Recent Advances in Amyloids Structural Studies and Thin Film Applications
Abstract
1. Introduction
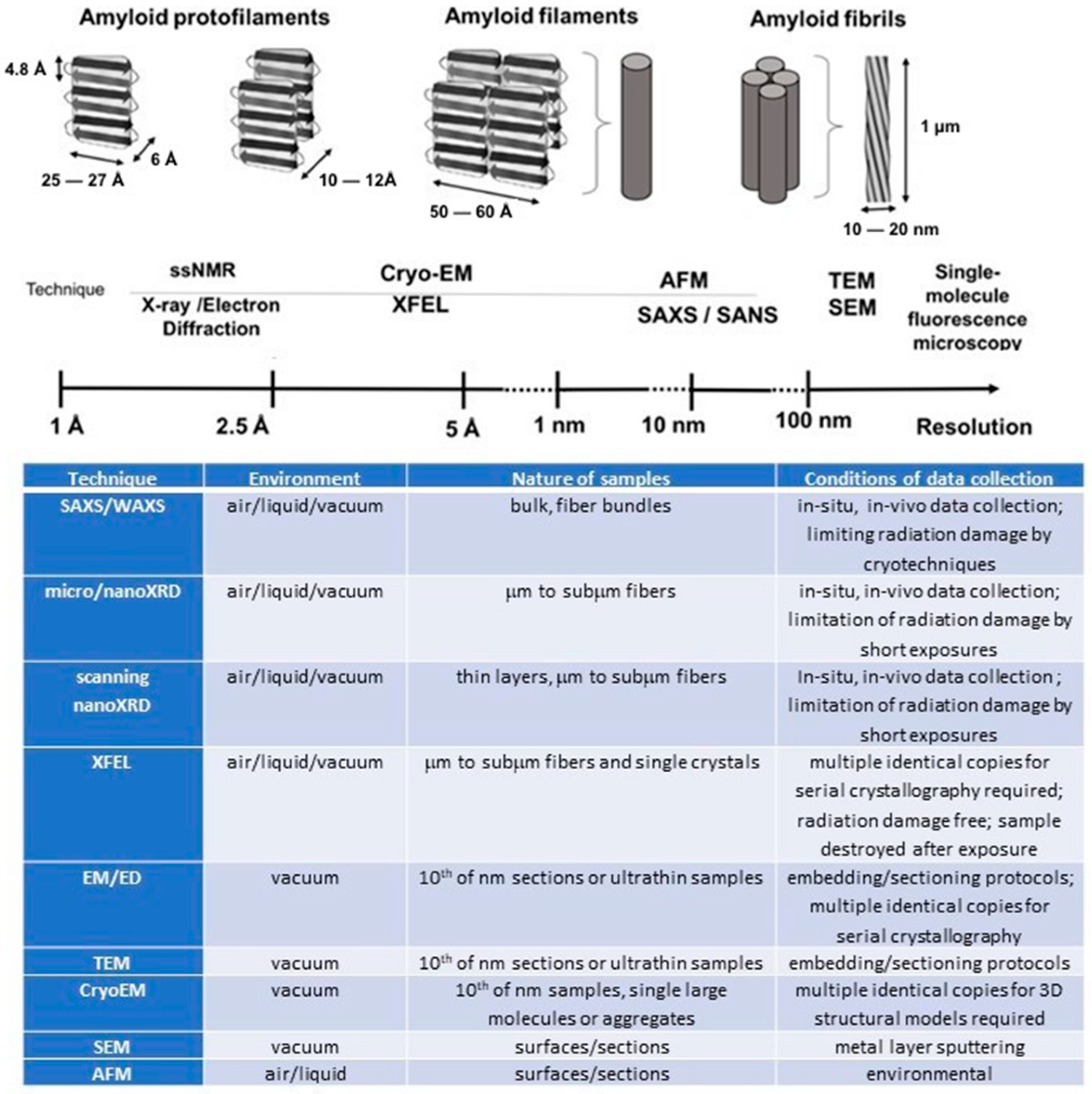
2. Amyloid’s Advanced Characterization Methods
3. Detection of Amyloids in Selected Recent Biomedical Research
4. Amyloids as Functional Materials for Selected Technological Applications
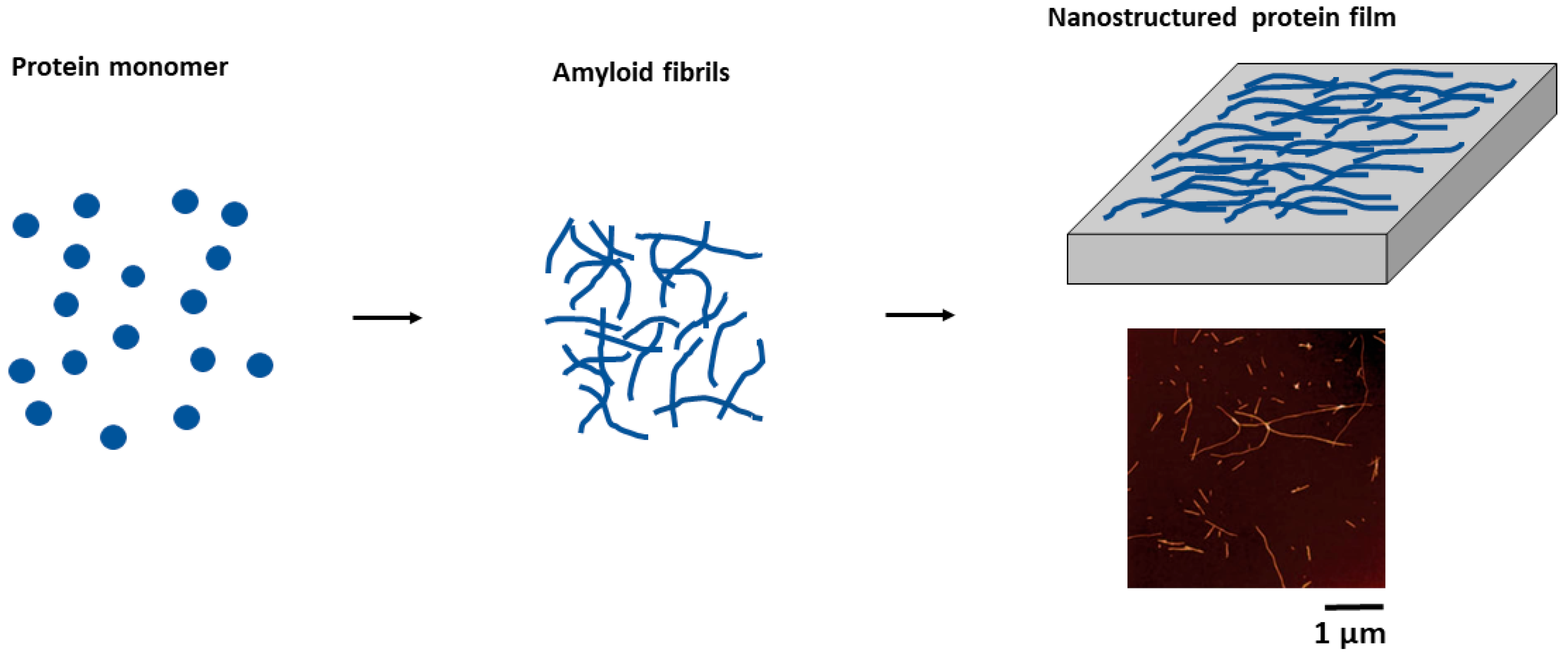
4.1. Amyloid Thin Film Production and Characterization
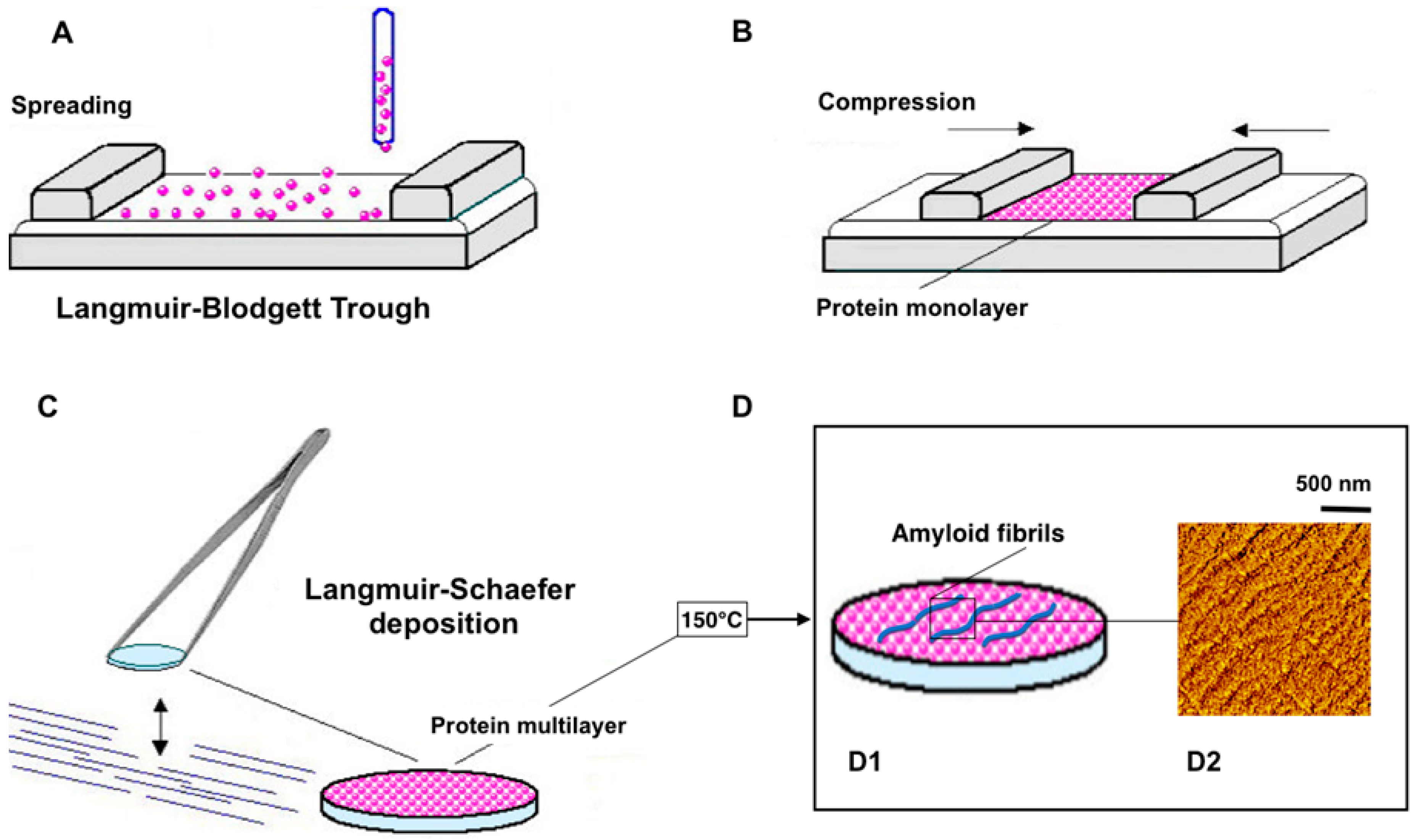
4.1.1. Immobilization of Amyloids
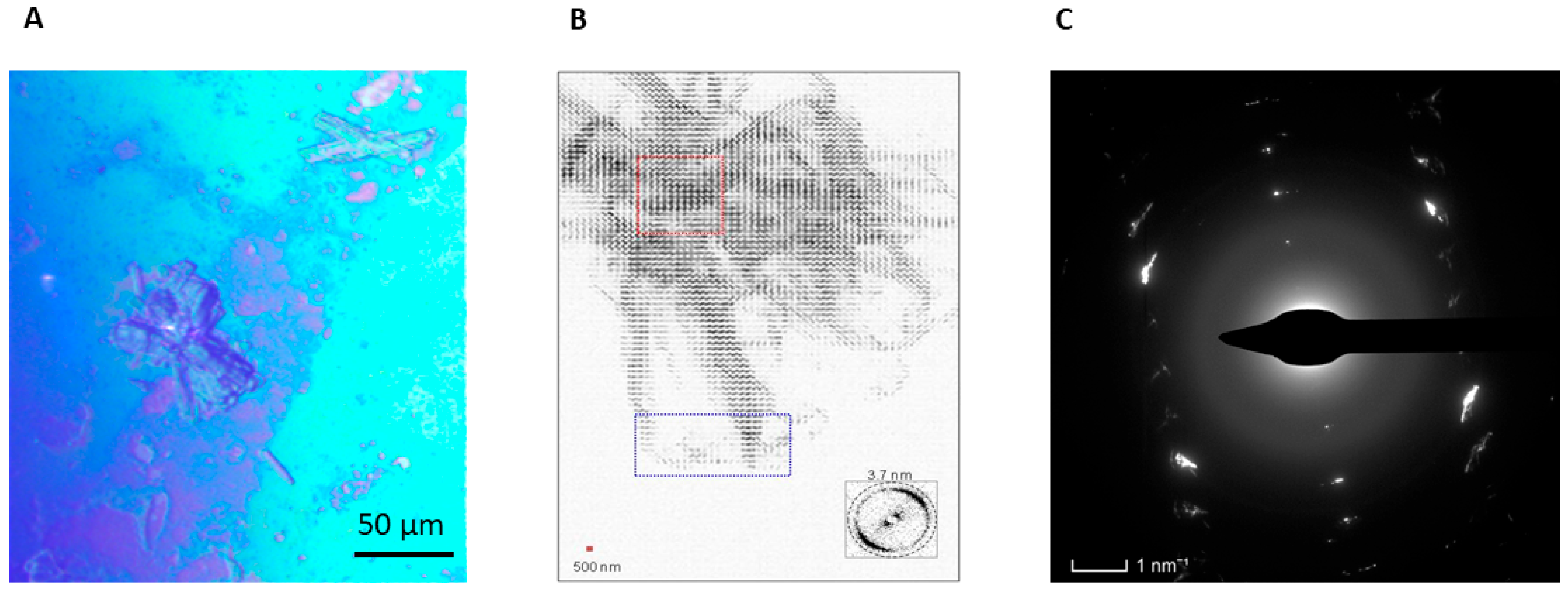
4.1.2. Appearance of Amyloid Motifs in Monomeric Protein Multilayers
5. Conclusions and Future Trends
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eisenberg, D.; Jucker, M. The amyloid state of proteins in human diseases. Cell 2012, 148, 1188–1203. [Google Scholar] [CrossRef] [PubMed]
- Sawaya, M.R.; Hughes, M.P.; Rodriguez, J.A.; Riek, R.; Eisenberg, D.S. The expanding amyloid family: Structure, stability, function, and pathogenesis. Cell 2021, 184, 4857–4873. [Google Scholar] [CrossRef] [PubMed]
- Fowler, D.M.; Koulov, A.V.; Balch, W.E.; Kelly, J.W. Functional amyloid—From bacteria to humans. Trends Biochem. Sci. 2007, 32, 217–224. [Google Scholar] [CrossRef]
- Li, J.; Zhang, F. Amyloids as Building Blocks for Macroscopic Functional Materials: Designs, Applications and Challenges. Int. J. Mol. Sci. 2021, 22, 10698. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.; Török, M. Functional amyloids in the human body. Bioorganic Med. Chem. Lett. 2021, 40, 127914. [Google Scholar] [CrossRef]
- Sunde, M.; Serpell, L.C.; Bartlam, M.; Fraser, P.E.; Pepys, M.B.; Blake, C.C. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 1997, 273, 729–739. [Google Scholar] [CrossRef]
