An Interpretable Deep Learning and Molecular Docking Framework for Repurposing Existing Drugs as Inhibitors of SARS-CoV-2 Main Protease
Abstract
1. Introduction
2. Results and Discussion
2.1. Framework Validation with Known Drug–Target Pairs
2.2. Predictive Application to COVID-19 Main Protease
3. Materials and Methods
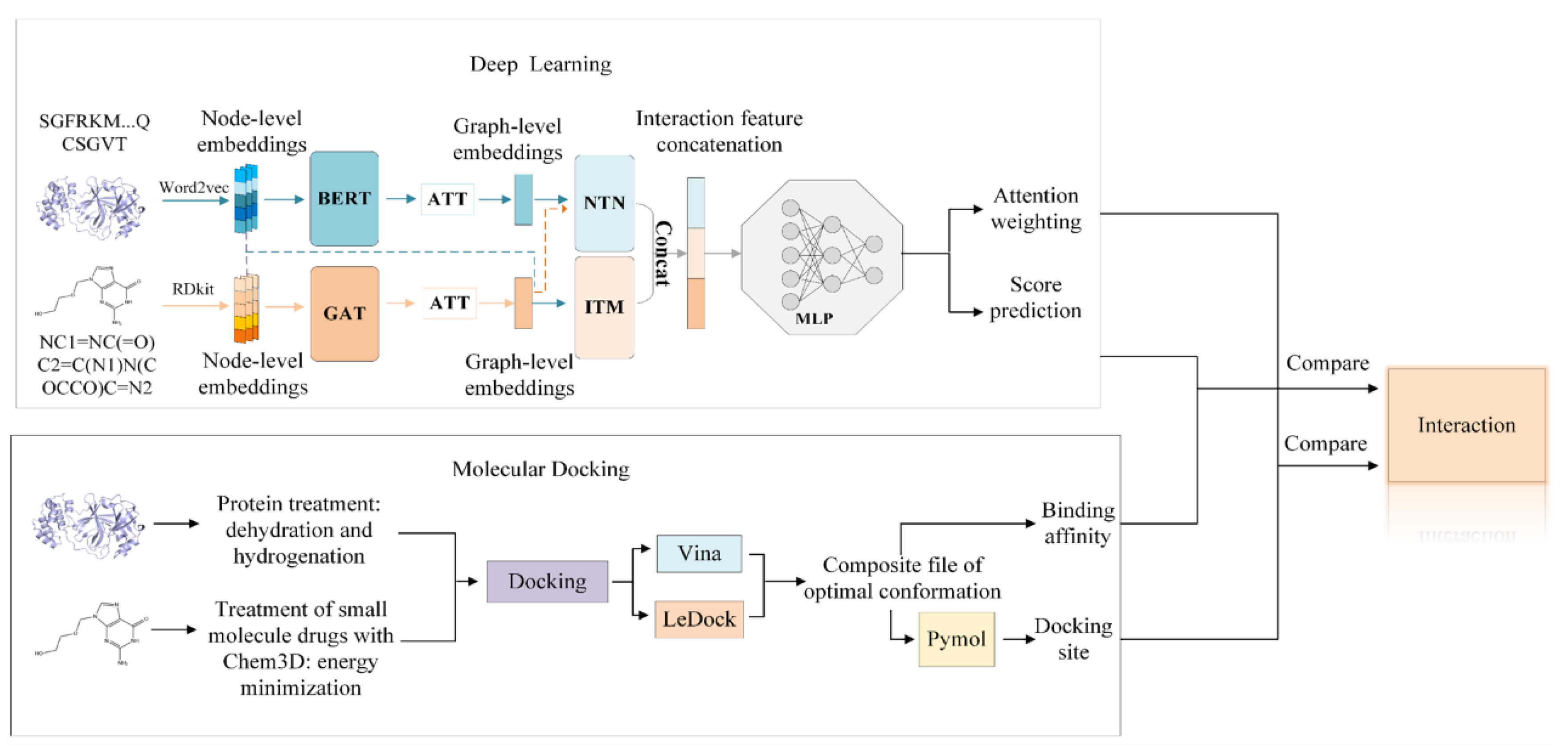
3.1. Deep Learning Model
3.2. Molecular Docking Programs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Machkovech, H.M.; Hahn, A.M.; Garonzik Wang, J.; Grubaugh, N.D.; Halfmann, P.J.; Johnson, M.C.; Lemieux, J.E.; O’Connor, D.H.; Piantadosi, A.; Wei, W.; et al. Persistent SARS-CoV-2 infection: Significance and implications. Lancet Infect. Dis. 2024, 24, e453–e462. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Huang, X.; Ma, Q.; Kuzmič, P.; Zhou, B.; Zhang, S.; Chen, J.; Xu, J.; Liu, B.; Jiang, H.; et al. Preclinical evaluation of the SARS-CoV-2 Mpro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir. Nat. Microbiol. 2024, 9, 1075–1088. [Google Scholar] [CrossRef] [PubMed]
- Narwal, M.; Armache, J.; Edwards, T.J.; Murakami, K.S. SARS-CoV-2 polyprotein substrate regulates the stepwise Mpro cleavage reaction. J. Biol. Chem. 2023, 299, 104697. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef]
- Ullrich, S.; Nitsche, C. The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 2020, 30, 127377. [Google Scholar] [CrossRef]
- Lamb, Y.N. Nirmatrelvir plus ritonavir: First approval. Drugs 2022, 82, 585–591. [Google Scholar] [CrossRef]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef]
- Yin, W.; Mao, C.; Luan, X.; Shen, D.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M.; Chang, S.; et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 2020, 368, 1499–1504. [Google Scholar] [CrossRef]
- Zak, M.; Hanan, E.J.; Lupardus, P.; Brown, D.G.; Robinson, C.; Siu, M.; Lyssikatos, J.P.; Romero, F.A.; Zhao, G.; Kellar, T.; et al. Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling. Bioorg. Med. Chem. Lett. 2019, 29, 1522–1531. [Google Scholar] [CrossRef]
- Davis, R.R.; Li, B.; Yun, S.Y.; Chan, A.; Nareddy, P.; Gunawan, S.; Ayaz, M.; Lawrence, H.R.; Reuther, G.W.; Lawrence, N.J.; et al. Structural insights into JAK2 inhibition by ruxolitinib, fedratinib, and derivatives thereof. J. Med. Chem. 2021, 64, 2228–2241. [Google Scholar] [CrossRef]
- Markham, A. Baricitinib: First global approval. Drugs 2017, 77, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Blair, H.A. Remdesivir: A review in COVID-19. Drugs 2023, 83, 1215–1237. [Google Scholar] [CrossRef] [PubMed]
- Bledsoe, R.K.; Montana, V.G.; Stanley, T.B.; Delves, C.J.; Apolito, C.J.; McKee, D.D.; Consler, T.G.; Parks, D.J.; Stewart, E.L.; Willson, T.M.; et al. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 2002, 110, 93–105. [Google Scholar] [CrossRef] [PubMed]
- The RECOVERY Collaborative Group. Dexamethasone in hospitalized patients with COVID-19. N. Engl. J. Med. 2021, 384, 693–704. [Google Scholar] [CrossRef]
- Deng, J.; Yang, Z.; Ojima, I.; Samaras, D.; Wang, F. Artificial intelligence in drug discovery: Applications and techniques. Brief. Bioinform. 2022, 23, bbab430. [Google Scholar] [CrossRef]
- Cavasotto, C.N.; Di Filippo, J.I. Artificial intelligence in the early stages of drug discovery. Arch. Biochem. Biophys. 2021, 698, 108730. [Google Scholar] [CrossRef]
- Chong, C.R.; Sullivan, D.J. New uses for old drugs. Nature 2007, 448, 645–646. [Google Scholar] [CrossRef]
- Ambrosio, F.A.; Costa, G.; Romeo, I.; Esposito, F.; Alkhatib, M.; Salpini, R.; Svicher, V.; Corona, A.; Malune, P.; Tramontano, E.; et al. Targeting SARS-CoV-2 main protease: A successful story guided by an in silico drug repurposing approach. J. Chem. Inf. Model. 2023, 63, 3601–3613. [Google Scholar] [CrossRef]
