Gallic, Aconitic, and Crocetin Acids as Potential TNF Modulators: An Integrated Study Combining Molecular Docking, Dynamics Simulations, ADMET Profiling, and Gene Expression Analysis
Abstract
1. Introduction
2. Results
2.1. Computational Investigation of Organic Acids as Potential Tumor Necrosis Factor (TNF) Inhibitors by Molecular Docking
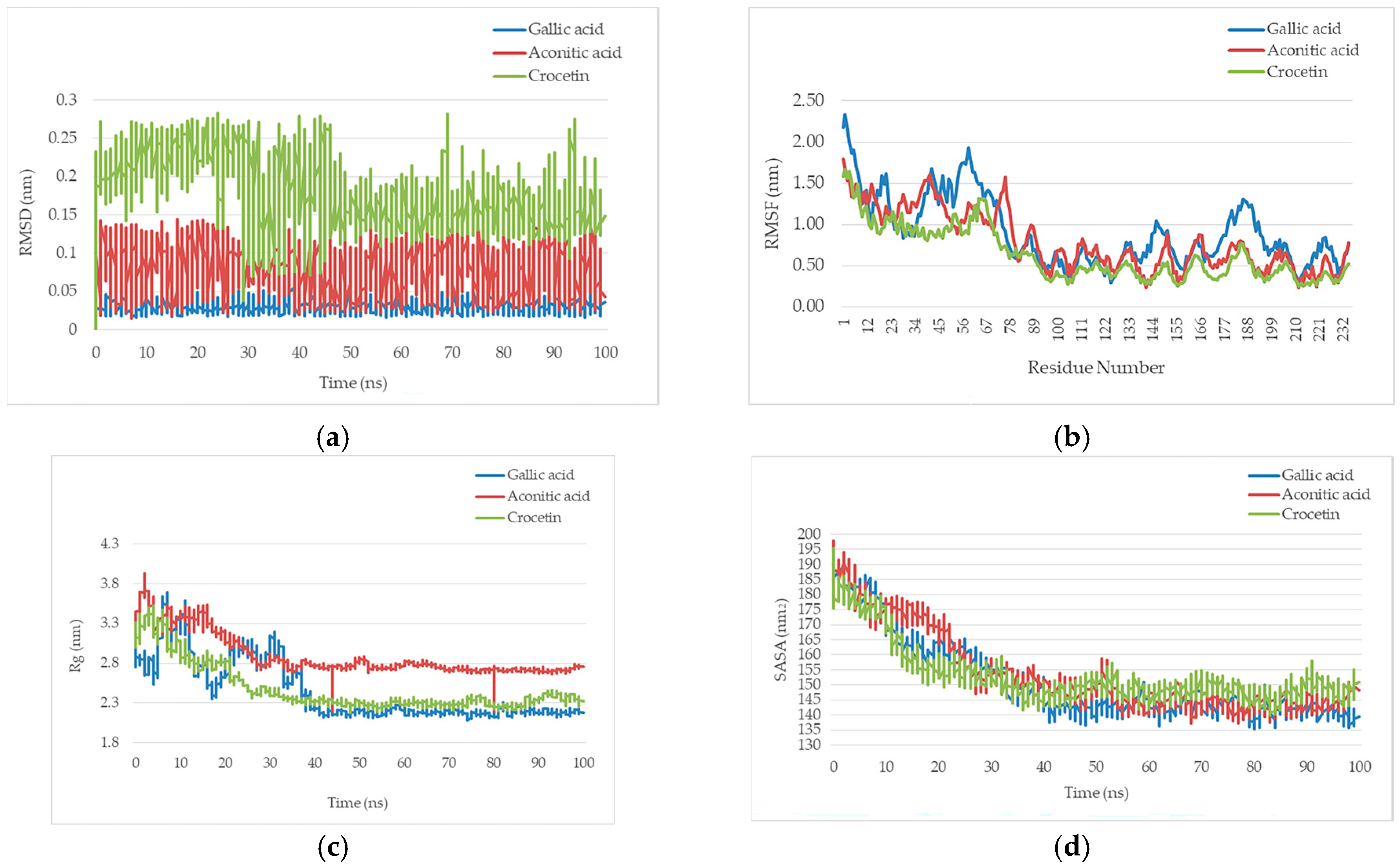
2.2. Molecular Dynamics Simulation and System Stability Analysis
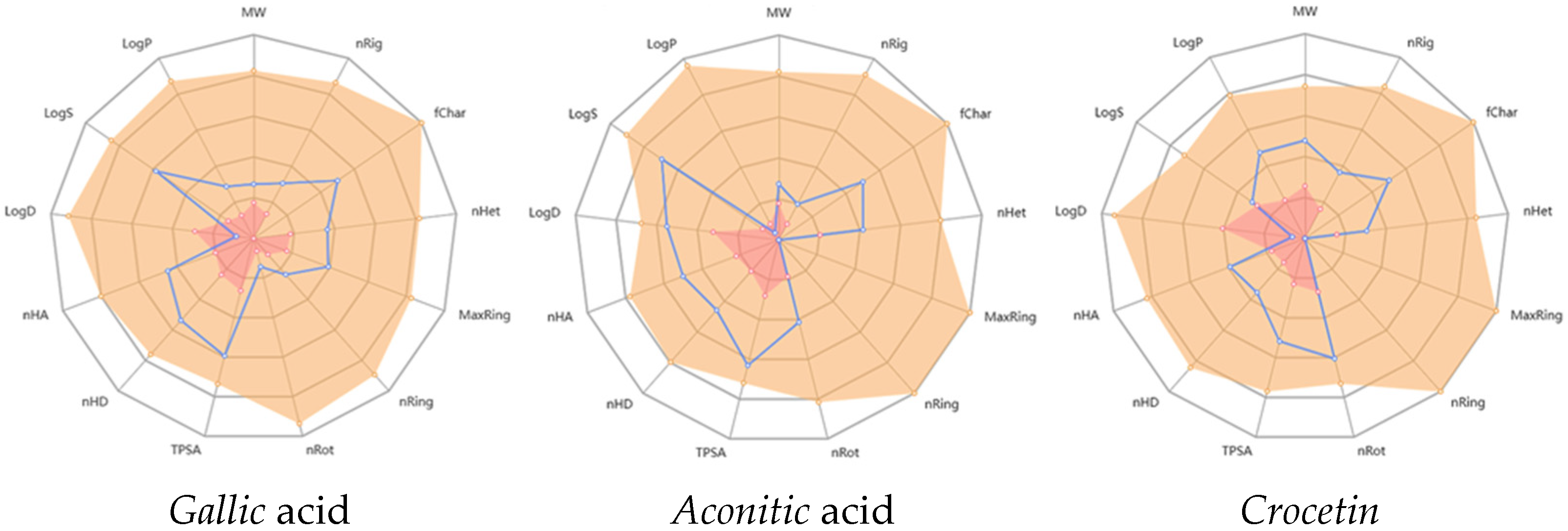
2.3. In Silico ADMET Analysis of Gallic Acid, Aconitic Acid, and Crocetin: Pharmacokinetic and Toxicity Profiling
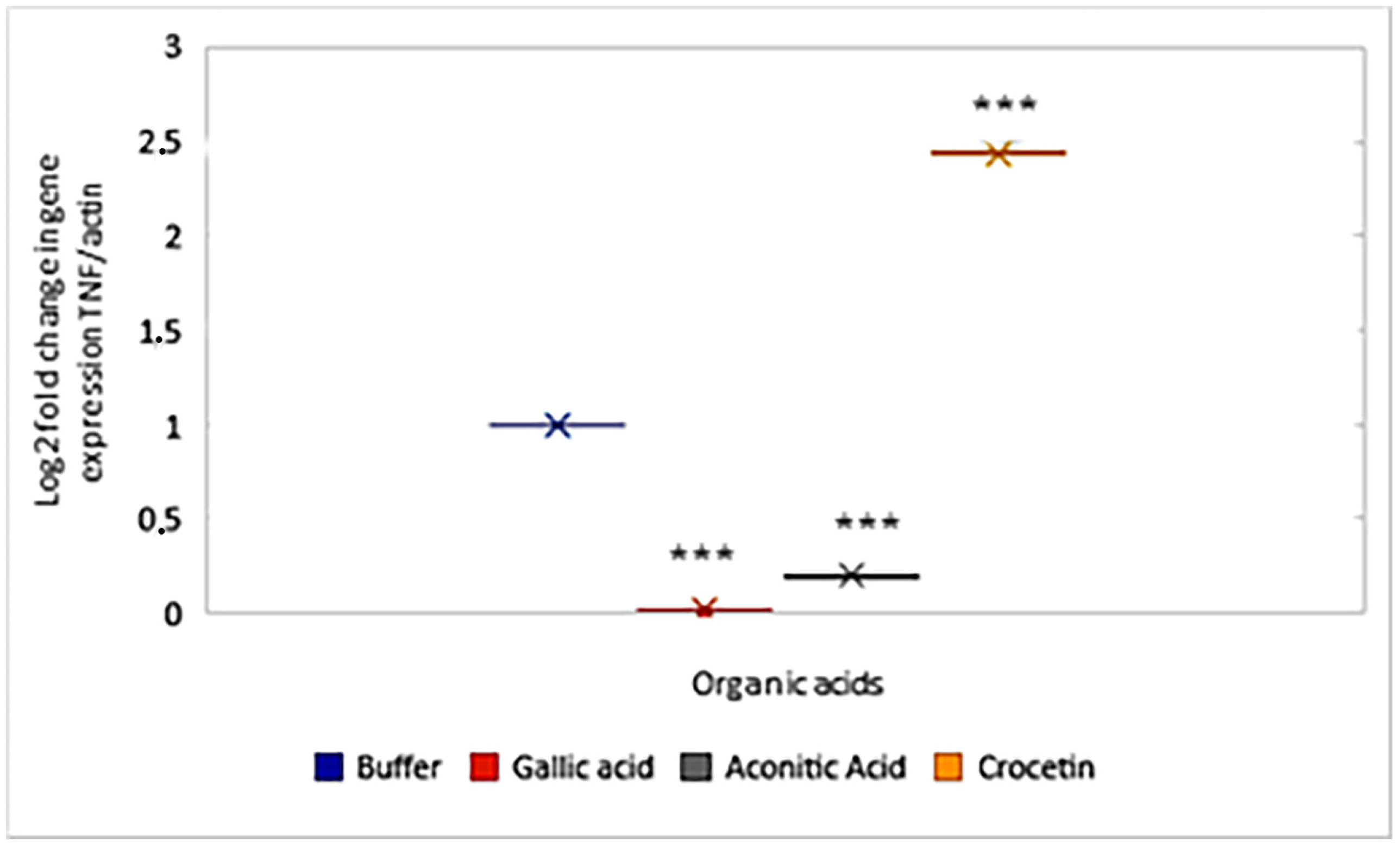
2.4. Influence of Organic Acids on the Expression of TNF
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Methods
4.2.1. Molecular Docking
4.2.2. ADMET Analysis
4.2.3. Molecular Dynamics Simulation
4.3. Animal Experiments
4.4. Isolation of RNA from Peritoneal Leukocytes
4.5. Reverse Transcription
4.6. Determination of Gene Expression Levels
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| TNF | Tumor necrosis factor |
| ADMET | Absorption, distribution, metabolism, excretion, and toxicity |
| MD | Molecular dynamic |
References
- Li, K.; Ma, S.; Zou, C.; Latif, J.; Jiang, Y.; Ni, Z.; Shen, S.; Feng, J.; Jia, H. Unrecognized Role of Organic Acid in Natural Attenuation of Pollutants by Mackinawite (FeS): The Significance of Carbon-Center Free Radicals. Environ. Sci. Technol. 2023, 57, 20871–20880. [Google Scholar] [CrossRef]
- Sindhu, S.S.; Sehrawat, A.; Glick, B.R. The involvement of organic acids in soil fertility, plant health and environment sustainability. Arch. Microbiol. 2022, 204, 720. [Google Scholar] [CrossRef]
- Wieërs, M.L.A.J.; Beynon-Cobb, B.; Visser, W.J.; Attaye, I. Dietary acid load in health and disease. Pflügers Arch.-Eur. J. Physiol. 2024, 476, 427–443. [Google Scholar] [CrossRef]
- Riaz, M.; Yan, L.; Wu, X.; Hussain, S.; Aziz, O.; Jiang, C. Mechanisms of organic acids and boron induced tolerance of aluminum toxicity: A review. Ecotoxicol. Environ. Saf. 2018, 165, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.U.; Naz, S.; Raziq, F.; Qudratullah, Q.; Khan, N.A.; Laudadio, V.; Tufarelli, V.; Ragni, M. Prospects of organic acids as safe alternative to antibiotics in broiler chickens diet. Environ. Sci. Pollut. Res. Int. 2022, 29, 32594–32604. [Google Scholar] [CrossRef]