- Alraawi, Z.; Banerjee, N.; Mohanty, S.; Kumar, T.K.S. Amyloidogenesis: What Do We Know So Far? Int. J. Mol. Sci. 2022, 23, 13970. [Google Scholar] [CrossRef]
- Smith, J.F.; Knowles, T.P.; Dobson, C.M.; Macphee, C.E.; Welland, M.E. Characterization of the nanoscale properties of individual amyloid fibrils. Proc. Natl. Acad. Sci. USA 2006, 103, 15806–15811. [Google Scholar] [CrossRef]
- Bucciantini, M.; Giannoni, E.; Chiti, F.; Baroni, F.; Formigli, L.; Zurdo, J.; Taddei, N.; Ramponi, G.; Dobson, C.M.; Stefani, M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 2002, 416, 507–511. [Google Scholar] [CrossRef]
- Winner, B.; Jappelli, R.; Maji, S.K.; Desplats, P.A.; Boyer, L.; Aigner, S.; Hetzer, C.; Loher, T.; Vilar, M.; Campioni, S.; et al. In vivo demonstration that α-synuclein oligomers are toxic. Proc. Natl. Acad. Sci. USA 2011, 108, 4194–4199. [Google Scholar] [CrossRef]
- Sulatsky, M.I.; Sulatskaya, A.I.; Stepanenko, O.V.; Povarova, O.I.; Kuznetsova, I.M.; Turoverov, K.K. Denaturant effect on amyloid fibrils: Declasterization, depolymerization, denaturation and reassembly. Int. J. Biol. Macromol. 2020, 150, 681–694. [Google Scholar] [CrossRef]
- Chuang, E.; Hori, A.M.; Hesketh, C.D.; Shorter, J. Amyloid assembly and disassembly. J Cell Sci. 2018, 131, jcs189928. [Google Scholar] [CrossRef] [PubMed]
- Taglialegna, A.; Lasa, I.; Valle, J. Amyloid Structures as Biofilm Matrix Scaffolds. J. Bacteriol. 2016, 198, 2579–2588. [Google Scholar] [CrossRef] [PubMed]
- Akbey, Ü.; Andreasen, M. Functional amyloids from bacterial biofilms—Structural properties and interaction partners. Chem. Sci. 2022, 13, 6457–6477. [Google Scholar] [CrossRef]