- Weth, F.R.; Hoggarth, G.B.; Weth, A.F.; Paterson, E.; White, M.P.J.; Tan, S.T.; Peng, L.; Gray, C. Unlocking hidden potential: Advancements, approaches, and obstacles in repurposing drugs for cancer therapy. Br. J. Cancer 2024, 130, 703–715. [Google Scholar] [CrossRef]
- He, H.; Chen, G.; Chen, C.Y. Machine learning and graph neural network for finding potential drugs related to multiple myeloma. New J. Chem. 2022, 46, 5188–5200. [Google Scholar] [CrossRef]
- Dhakal, A.; McKay, C.; Tanner, J.J.; Cheng, J. Artificial intelligence in the prediction of protein-ligand interactions: Recent advances and future directions. Brief. Bioinform. 2022, 23, bbab476. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, S.; Meng, K.; Sun, S. Machine learning for sequence and structure-based protein−ligand interaction prediction. J. Chem. Inf. Model. 2024, 64, 1456–1472. [Google Scholar] [CrossRef]
- Velazquez, M.; Anantharaman, R.; Velazquez, S.; Lee, Y. Alzheimer’s Disease Neuroimaging Initiative. RNN-based Alzheimer’s disease prediction from prodromal stage using diffusion tensor imaging. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; pp. 1665–1672. [Google Scholar]
- Lee, I.; Keum, J.; Nam, H. DeepConv-DTI: Prediction of drug–target interactions via deep learning with convolution on protein sequences. PLoS Comput. Biol. 2019, 15, e1007129. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, L.; Zhong, F.; Wang, D.; Jiang, J.; Zhang, S.; Jiang, H.; Zheng, M.; Li, X. Graph neural network approaches for drug-target interactions. Curr. Opin. Struc. Biol. 2022, 73, 102327. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017; p. 30. [Google Scholar]
- Chen, L.; Tan, X.; Wang, D.; Zhong, F.; Liu, X.; Yang, T.; Luo, X.; Chen, K.; Jiang, H.; Zheng, M. TransformerCPI: Improving compound–protein interaction prediction by sequence-based deep learning with self-attention mechanism and label reversal experiments. Bioinformatics 2020, 36, 4406–4414. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Zhao, H.; Zheng, K.; Wang, J. HyperAttentionDTI: Improving drug–protein interaction prediction by sequence-based deep learning with attention mechanism. Bioinformatics 2022, 38, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Morrison, J.L.; Breitling, R.; Higham, D.J.; Gilbert, D.R. A lock-and-key model for protein–protein interactions. Bioinformatics 2006, 22, 2012–2019. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2010, 30, 2785–2791. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New docking methods, expanded force field, and Python bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Zhao, H.; Dong, J.; Lafleur, K.; Nevado, C.; Caflisch, A. Discovery of a novel chemotype of tyrosine kinase inhibitors by fragment-based docking and molecular dynamics. ACS Med. Chem. Lett. 2012, 3, 834–838. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: a new approach for rapid, accurate docking and scoring. 1. method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: a new approach for rapid, accurate docking and scoring. 2. enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef] [PubMed]
- Rarey, M.; Kramer, B.; Lengauer, T.; Klebe, G. A fast flexible docking method using an incremental construction algorithm. J. Mol. Biol. 1996, 261, 470–489. [Google Scholar] [CrossRef]
- Zhao, H.; Jiang, D.; Shen, C.; Zhang, J.; Zhang, X.; Wang, X.; Nie, D.; Hou, T.; Kang, Y. Comprehensive evaluation of 10 docking programs on a diverse set of protein−cyclic peptide complexes. J. Chem. Inf. Model. 2024, 64, 2112–2124. [Google Scholar] [CrossRef]