- Xia, W.; Dai, X.Y.; Ding, L.K.; Xi, Y.; Yan, M.; Zhang, M.; Wu, L.; Yi, C.X.; Xu, H.X. Three main short-chain fatty acids inhibit the activation of THP-1 cells by Mycoplasma pneumoniae. Biosci. Biotechnol. Biochem. 2021, 85, 923–930. [Google Scholar]
- Wang, F.; Liu, J.; Weng, T.; Shen, K.; Chen, Z.; Yu, Y.; Huang, Q.; Wang, G.; Liu, Z.; Jin, S. The inflammation induced by lipopolysaccharide can be mitigated by short-chain fatty acid, butyrate, through upregulation of il-10 in septic shock. Scand. J. Immunol. 2017, 85, 258–263. [Google Scholar] [CrossRef]
- Rieira, A.T.; Galvao, I.; Macia, L.M.; Sernaglia, E.M.; Vinolo, M.A.R.; Garcia, C.C.; Tavares, L.P.; Amaral, F.A.; Sousa, L.P.; Martins, F.S.; et al. Dietary fiber and the short-chain fatty acid acetate promote resolution of neutrophilic inflammation in a model of gout in mice. J. Leukoc. Biol. 2017, 101, 275–284. [Google Scholar]
- Olsson, A.; Gustavsen, S.; Nguyen, T.D.; Nyman, M.; Langkilde, A.R.; Hansen, T.H.; Sellebjerg, F.; Oturai, A.B.; Sondergaard, H.B. Serum short-chain fatty acids and associations with inflsunammation in newly diagnosed patients with multiple sclerosis and healthy controls. Front. Immunol. 2021, 12, 661493. [Google Scholar] [CrossRef] [PubMed]
- Chédotal, H.; Narayanan, D.; Povlsen, K.; Gotfredsen, C.H.; Brambilla, R.; Gajhede, M.; Bach, A.; Clausen, M.H. Small-molecule modulators of tumor necrosis factor signaling. Drug Discov. Today 2023, 28, 103575. [Google Scholar] [CrossRef]
- Richmond, V.; Michelini, F.M.; Bueno, C.A.; Alche, L.E.; Ramirez, J.A. Small molecules as anti-TNF drugs. Curr. Med. Chem. 2015, 22, 2920–2942. [Google Scholar] [CrossRef]
- Fischer, R.; Kontermann, R.E.; Pfizenmaier, K. Selective targeting of TNF receptors as a novel therapeutic approach. Front. Cell Dev. Biol. 2020, 8, 401. [Google Scholar] [CrossRef]