- Duran-Meza, E.; Diaz-Espinoza, R. Catalytic Amyloids as Novel Synthetic Hydrolases. Int. J. Mol. Sci. 2021, 22, 9166. [Google Scholar] [CrossRef]
- Arad, E.; Baruch Leshem, A.; Rapaport, H.; Jelinek, R. β-Amyloid fibrils catalyze neurotransmitter degradation. Chem. Catal. 2021, 1, 908–922. [Google Scholar] [CrossRef]
- Wittung-Stafshede, P. Chemical catalysis by biological amyloids. Biochem. Soc. Trans. 2023, 51, 1967–1974. [Google Scholar] [CrossRef]
- Li, Y.; Li, K.; Wang, X.; Cui, M.; Ge, P.; Zhang, J.; Qiu, F.; Zhong, C. Conformable self-assembling amyloid protein coatings with genetically programmable functionality. Sci. Adv. 2020, 6, eaba1425. [Google Scholar] [CrossRef]
- Jin, T.; Peydayesh, M.; Li, M.; Yao, Y.; Wu, D.; Mezzenga, R. Functional Coating from Amyloid Superwetting Films. Adv. Mater. 2022, 34, 2205072. [Google Scholar] [CrossRef]
- Kobayashi, S.; Tanaka, Y.; Kiyono, M.; Chino, M.; Chikuma, T.; Hoshi, K.; Ikeshima, H. Dependence pH and proposed mechanism for aggregation of Alzheimer’s disease-related amyloid-β(1–42) protein. J. Mol. Struct. 2015, 1094, 109–117. [Google Scholar] [CrossRef]
- Novo, M.; Freire, S.; Al-Soufi, W. Critical aggregation concentration for the formation of early Amyloid-β (1–42) oligomers. Sci. Rep. 2018, 8, 1783. [Google Scholar] [CrossRef] [PubMed]
- Ziaunys, M.; Sakalauskas, A.; Mikalauskaite, K.; Snieckute, R.; Smirnovas, V. Temperature-Dependent Structural Variability of Prion Protein Amyloid Fibrils. Int. J. Mol. Sci. 2021, 22, 5075. [Google Scholar] [CrossRef]