- Sharma, G.; Kumar, N.; Sharma, C.S.; Alqahtani, T.; Tiruneh, Y.K.; Sultana, S.; Rolim Silva, G.V.; de Lima Menezes, G.; Zaki, M.E.A.; Oliveira, J.I.N. Identification of promising SARSCoV-2 main protease inhibitor through molecular docking, dynamics simulation, and ADMET analysis. Sci. Rep. 2025, 15, 2830. [Google Scholar]
- Zhang, H.; Liang, B.; Sang, X.; An, J.; Huang, Z. Discovery of potential inhibitors of SARS-CoV-2 main protease by a transfer learning method. Viruses 2023, 15, 891. [Google Scholar] [CrossRef]
- Su, Q.; Wang, J.; Gou, Q.; Hu, R.; Jiang, L.; Zhang, H.; Wang, T.; Liu, Y.; Shen, C.; Kang, Y.; et al. Robust protein–ligand interaction modeling through integrating physical laws and geometric knowledge for absolute binding free energy calculation. Chem. Sci. 2025, 16, 5043–5057. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, Y.; Chen, Q. AMMVF-DTI: A novel model predicting drug-target interactions based on attention mechanism and multi-view fusion. Int. J. Mol. Sci. 2023, 24, 14142. [Google Scholar] [CrossRef]
- Yang, Z.; Zhong, W.; Lv, Q.; Dong, T.; Chen, C.Y. Geometric interaction graph neural network for predicting protein-ligand binding affinities from 3D structures (GIGN). J. Phys. Chem. Lett. 2023, 14, 2020–2033. [Google Scholar] [CrossRef]
- Manivannan, E.; Karthikeyan, C.; Moorthy, N.S.H.N.; Chaturvedi, S.C. The rise and fall of chloroquine/hydroxychloroquine as compassionate therapy of COVID-19. Front. Pharmacol. 2021, 12, 584940. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Wang, J. Dipyridamole, chloroquine, montelukast sodium, candesartan, oxytetracycline, and atazanavir are not SARS-CoV-2 main protease inhibitors. Proc. Natl. Acad. Sci. USA 2021, 118, e2024420118. [Google Scholar] [CrossRef] [PubMed]
- Cong, W.; Poudel, A.N.; Alhusein, N.; Wang, H.; Yao, G.; Lambert, H. Antimicrobial use in COVID-19 patients in the first phase of the SARS-CoV-2 pandemic: A scoping review. Antibiotics 2021, 10, 745. [Google Scholar] [CrossRef] [PubMed]
- Ghahremanpour, M.M.; Tirado-Rives, J.; Deshmukh, M.; Ippolito, J.A.; Zhang, C.H.; Cabeza de Vaca, I.; Liosi, M.E.; Anderson, K.S.; Jorgensen, W.L. Identification of 14 known drugs as inhibitors of the main protease of SARS-CoV-2. ACS Med. Chem. Lett. 2020, 11, 2526–2533. [Google Scholar] [CrossRef]
- Zheng, S.; Li, Y.; Chen, S.; Xu, J.; Yang, Y. Predicting drug–protein interaction using quasi-visual question answering system. Nat. Mach. Intell. 2020, 2, 134–140. [Google Scholar] [CrossRef]
- Öztürk, H.; Özgür, A.; Ozkirimli, E. DeepDTA: Deep drug–target binding affinity prediction. Bioinformatics 2018, 34, i821–i829. [Google Scholar] [CrossRef]
- Antypenko, L.; Shabelnyk, K.; Antypenko, O.; Arisawa, M.; Kamyshnyi, O.; Oksenych, V.; Kovalenko, S. In silico identification and characterization of spiro[1,2,4]triazolo[1,5-c]quinazolines as diacylglycerol kinase α modulators. Molecules 2025, 30, 2324. [Google Scholar] [CrossRef]
- Cosconati, S.; Forli, S.; Perryman, A.L.; Harris, R.; Goodsell, D.S.; Olson, A.J. Virtual screening with AutoDock: Theory and practice. Expert. Opin. Drug Discov. 2010, 5, 597–607. [Google Scholar] [CrossRef]
- Fischer, A.; Sellner, M.; Neranjan, S.; Smieško, M.; Lill, M.A. Potential inhibitors for novel coronavirus protease identified by virtual screening of 606 million compounds. Int. J. Mol. Sci. 2020, 21, 3626. [Google Scholar] [CrossRef]
- Owen, D.R.; Allerton, C.M.N.; Anderson, A.S.; Aschenbrenner, L.; Avery, M.; Berritt, S.; Boras, B.; Cardin, R.D.; Carlo, A.; Coffman, K.J.; et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 2021, 374, 1586–1593. [Google Scholar] [CrossRef]