- Sun, W.; Wu, Y.; Zheng, M.; Yang, Y.; Liu, Y.; Wu, C.; Zhou, Y.; Zhang, Y.; Chen, L.; Li, H. Discovery of an Orally Active Small-Molecule Tumor Necrosis Factor-α Inhibitor. J. Med. Chem. 2020, 63, 8146–8156. [Google Scholar] [CrossRef] [PubMed]
- de Castro Barbosa, M.L.; Fumian, M.M.; de Miranda, A.L.P.; Barreiro, E.J.; Lima, L.M. Therapeutic approaches for tumor necrosis factor inhibition. Braz. J. Pharm. Sci. 2011, 47, 427–446. [Google Scholar] [CrossRef]
- O’Connell, J.; Porter, J.; Kroeplien, B.; Norman, T.; Rapecki, S.; Davis, R.; McMillan, D.; Arakaki, T.; Burgin, A.; Fox Iii, D.; et al. Small molecules that inhibit TNF signalling by stabilising an asymmetric form of the trimer. Nat. Commun. 2019, 10, 5795. [Google Scholar] [CrossRef] [PubMed]
- Roshanravan, N.; Seif, F.; Ostadrahimi, A.; Pouraghaei, M.; Ghaffari, S. Targeting cytokine storm to manage patients with COVID-19: A mini-review. Arch. Med. Res. 2020, 51, 608–612. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, S.; Sharma, K.; Silakari, O. The interplay between inflammatory pathways and COVID-19: A critical review on pathogenesis and therapeutic options. Microb. Pathog. 2021, 150, 104673. [Google Scholar] [CrossRef]
- Patel, S.; Wadhwa, M. Therapeutic use of specific tumour necrosis factor inhibitors in inflammatory diseases including COVID-19. Biomed. Pharmacother. 2021, 140, 111785. [Google Scholar] [CrossRef]
- Chukwuma, I.F.; Apeh, V.O.; Nwodo, O.F.C. Mechanisms and potential therapeutic targets of hyperinflammatory responses in SARS-CoV-2. Acta Virol. 2021, 65, 3–9. [Google Scholar] [CrossRef]
- Chan, H.C.S.; Shan, H.; Dahoun, T.; Vogel, H.; Yuan, S. Advancing drug discovery via artificial intelligence. Trends Pharmacol. Sci. 2019, 40, 592–604. [Google Scholar] [CrossRef]
- Veljkovic, V.; Veljkovic, N.; Este, J.A.; Huther, A.; Dietrich, U. Application of the EIIP/ISM bioinformatics concept in development of new drugs. Curr. Med. Chem. 2007, 14, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Verbanac, D. Predictive methods as a powerful tool in drug discovery. Biochem. Medica 2010, 20, 314–318. [Google Scholar] [CrossRef]
- Singh, S.; Shingatgeri, V.; Srivastava, P. Revolutionizing new drug discovery: Harnessing AI and machine learning to overcome traditional challenges and accelerate targeted therapies. Artif. Intell. Health 2024, 2, 29–40. [Google Scholar] [CrossRef]