- Vugmeyster, L.; Au, D.F.; Ostrovsky, D.; Kierl, B.; Fu, R.; Hu, Z.W.; Qiang, W. Effect of Post-Translational Modifications and Mutations on Amyloid-β Fibrils Dynamics at N Terminus. Biophys. J. 2019, 117, 1524–1535. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef]
- Pansieri, J.; Halim, M.A.; Vendrely, C.; Dumoulin, M.; Legrand, F.; Moulin Sallanon, M.; Chierici, S.; Denti, S.; Dagany, X.; Dugourd, P.; et al. Mass and charge distributions of amyloid fibers involved in neurodegenerative diseases: Mapping heterogeneity and polymorphism. Chem. Sci. 2018, 9, 2791–2796. [Google Scholar] [CrossRef] [PubMed]
- Galzitskaya, O. New Mechanism of Amyloid Fibril Formation. Curr. Protein Pept. Sci. 2019, 20, 630–640. [Google Scholar] [CrossRef]
- Kaur, A.; Adair, L.D.; Ball, S.R.; New, E.J.; Sunde, M. A fluorescent sensor for quantitative super-resolution imaging of amyloid fibril assembly. Angew. Chem. Int. Ed. 2022, 61, e202112832. [Google Scholar] [CrossRef]
- Horrocks, M.H.; Lee, S.F.; Gandhi, S.; Magdalinou, N.K.; Chen, S.W.; Devine, M.J.; Tosatto, L.; Kjaergaard, M.; Beckwith, J.S.; Zetterberg, H.; et al. Single-molecule imaging of individual amyloid protein aggregates in human biofluids. ACS Chem. Neurosci. 2016, 7, 399–406. [Google Scholar] [CrossRef]
- Rice, L.J.; Ecroyd, H.; Antoine, M.; van Oijen, A.M. Illuminating amyloid fibrils: Fluorescence-based single-molecule approaches. Comput. Struct. Biotechnol. J. 2021, 19, 4711–4724. [Google Scholar] [CrossRef]
- Drolle, E.; Hane, F.; Lee, B.; Leonenko, Z. Atomic force microscopy to study molecular mechanisms of amyloid fibril formation and toxicity in Alzheimer’s disease. Drug Metab. Rev. 2014, 46, 207–223. [Google Scholar] [CrossRef]