- Pant, S.; Jena, N.R. Inhibition of the RNA-dependent RNA polymerase of the SARS-CoV-2 by short peptide inhibitors. Eur. J. Pharm. Sci. 2021, 167, 106012. [Google Scholar] [CrossRef]
- Chang, Y.; Min, J.; Jarusiewicz, J.A.; Actis, M.; Bradford, S.Y.; Mayasundari, A.; Yang, L.; Chepyala, D.; Alcock, L.J.; Roberts, K.G.; et al. Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia. Blood 2021, 138, 2313–2326. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Sun, H.; Yao, X.; Li, D.; Xu, L.; Li, Y.; Tian, S.; Hou, T. Comprehensive evaluation of ten Docking programs on a diverse set of protein–ligand complexes: The prediction accuracy of sampling power and scoring power. Phys. Chem. Chem. Phys. 2016, 18, 12964. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Wang, Z.; Yao, X.; Li, Y.; Lei, T.; Wang, E.; Xu, L.; Zhu, F.; Li, D.; Hou, T. Comprehensive assessment of nine docking programs on type II kinase inhibitors: Prediction accuracy of sampling power, scoring power and screening power. Brief. Bioinform. 2020, 21, 282–297. [Google Scholar] [CrossRef] [PubMed]
- Soy, M.; Keser, G.; Atagündüz, P.; Tabak, F.; Atagündüz, I.; Kayhan, S. Cytokine storm in COVID-19: Pathogenesis and overview of anti-inflammatory agents used in treatment. Clin. Rheumatol. 2020, 39, 2085–2094. [Google Scholar] [CrossRef]
- Stebbing, J.; Phelan, A.; Griffinb, I.; Tucker, C.; Oechsle, O.; Smith, D.; Richardson, P. COVID-19: Combining antiviral and anti-inflammatory treatments. Lancet Infect. Dis. 2020, 20, 400–402. [Google Scholar] [CrossRef]
- Perico, N.; Cortinovis, M.; Suter, F.; Remuzzi, G. Home as the new frontier for the treatment of COVID-19: The case for anti-inflammatory agents. Lancet Infect. Dis. 2023, 23, e22–e33. [Google Scholar] [CrossRef]
- Micallefa, J.; Soeiro, T.; Jonville-Béra, A.P.; French Society of Pharmacology. Non-steroidal anti-inflammatory drugs, pharmacology, and COVID-19 infection. Therapies 2020, 75, 355–362. [Google Scholar] [CrossRef]
- Borcherding, N.; Jethava, Y.; Vikas, P. Repurposing anti-cancer drugs for COVID-19 treatment. Drug Des. Dev. Ther. 2020, 14, 5045–5058. [Google Scholar] [CrossRef]
- Anwaar, M.U.; Adnan, F.; Abro, A.; Khan, R.A.; Rehman, A.U.; Osama, M.; Rainville, C.; Kumar, S.; Sterner, D.E.; Javed, S.; et al. Combined deep learning and molecular docking simulations approach identifies potentially effective FDA approved drugs for repurposing against SARS-CoV-2. Comput. Biol. Med. 2022, 141, 105049. [Google Scholar] [CrossRef]
- Huang, S.; Zhang, Y.; Wang, Z.; Xiong, F. Computational discovery of SARS-CoV-2 main protease inhibitors via a virtual screening, molecular docking, molecular Dynamics and MM/PBSA calculation-driven approach. New J. Chem. 2024, 48, 19650. [Google Scholar] [CrossRef]
- Fu, H.; Zhu, Y.; Chen, Q. Free energy calculations in biomolecule-nanomaterial interactions. Front. Phys. 2024, 12, 1469515. [Google Scholar] [CrossRef]
- Khan, M.A.; Mutahir, S.; Jabar, G.; Wenwei, Z.; Tariq, M.A.; Almehizia, A.A.; Mustafa, M. DFT, Molecular Docking, ADME, and Cardiotoxicity Studies of Persuasive Thiazoles as Potential Inhibitors of the Main Protease of SARS-CoV-2. Chem. Biodiversity 2024, 21, e202401775. [Google Scholar] [CrossRef]
- Sonadevi, S.; Rajaraman, D.; Saritha, M.; Solo, P.; Anthony, L.A. 2,4-dichloro-6-(1,4,5-triphenyl-1 H -imidazol-2-yl) phenol: Synthesis, DFT analysis, Molecular docking, molecular dynamics, ADMET properties against COVID-19 main protease (Mpro: 6WCF/6Y84/6LU7). Mol. Phys. 2025, 123. [Google Scholar] [CrossRef]
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- RDKit: Open-Source Cheminformatics Software. Available online: https://www.rdkit.org (accessed on 10 December 2024).