- Wu, F.; Zhou, Y.; Li, L.; Shen, X.; Chen, G.; Wang, X.; Liang, X.; Tan, M.; Huang, Z. Computational approaches in preclinical studies on drug discovery and development. Front. Chem. 2020, 8, 726. [Google Scholar] [CrossRef]
- Xia, S.; Chen, E.; Zhang, Y. Integrated molecular modeling and machine learning for drug design. J. Chem. Theory Comput. 2023, 19, 7478–7495. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2025 update. Nucleic Acids Res. 2025, 53, D1516–D1525. [Google Scholar] [CrossRef]
- Remy, G.; Risco, A.M.; Inesta-Vaquera, F.A.; Gonzalez-Teran, B.; Sabio, G.; Davis, R.J.; Cuenda, A. Differential activation of p38MAPK isoforms by MKK6 and MKK3. Cell. Signal. 2010, 22, 660–667. [Google Scholar] [CrossRef]
- Sabio, G.; Davis, R.J. TNF and MAP kinase signalling pathways. Semin. Immunol. 2014, 26, 237–245. [Google Scholar] [CrossRef]
- Lee, C.-K.; Lee, E.Y.; Kim, Y.G.; Mun, S.H.; Moon, H.-B.; Yoo, B. Alpha-lipoic acid inhibits TNF-α induced NF-κB activation through blocking of MEKK1–MKK4–IKK signaling cascades. Int. Immunopharmacol. 2008, 8, 362–370. [Google Scholar] [CrossRef]
- Malemud, C.J.; Pearlman, E. Targeting JAK/STAT signaling pathway in inflammatory diseases. Curr. Signal Transduct. Ther. 2009, 4, 201–221. [Google Scholar] [CrossRef]
- Kilani-Jaziri, S.; Mokdad-Bzeouich, I.; Krifa, M.; Nasr, N.; Ghedira, K.; Chekir-Ghedira, L. Immunomodulatory and cellular anti-oxidant activities of caffeic, ferulic, and p-coumaric phenolic acids: A structure–activity relationship study. Drug Chem. Toxicol. 2016, 40, 416–424. [Google Scholar] [CrossRef]
- Muhammad Abdul Kadar, N.N.; Ahmad, F.; Teoh, S.L.; Yahaya, M.F. Caffeic Acid on Metabolic Syndrome: A Review. Molecules 2021, 26, 5490. [Google Scholar] [CrossRef]
- Bai, J.; Zhang, Y.; Tang, C.; Hou, Y.; Ai, X.; Chen, X.; Zhang, Y.; Wang, X.; Meng, X. Gallic acid: Pharmacological activities and molecular mechanisms involved in inflammation-related diseases. Biomed. Pharmacother. 2020, 133, 110985. [Google Scholar] [CrossRef]