- Charnley, M.; Gilbert, J.; Jones, O.G.; Reynolds, N.P. Characterization of Amyloid Fibril Networks by Atomic Force Microscopy. Bio Protoc. 2018, 8, e2732. [Google Scholar] [CrossRef]
- Ruggeri, F.S.; Benedetti, F.; Knowles, T.P.J.; Lashuel, H.A.; Sekatskii, S.; Dietler, G. Identification and nanomechanical characterization of the fundamental single-strand protofilaments of amyloid α-synuclein fibrils. Proc. Natl. Acad. Sci. USA 2018, 115, 7230–7235. [Google Scholar] [CrossRef]
- Ruggeri, F.S.; Šneideris, T.; Vendruscolo, M.; Knowles, T.P.J. Atomic force microscopy for single molecule characterisation of protein aggregation. Arch. Biochem. Biophys. 2019, 664, 134–148. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; He, G. Toward a Rational Design to Regulate β-Amyloid Fibrillation for Alzheimer’s Disease Treatment. ACS Chem Neurosci. 2018, 9, 198–210. [Google Scholar] [CrossRef]
- Almeida, Z.L.; Brito, R.M.M. Structure and Aggregation Mechanisms in Amyloids. Molecules 2020, 25, 1195. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.W.; Kelly, J.E.; Kelz, J.I. Advances in instrumentation and methodology for solid-state NMR of biological assemblies. J. Struct. Biol. 2019, 206, 73–89. [Google Scholar] [CrossRef] [PubMed]
- Meier, B.H.; Riek, R.; Böckmann, A. Emerging structural understanding of amyloid fibrils by solid-state NMR. Trends Biochem. Sci. 2017, 42, 777–787. [Google Scholar] [CrossRef]
- Tycko, R. Solid-state NMR studies of amyloid fibril structure. Annu. Rev. Phys. Chem. 2011, 62, 279–299. [Google Scholar] [CrossRef]
- Lee, M.; Wang, T.; Makhlynets, O.V. Zinc-binding structure of a catalytic amyloid from solid-state NMR. Proc. Natl. Acad. Sci. USA 2017, 114, 6191–6196. [Google Scholar] [CrossRef]