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient estimation of word representations in vector space. In Proceedings of the 1st International Conference on Learning Representations, ICLR 2013, Scottsdale, AZ, USA, 2–4 May 2013. [Google Scholar]
- Veličković, P.; Cucurull, G.; Casanova, A.; Romero, A.; Lio, P.; Bengio, Y. Graph attention networks. arXiv 2017, arXiv:1710.10903. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Minneapolis, MN, USA, 2–7 June 2019; Association for Computational Linguistics: Minneapolis, MN, USA, 2019; Volume 1, pp. 4171–4186. [Google Scholar]
- Tolstikhin, I.O.; Houlsby, N.; Kolesnikov, A.; Beyer, L.; Zhai, X.; Unterthiner, T.; Yung, J.; Steiner, A.; Keysers, D.; Uszkoreit, J.; et al. Mlp-mixer: An all-mlp architecture for vision. Adv. Neural Inf. Process. Syst. 2021, 34, 24261–24272. [Google Scholar]
- Cheng, Z.; Yan, C.; Wu, F.; Wang, J. Drug-target interaction prediction using multi-head self-attention and graph attention network. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 19, 2208–2218. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Long, W.; Wei, L. MDL-CPI: Multi-view deep learning model for compound-protein interaction prediction. Methods 2022, 204, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Tsubaki, M.; Tomii, K.; Sese, J. Compound-protein interaction prediction with end-to-end learning of neural networks for graphs and sequences. Bioinformatics 2018, 35, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Le, H.; Quinn, T.P.; Nguyen, T.; Le, T.D.; Venkatesh, S. GraphDTA: Predicting drug-target binding affinity with graph neural networks. Bioinformatics 2021, 37, 1140–1147. [Google Scholar] [CrossRef]
- Lee, I.; Nam, H. Identification of drug-target interaction by a random walk with restart method on an interactome network. BMC Bioinf. 2018, 19, 208. [Google Scholar] [CrossRef]
- Yuan, Q.; Gao, J.; Wu, D.; Zhang, S.; Mamitsuka, H.; Zhu, S. DrugE-rank: Improving drug-target interaction prediction of new candidate drugs or targets by ensemble learning to rank. Bioinformatics 2016, 32, i18–i27. [Google Scholar] [CrossRef]