- Jiang, R.-W.; Lau, K.-M.; Hon, P.-M.; Mak, T.C.W.; Woo, K.-S.; Fung, K.-P. Chemistry and biological activities of caffeic acid derivatives from Salvia miltiorrhiza. Curr. Med. Chem. 2005, 12, 237–246. [Google Scholar] [CrossRef]
- Manhas, D.; Dhiman, S.; Kour, H.; Kour, D.; Sharma, K.; Wazir, P.; Vij, B.; Kumar, A.; Sawant, S.D.; Ahmed, Z.; et al. ADME/PK Insights of Crocetin: A Molecule Having an Unusual Chemical Structure with Druglike Features. ACS Omega 2024, 9, 21494–21509. [Google Scholar] [CrossRef]
- Hashemi, M.; Hosseinzadeh, H. A comprehensive review on biological activities and toxicology of crocetin. Food Chem. Toxicol. 2019, 130, 44–60. [Google Scholar] [CrossRef]
- Guo, Z.L.; Li, M.X.; Li, X.L.; Wang, P.; Wang, W.G.; Du, W.Z.; Yang, Z.Q.; Chen, S.F.; Wu, D.; Tian, X.Y. Crocetin: A Systematic Review. Front. Pharmacol. 2022, 12, 745683. [Google Scholar] [CrossRef]
- Giaccio, M. Crocetin from Saffron: An Active Component of an Ancient Spice. Crit. Rev. Food Sci. Nutr. 2004, 44, 155–172. [Google Scholar] [CrossRef] [PubMed]
- Pfendt, L.; Dražić, B.; Popović, G.; Drakulić, B.; Vitnik, Ž; Juranić, I. Determination of all pKa values of some di- and tri-carboxylic unsaturated and epoxy acids and their polylinear correlation with the carboxylic group atomic charges. J. Chem. Res. 2003, 5, 247–248. [Google Scholar] [CrossRef]
- Srinivasulu, C.; Ramgopal, M.; Ramanjaneyulu, G.; Anuradha, C.M.; Suresh Kumar, C. Syringic acid (SA)—A Review of Its Occurrence, Biosynthesis, Pharmacological and Industrial Importance. Biomed. Pharmacother. 2018, 108, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Mohamad, H.; Wan Abdul Jamil, W.A.N.; Abas, F.; Mohamad, K.S.; Ali, A.M. Octacosanoic acid, long chains saturated fatty acid from the marine sponges Xestospongia sp. Pertanika J. Trop. Agric. Sci. 2009, 32, 51–55. [Google Scholar]
- Pence, H.E.; Williams, A. ChemSpider: An Online Chemical Information Resource. J. Chem. Educ. 2010, 87, 1123. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Wua, R.; Dinga, F.; Wanga, R.; Shena, R.; Zhanga, X.; Luoa, S.; Sua, C.; Wua, Z.; Xieb, Q.; Bergerc, B.; et al. High-resolution de novo structure prediction from primary sequence. bioRxiv 2022. [Google Scholar] [CrossRef]