- Makin, O.S.; Atkins, E.; Sikorski, P.; Johansson, J.; Serpell, L.C. Molecular basis for amyloid fibril formation and stability. Proc. Natl. Acad. Sci. USA 2005, 102, 315. [Google Scholar] [CrossRef]
- Bowler, J.T.; Sawaya, M.R.; Boyer, D.R.; Cascio, D.; Bali, M.; Eisenberg, D.S. Micro-electron diffraction structure of the aggregation-driving N terminus of Drosophila neuronal protein Orb2A reveals amyloid-like beta-sheets. J. Biol. Chem. 2022, 298, 102396. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.; Sawaya, M.R.; Balbirnie, M.; Madsen, A.Ø.; Riekel, C.; Grothe, R.; Eisenberg, D. Structure of the cross-β spine of amyloid-like fibrils. Nature 2005, 435, 773–778. [Google Scholar] [CrossRef]
- Morris, K.L.; Serpell, L.C. X-Ray Fiber Diffraction Studies of Amyloid Fibrils. In Amyloid Proteins; Humana Press: Totowa, NJ, USA, 2012; pp. 121–135. [Google Scholar]
- Matsuo, T.; Peters, J. Fiber Diffraction and Small-Angle Scattering for Structural Investigation of Bacterial Amy-loids. Methods Mol. Biol. 2022, 2538, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Dahal, E.; Choi, M.; Alam, N.; Bhirde, A.A.; Beaucage, S.L.; Badano, A. Structural evaluation of an amyloid fibril model using small-angle X-ray scattering. Phys. Biol. 2017, 14, 046001. [Google Scholar] [CrossRef]
- Brewster, A.S.; Sawaya, M.R.; Rodriguez, J.; Hattne, J.; Echols, N.; McFarlane, H.T.; Cascio, D.; Adams, P.D.; Eisenberg, D.S.; Sauter, N.K. Indexing amyloid peptide diffraction from serial femtosecond crystallography: New algorithms for sparse patterns. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 357–366. [Google Scholar] [CrossRef]