- Wan, F.; Zhu, Y.; Hu, H.; Dai, A.; Cai, X.; Chen, L.; Gong, H.; Xia, T.; Yang, D.; Wang, M.; et al. DeepCPI: A deep learning-based framework for large-scale in silico drug screening. Genom. Proteom. Bioinf. 2019, 17, 478–495. [Google Scholar] [CrossRef]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Li, Z.; Wan, H.; Shi, Y.; Ouyang, P. Personal experience with four kinds of chemical structure drawing software: Review on ChemDraw, ChemWindow, ISIS/Draw, and ChemSketch. J. Chem. Inf. Comput. Sci. 2004, 44, 1886–1890. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W. PyMOL, version 2.6; Schrödinger, LLC: New York, NY, USA, 2020; Available online: http://www.pymol.org/pymol (accessed on 3 August 2025).
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmuller, H.; MacKerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered Proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Bugnon, M.; Goullieux, M.; Röhrig, U.F.; Perez, M.A.S.; Daina, A.; Michielin, O.; Zoete, V. SwissParam 2023: A Modern Web-Based Tool for Efficient Small Molecule Parametrization. J. Chem. Inf. Model. 2023, 63, 6469–6475. [Google Scholar] [CrossRef]
- Allen, M.P.; Tildesley, D.J. Computer Simulation of Liquids, 1st ed.; Clarendon: Oxford, UK, 1989. [Google Scholar]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Van der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]

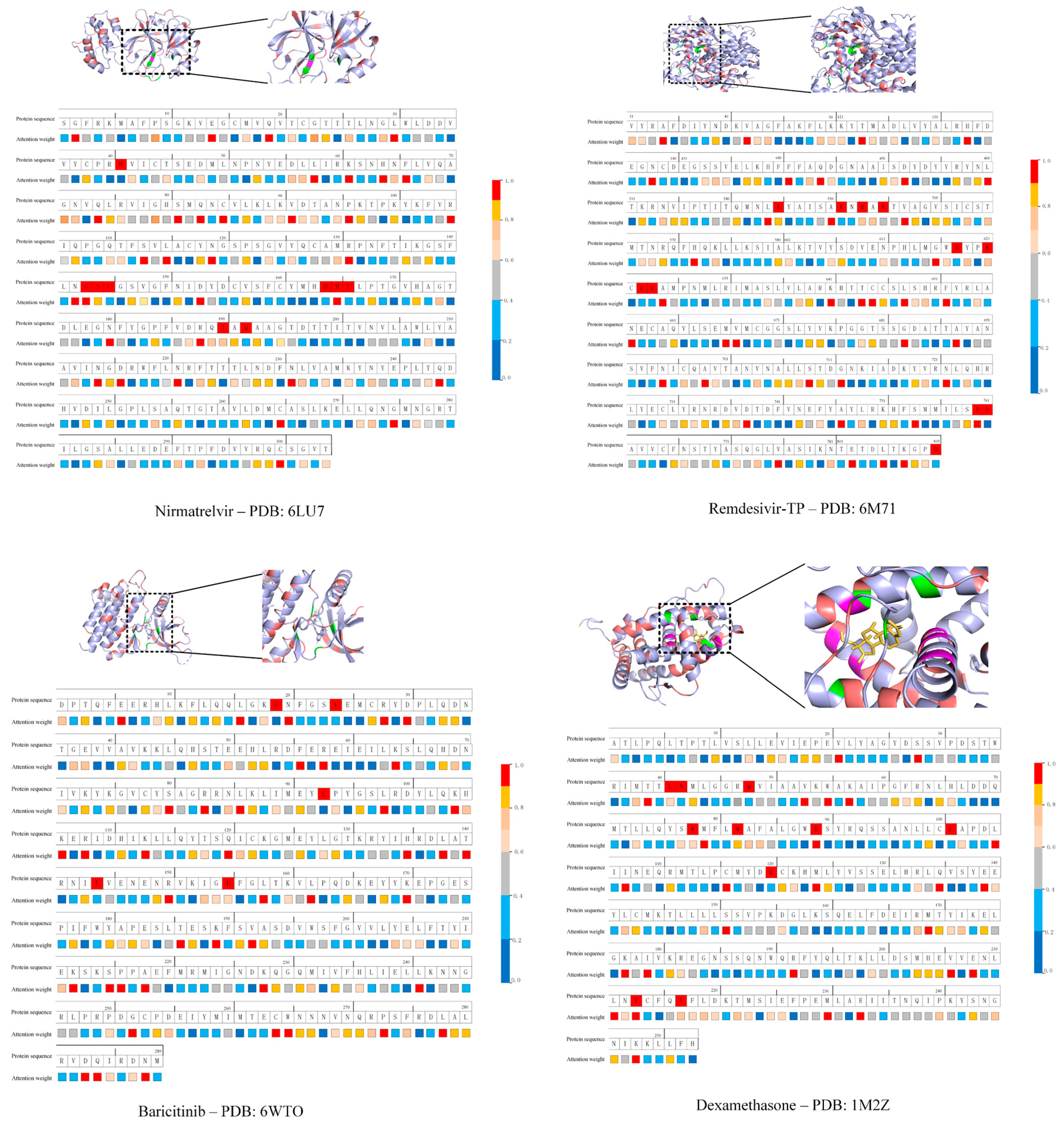
| DrugID | PDBID | DL Score | Docking (kcal·mol−1) | |
|---|---|---|---|---|
| AutoDock Vina | LeDock | |||
| Nirmatrelvir | 6LU7 | 0.994 | −6.6 | −7.26 |
| Remdesivir-TP | 6M71 | 0.991 | −8.0 | −9.26 |
| Baricitinib | 6WTO | 0.828 | −8.8 | −7.52 |
| Dexamethasone | 1M2Z | 0.972 | −8.0 | −5.25 |
| Chloroquine | 6LU7 | 0.065 | −5.6 | −4.92 |
| Amoxicillin | 6LU7 | 0.001 | −7.4 | −5.89 |
| Cephradine | 6LU7 | 0.007 | −7.4 | −5.81 |
| DrugID | H-Bonding Interactions | Hydrophobic Interactions | Salt Bridge | π-Stacking | ||
|---|---|---|---|---|---|---|
| Vina | LeDock | Vina | LeDock | Vina | Vina | |
| Chloroquine | n/a | Gln110 Asn151 | Phe8 Ile106 Gln110 Asn151 Phe294 | Phe8 Asn151 Phe294 | Asp153 | n/a |
| Amoxicillin | Gln110 Thr111 Asp295 | Thr26 Leu141 Gly143 Ser144 Cys145 Glu166 | Val104 Asn151 | Thr25 Glu166 | n/a | Phe294 |
| Cephradine | Gln110 Thr111 Asn151 Asp295 | Asn142 Gly143 Ser144 Cys145 His172 | Val104 Asn151 | Glu166 | n/a | Phe294 |
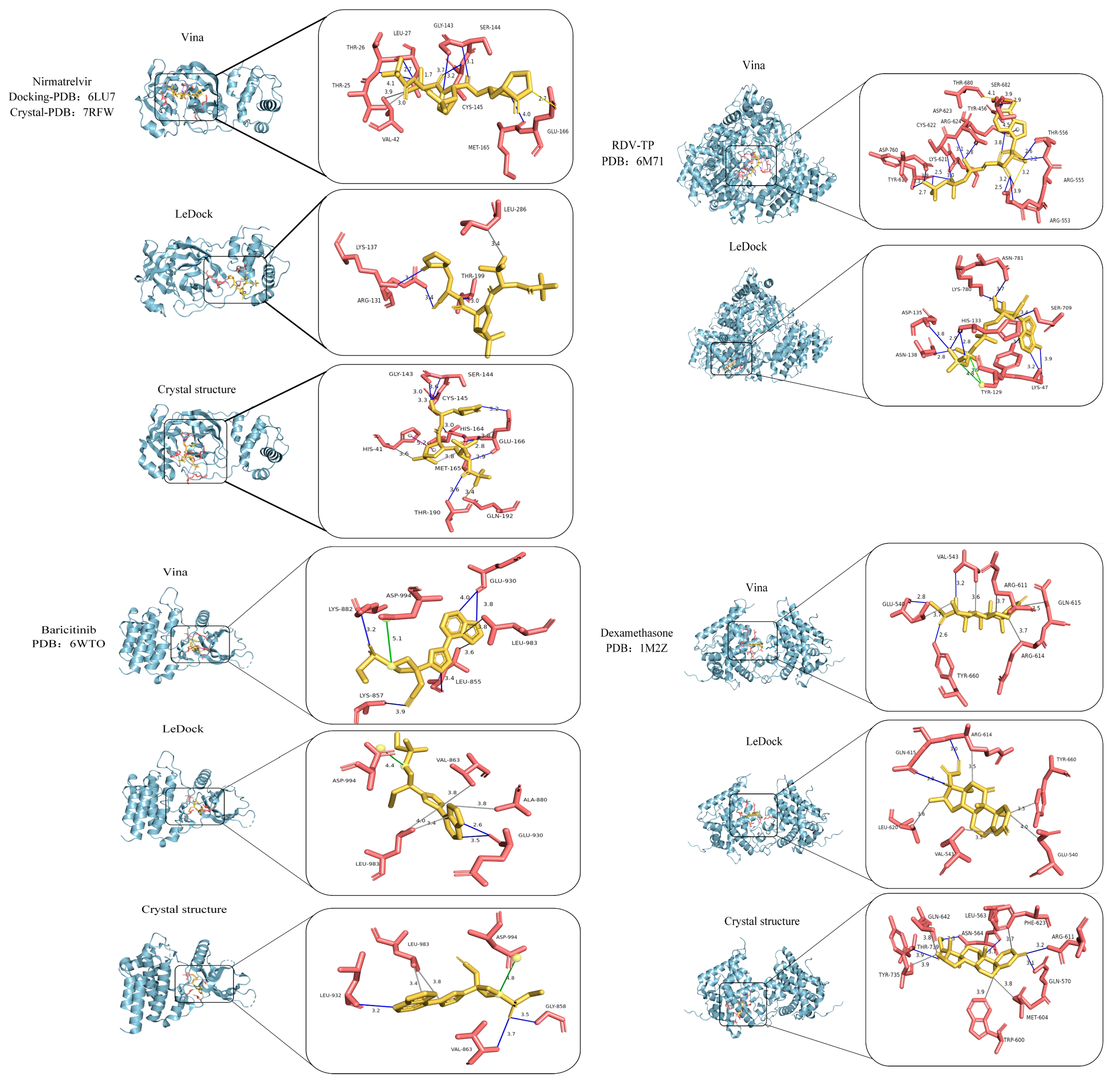
| DrugID | PDBID | AutoDock Vina | LeDock | Crystal Structure |
|---|---|---|---|---|