- Kaur, T.; Madgulkar, A.; Bhalekar, M.; Asgaonkar, K. Molecular Docking in Formulation and Development. Curr. Drug Discov. Technol. 2019, 16, 30–39. [Google Scholar] [CrossRef]
- Vugler, A.; O’Connell, J.; Nguyen, M.A.; Weitz, D.; Leeuw, T.; Hickford, E.; Verbitsky, A.; Ying, X.; Rehberg, M.; Carrington, B.; et al. An orally available small molecule that targets soluble TNF to deliver anti-TNF biologic-like efficacy in rheumatoid arthritis. Front Pharmacol. 2022, 13, 1037983, Erratum in Front Pharmacol. 2024, 15, 1479557. [Google Scholar] [CrossRef]
- Kagami, L.; Wilter, A.; Diaz, A.; Vranken, W. The ACPYPE web server for small-molecule MD topology generation. Bioinformatics 2023, 39, btad350. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| No. | Compound | Structure | 3D Structure | Binding Affinity, (kcal/mol) | |
|---|---|---|---|---|---|
| Max | Min | ||||
| 1 | Gallic acid | C7H6O5 |  | −4.7 | −4.2 |
| 2 | Syringic acid | C9H10O5 |  | −5.2 | −3.8 |
| 3 | Caffeic acid | C9H8O4 |  | −4.8 | −3.9 |
| 4 | Aconitic acid | C6H6O6 |  | −3.9 | −3.5 |
| 5 | Crocetin | C20H24O4 |  | −5.6 | −4.6 |
| 6 | Octacosanoic acid | C28H56O2 |  | −3.2 | −2.6 |
| 7 | Pentatriacontanoic acid | C35H70O2 |  | −3.2 | −2.7 |
| 8 | Ferulic acid | C10H10O4 |  | −4.8 | −4.0 |
| 9 | Vanillic acid | C8H8O4 |  | −5.0 | −3.8 |
| 10 | Isocitric acid | C6H8O7 |  | −4.3 | −3.7 |
| 11 | Curcumin | C21H20O6 |  | −4.9 | −4.7 |
| 12 | Thymoquinone | C10H12O2 |  | −4.5 | −4.0 |
| No. | Compound | H-Bonds | Van Der Waals Force | 2D Visualization in LigPlot |
|---|---|---|---|---|
| 1 | Gallic acid | His99, Gly226, Ala112 | Ser225, Val96, Arg111 | 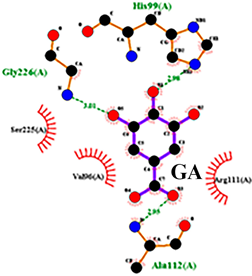 |
| 2 | Aconitic acid | Asn125, Leu105, Val214 | Glu213, Gln104, Pro217 |  |
| 3 | Crocetin | Tyr197 | Tyr138, Tyr229, His94, Gly226, Ala112, Val96, Ser225, His99, Arg111 |  |
| Property | Model Name | GA | AA | Cr | Unit |
|---|---|---|---|---|---|
| Absorption | Caco-2 Permeability | −5.728 | −6.055 | −5.511 | Numeric (log Papp in 10−6 cm/s) |
| HIA | 0.085 | 0.896 | 0.015 | Probability (0–1) | |