- Seuring, C.; Ayyer, K.; Filippaki, E.; Barthelmess, M.; Longchamp, J.N.; Ringler, P.; Pardini, T.; Wojtas, D.H.; Coleman, M.A.; Dörner, K.; et al. Femtosecond X-ray coherent diffraction of aligned amyloid fibrils on low background graphene. Nat. Commun. 2018, 9, 1836. [Google Scholar] [CrossRef]
- Wojtas, D.H.; Ayyer, K.; Liang, M.; Mossou, E.; Romoli, F.; Seuring, C.; Beyerlein, K.R.; Bean, R.J.; Morgan, A.J.; Oberthuer, D.; et al. Analysis of XFEL serial diffraction data from individual crystalline fibrils. IUCrJ 2017, 4, 795–811. [Google Scholar] [CrossRef]
- Botha, S.; Fromme, P. Review of serial femtosecond crystallography including the COVID-19 pandemic impact and future outlook. Structure 2023, 31, 1306–1319. [Google Scholar] [CrossRef]
- Marinaro, G.; Graceffa, R.; Riekel, C. Wall-free droplet microfluidics for probing biological processes by high-brilliance X-ray scattering techniques. Front. Mol. Biosci. 2022, 9, 1049327. [Google Scholar] [CrossRef]
- Kühlbrandt, W. The Resolution Revolution. Science 2014, 343, 1443–1444. [Google Scholar] [CrossRef]
- Li, X.; Zhang, S.; Liu, Z. Subtle change of fibrillation condition leads to substantial alteration of recombinant Tau fibril structure. iScience 2022, 25, 105645. [Google Scholar] [CrossRef]
- Yang, Y.; Arseni, D.; Zhang, W.; Huang, M.; Lovestam, S.; Schweighauser, M.; Kotecha, A.; Murzin, A.G.; Peak-Chew, S.Y.; Macdonald, J.; et al. Cryo-EM structures of amyloid-beta 42 filaments from human brains. Science 2022, 375, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Babinchak, W.M.; Surewicz, W.K. Cryo-EM structure of amyloid fibrils formed by the entire low complexity domain of TDP-43. Nat. Commun. 2021, 12, 1620. [Google Scholar] [CrossRef] [PubMed]