| Nirmatrelvir | 6LU7 | Thr24, Thr25, Thr26, Leu27, Val42, Gly143, Ser144, Cys145, Glu166 | Arg131, Lys137, Thr199, Leu286 | His41, Gly143, Ser144, Cys145, His164, Met165, Glu166, Thr190, Gln192 |
| Remdesivir-TP | 6M71 | Lys47, Tyr129, His133, Asp135, Asn138, Ser709, Lys780, Asn781 | Tyr456, Arg553, Arg555, Thr556, Tyr619, Lys621, Cys622, Asp623, Arg624, Thr680, Ser682, Asp760 | Lys545, Lys551, Arg553, Arg555, Asp618, Lys621, Asp623, Arg624, Asp760, Asp761 His810 (predicted) |
| Baricitinib | 6WTO | Leu855, Lys857, Lys882, Glu930, Leu983, Asp994 | Val863, Ala880, Glu930, Leu983, Asp994 | Gly858, Val863, Leu932, Leu983, Asp994 |
| Dexamethasone | 1M2Z | Glu540, Val543, Arg611, Arg614, Gln615, Leu620, Tyr660 | Glu540, Val543, Arg614, Gln615, Leu620, Tyr660 | Leu563, Asn564, Gln570, Trp600, Met604, Arg611, Phe623, Gln642, Tyr735, Thr739 |
| DrugID | Drug Action | DL Score | Docking (kcal·mol−1) | |
|---|---|---|---|---|
| AutoDock Vina | LeDock | |||
| Favipiravir | antiviral | 0.003 | −5.1 | −4.5 |
| Oseltamivir | antiviral | 0.115 | −5.3 | −4.8 |
| Zanamivir | antiviral | 0.836 | −6.7 | −6.8 |
| Peramivir | antiviral | 0.882 | −6.1 | −5.3 |
| Lamivudine | antiviral | 0.991 | −5.7 | −5.5 |
| Efavirenz | antiviral | 0.028 | −7.2 | −4.5 |
| Lopinavir | antiviral | 0.558 | −7.2 | −6.9 |
| Ritonavir | antiviral | 0.451 | −7.2 | −7.9 |
| Sofosbuvir | antiviral | 0.922 | −4.8 | −6.6 |
| Velpatasvir | antiviral | 0.781 | −8.9 | −7.7 |
| Voxilaprevir | antiviral | 0.924 | −8.8 | −6.8 |
| Daclatasvir | antiviral | 0.223 | −8.3 | −8.1 |
| Glecaprevir | antiviral | 0.875 | −8.6 | −6.5 |
| Pibrentasvir | antiviral | 0.839 | −7.7 | −7.4 |
| Ribavirin | antiviral | 0.003 | −6.4 | −5.7 |
| Acyclovir | antiviral | 0.572 | −5.3 | −6.1 |
| Tofacitinib | anti-inflammatory | 0.968 | −6.1 | −5.8 |
| Sirolimus | anti-inflammatory | 0.388 | −8.9 | −6.8 |
| Imiquimod | anti-inflammatory | 0.005 | −6.8 | −4.6 |
| Ibuprofen | anti-inflammatory | 0.141 | −6.0 | −3.6 |
| Diclofenac | anti-inflammatory | 0.001 | −6.4 | −4.7 |
| Naproxen | anti-inflammatory | 0.335 | −6.5 | −4.0 |
| Indomethacin | anti-inflammatory | 0.005 | −6.9 | −5.0 |
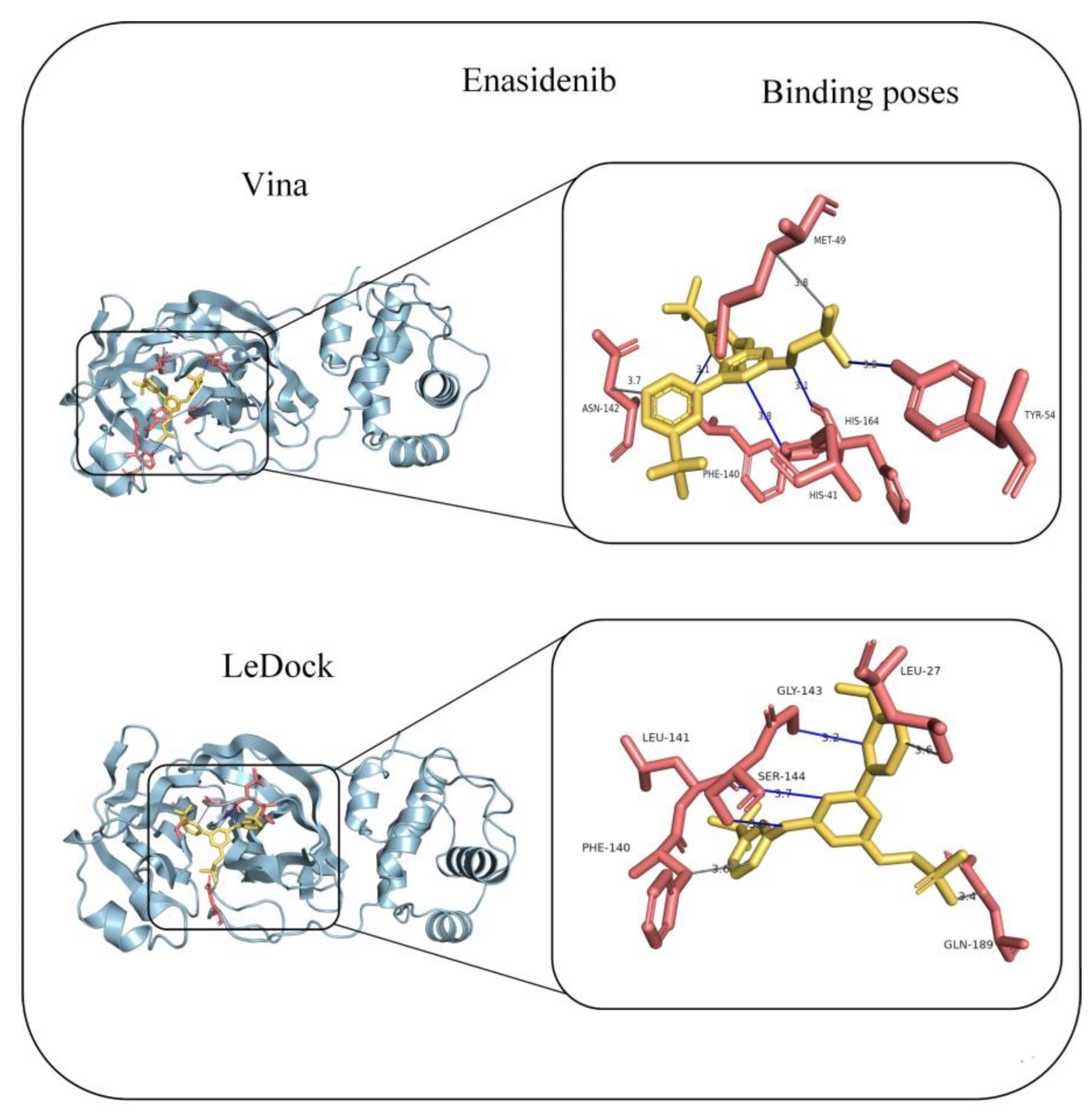
| Enasidenib | anti-cancer | 0.878 | −9.2 | −7.6 |
| Carmofur | anti-cancer | 0.956 | −5.6 | −5.2 |
| Masitinib | anti-cancer | 0.990 | −8.2 | −7.42 |