| Distribution | PPB | 53.49 | 26.98 | 92.92 | % |
| BBB permeability | 0.099 | 0.05 | 0.083 | Numeric (log BB) | |
| Metabolism | CYP2C9 substrate | 0.061 | 0.192 | 0.996 | Probability (0–1) |
| CYP2D6 inhibitor | 0.008 | 0.018 | 0.776 | Probability (0–1) | |
| Excretion | CL (mL/min/kg) | 10.108 | 1.776 | 0.744 | Numeric (mL/min/kg) |
| T1/2 | 0.947 | 0.924 | 0.738 | Numeric (h) | |
| Toxicity | hERG Blockers | 0.017 | 0.002 | 0.005 | Probability (0–1) |
| H-HT | 0.433 | 0.481 | 0.815 | Probability (0–1) | |
| Carcinogenicity | 0.024 | 0.016 | 0.406 | Probability (0–1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manakbayeva, A.; Bogoyavlenskiy, A.; Kerimov, T.; Yershov, I.; Alexyuk, P.; Alexyuk, M.; Berezin, V.; Dushenkov, V. Gallic, Aconitic, and Crocetin Acids as Potential TNF Modulators: An Integrated Study Combining Molecular Docking, Dynamics Simulations, ADMET Profiling, and Gene Expression Analysis. Molecules 2025, 30, 3175. https://doi.org/10.3390/molecules30153175
Manakbayeva A, Bogoyavlenskiy A, Kerimov T, Yershov I, Alexyuk P, Alexyuk M, Berezin V, Dushenkov V. Gallic, Aconitic, and Crocetin Acids as Potential TNF Modulators: An Integrated Study Combining Molecular Docking, Dynamics Simulations, ADMET Profiling, and Gene Expression Analysis. Molecules. 2025; 30(15):3175. https://doi.org/10.3390/molecules30153175
Chicago/Turabian StyleManakbayeva, Adolat, Andrey Bogoyavlenskiy, Timur Kerimov, Igor Yershov, Pavel Alexyuk, Madina Alexyuk, Vladimir Berezin, and Vyacheslav Dushenkov. 2025. "Gallic, Aconitic, and Crocetin Acids as Potential TNF Modulators: An Integrated Study Combining Molecular Docking, Dynamics Simulations, ADMET Profiling, and Gene Expression Analysis" Molecules 30, no. 15: 3175. https://doi.org/10.3390/molecules30153175
APA StyleManakbayeva, A., Bogoyavlenskiy, A., Kerimov, T., Yershov, I., Alexyuk, P., Alexyuk, M., Berezin, V., & Dushenkov, V. (2025). Gallic, Aconitic, and Crocetin Acids as Potential TNF Modulators: An Integrated Study Combining Molecular Docking, Dynamics Simulations, ADMET Profiling, and Gene Expression Analysis. Molecules, 30(15), 3175. https://doi.org/10.3390/molecules30153175







