; Weng, H.; Li, D.; Cai, W.; Zhu, H.; Niu, Y.; et al. PRR11 is a novel gene implicated in cell cycle progression and lung cancer. Int. J. Biochem. Cell Biol. 2013, 45, 645–656. [Google Scholar] [CrossRef]
- Ni, W.; Yi, L.; Dong, X.; Cao, M.; Zheng, J.; Wei, Q.; Yuan, C. PRR11 is a prognostic biomarker and correlates with immune infiltrates in bladder urothelial carcinoma. Sci. Rep. 2023, 13, 2051. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Wu, X.; Zheng, G.; Jin, J.; Li, C.; Yu, G.; Li, W. Proline-rich protein 11 overexpression is associated with a more aggressive phenotype and poor overall survival in ovarian cancer patients. World J. Surg. Oncol. 2020, 18, 318. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Liu, Y.; Zhang, T.; Huang, Z.; Xu, X.; Zhao, Z.; Liu, J.; Zhai, E.; Cai, S.; Chen, J. Spindle function and Wnt pathway inhibition by PBX1 to suppress tumor progression via downregulating DCDC2 in colorectal cancer. Oncogenesis 2023, 12, 3. [Google Scholar] [CrossRef]
- Maclean, A.; Adishesh, M.; Button, L.; Richards, L.; Alnafakh, R.; Newton, E.; Drury, J.; Hapangama, D.K. The effect of pre-analytical variables on downstream application and data analysis of human endometrial biopsies. Hum. Reprod. Open 2022, 2022, hoac026. [Google Scholar] [CrossRef]
- Medina-Gutierrez, E.; Céspedes, M.V.; Gallardo, A.; Rioja-Blanco, E.; Pavon, M.A.; Asensio-Puig, L.; Farre, L.; Alba-Castellon, L.; Unzueta, U.; Villaverde, A.; et al. Novel Endometrial Cancer Models Using Sensitive Metastasis Tracing for CXCR4-Targeted Therapy in Advanced Disease. Biomedicines 2022, 10, 1680. [Google Scholar] [CrossRef]
- Ueda, H.; Ishiguro, T.; Mori, Y.; Yamawaki, K.; Okamoto, K.; Enomoto, T.; Yoshihara, K. Glycolysis-mTORC1 crosstalk drives proliferation of patient-derived endometrial cancer spheroid cells with ALDH activity. Cell Death Discov. 2024, 10, 435. [Google Scholar] [CrossRef]
- Kim, M.; Lee, S.; Park, W.H.; Suh, D.H.; Kim, K.; Kim, Y.B.; No, J.H. Silencing Bmi1 expression suppresses cancer stemness and enhances chemosensitivity in endometrial cancer cells. Biomed. Pharmacother. 2018, 108, 584–589. [Google Scholar] [CrossRef]
- Faruqi, S.A.; Satyaswaroop, P.G.; LiVolsi, V.A.; Deger, R.B.; Noumoff, J.S. Establishment and characterization of a poorly differentiated lethal human endometrial carcinoma cell line (NOU-1) with karyotype 46,XX. Cancer Genet. Cytogenet. 2002, 138, 44–49. [Google Scholar] [CrossRef]
- Tang, G.; Cho, M.; Wang, X. OncoDB: An interactive online database for analysis of gene expression and viral infection in cancer. Nucleic Acids Res. 2022, 50, D1334–D1339. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Akhmedov, M.; Martinelli, A.; Geiger, R.; Kwee, I. Omics Playground: A comprehensive self-service platform for visualization, analytics and exploration of Big Omics Data. NAR Genom. Bioinform. 2020, 2, lqz019. [Google Scholar] [CrossRef]
- Van Der Maaten, L.; Courville, A.; Fergus, R.; Manning, C. Accelerating t-SNE Using Tree-Based Algorithms. 2014. Available online: https://jmlr.org/papers/v15/vandermaaten14a.html (accessed on 15 July 2025).
- Witten, D.M.; Tibshirani, R.; Hastie, T. A penalized matrix decomposition, with applications to sparse principal components and canonical correlation analysis. Biostatistics 2009, 10, 515–534. [Google Scholar] [CrossRef]
- Wang, X.; Spandidos, A.; Wang, H.; Seed, B. PrimerBank: A PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012, 40, D1144–D1149. [Google Scholar] [CrossRef]

| TOP DEGs | Function in Endometrial Cancer | Reference |
|---|---|---|
| DDIT4 | High expression correlated to favourable prognosis in progression-free survival and overall survival | [45] |
| GPR157 | Potential biomarker for endometrioid endometrial carcinoma | [38] |
| CA9 | Increased gene expression when compared to controls | [51] |
| CXCR4 | Overexpression associated with enhanced metastatic dissemination | [52] |
| LDAH | Inhibition results in a reduction of cell growth | [53] |
| BM1 | Silencing suppresses cancer stemness and enhances chemosensitivity | [54] |
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| YWHAZ | AGACGGAAGGTGCTGAGAAA | GAAGCATTGGGGATCAAGAA |
| B-ACTIN | CAAGATGAGATTGGATGGC | CACATTGTGAACTTTGGGG |
| TLR4 | AGTTGATCTACCAAGCCTTGAGT | GCTGGTTGTCCCAAAATCACTTT |
| PTPRD | CAGGCGGAAGCGTTAATATCA | TTGGCATATCATCTTCAGGTGTC |
| OR4M1 | TCTGTTAATGTCCTATGCCTTCC | AATGTGGGAATAGCAGGTGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karkia, R.; Sisu, C.; Saravi, S.; Kyrou, I.; Randeva, H.S.; Chatterjee, J.; Karteris, E. Effects of Asprosin and Role of TLR4 as a Biomarker in Endometrial Cancer. Molecules 2025, 30, 3410. https://doi.org/10.3390/molecules30163410
Karkia R, Sisu C, Saravi S, Kyrou I, Randeva HS, Chatterjee J, Karteris E. Effects of Asprosin and Role of TLR4 as a Biomarker in Endometrial Cancer. Molecules. 2025; 30(16):3410. https://doi.org/10.3390/molecules30163410
Chicago/Turabian StyleKarkia, Rebecca, Cristina Sisu, Sayeh Saravi, Ioannis Kyrou, Harpal S. Randeva, Jayanta Chatterjee, and Emmanouil Karteris. 2025. "Effects of Asprosin and Role of TLR4 as a Biomarker in Endometrial Cancer" Molecules 30, no. 16: 3410. https://doi.org/10.3390/molecules30163410
APA StyleKarkia, R., Sisu, C., Saravi, S., Kyrou, I., Randeva, H. S., Chatterjee, J., & Karteris, E. (2025). Effects of Asprosin and Role of TLR4 as a Biomarker in Endometrial Cancer. Molecules, 30(16), 3410. https://doi.org/10.3390/molecules30163410