| Nilotinib | anti-cancer | 0.979 | −9.3 | −7.4 |
| Dasatinib | anti-cancer | 0.997 | −7.1 | −7.4 |
| Imatinib | anti-cancer | 0.991 | −8.3 | −7.4 |
| Model | AUC | Precision | Recall |
|---|---|---|---|
| KNN | 0.860 | 0.927 | 0.798 |
| RF | 0.940 | 0.897 | 0.861 |
| L2 | 0.911 | 0.913 | 0.867 |
| SVM | 0.910 | 0.966 | 0.969 |
| MDL-CPI | 0.959 | 0.924 | 0.905 |
| GNN | 0.970 | 0.923 | 0.918 |
| GCN | 0.956 | 0.862 | 0.928 |
| GraphDTA | 0.960 | 0.882 | 0.912 |
| DrugVQA(VQA-seq) | 0.964 | 0.897 | 0.948 |
| TransformerCPI | 0.973 | 0.916 | 0.925 |
| This work | 0.989 | 0.958 | 0.987 |
| Model | AUC | Precision | Recall |
|---|---|---|---|
| KNN | 0.858 | 0.801 | 0.827 |
| RF | 0.902 | 0.821 | 0.844 |
| L2 | 0.892 | 0.890 | 0.877 |
| SVM | 0.894 | 0.785 | 0.818 |
| MDL-CPI | 0.975 | 0.943 | 0.929 |
| GNN | 0.978 | 0.938 | 0.929 |
| GCN | 0.975 | 0.921 | 0.927 |
| GraphDTA | 0.974 | 0.927 | 0.912 |
| TransformerCPI | 0.988 | 0.952 | 0.953 |
| This work | 0.989 | 0.981 | 0.963 |
| Model | AUC | Precision | Recall |
|---|---|---|---|
| RWR | 0.760 | 0.705 | 0.651 |
| DrugE-Rank | 0.759 | 0.707 | 0.629 |
| GNN | 0.802 | 0.737 | 0.716 |
| DeepCPI | 0.700 | 0.700 | 0.556 |
| GraphDTA | 0.821 | 0.751 | 0.769 |
| This work | 0.815 | 0.775 | 0.760 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Gao, J.; Chen, Q. An Interpretable Deep Learning and Molecular Docking Framework for Repurposing Existing Drugs as Inhibitors of SARS-CoV-2 Main Protease. Molecules 2025, 30, 3409. https://doi.org/10.3390/molecules30163409
Huang J, Gao J, Chen Q. An Interpretable Deep Learning and Molecular Docking Framework for Repurposing Existing Drugs as Inhibitors of SARS-CoV-2 Main Protease. Molecules. 2025; 30(16):3409. https://doi.org/10.3390/molecules30163409
Chicago/Turabian StyleHuang, Juan, Jialong Gao, and Qu Chen. 2025. "An Interpretable Deep Learning and Molecular Docking Framework for Repurposing Existing Drugs as Inhibitors of SARS-CoV-2 Main Protease" Molecules 30, no. 16: 3409. https://doi.org/10.3390/molecules30163409
APA StyleHuang, J., Gao, J., & Chen, Q. (2025). An Interpretable Deep Learning and Molecular Docking Framework for Repurposing Existing Drugs as Inhibitors of SARS-CoV-2 Main Protease. Molecules, 30(16), 3409. https://doi.org/10.3390/molecules30163409