- Kollmer, M.; Close, W.; Funk, L.; Rasmussen, J.; Bsoul, A.; Schierhorn, A.; Schmidt, M.; Sigurdson, C.J.; Jucker, M.; Fändrich, M. Cryo-EM structure and polymorphism of Aβ amyloid fibrils purified from Alzheimer’s brain tissue. Nat. Commun. 2019, 10, 4760. [Google Scholar] [CrossRef]
- Hervas, R.; Rau, M.J.; Park, Y.; Zhang, W.; Murzin, A.G.; Fitzpatrick, J.A.J.; Scheres, S.H.W.; Si, K. Cryo-EM structure of a neuronal functional amyloid implicated in memory persistence in Drosophila. Science 2020, 367, 1230–1234. [Google Scholar] [CrossRef]
- Lashuel, H.A.; LaBrenz, S.R.; Woo, L.; Serpell, L.C.; Kelly, J.W. Protofilaments, Filaments, Ribbons, and Fibrils from Peptidomimetic Self-Assembly: Implications for Amyloid Fibril Formation and Materials Science. J. Am. Chem. Soc. 2000, 122, 5262–5277. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, D.S.; Sawaya, M.R. Structural Studies of Amyloid Proteins at the Molecular Level. Annu. Rev. Biochem. 2017, 86, 69–95. [Google Scholar] [CrossRef]
- Müller, U.; Deller, T.; Korte, M. Not just amyloid: Physiological functions of the amyloid precursor protein family. Nat. Rev. Neurosci. 2017, 18, 281–298. [Google Scholar] [CrossRef]
- Carneiro, P.; Loureiro, J.; Delerue-Matos, C.; Morais, S.; Pereira, M.D.C. Alzheimer’s disease: Development of a sensitive label-free electrochemical immunosensor for detection of amyloid beta peptide. Sens. Actuators B Chem. 2017, 239, 157–165. [Google Scholar] [CrossRef]
- Jra-Guajardo, P.; Cabrera, P.; Celis, F.; Soler, M.; Berlanga, I.; Parra-Munoz, N.; Acosta, G.; Albericio, F.; Guz-man, F.; Campos, M.; et al. Gold nanoparticles mediate improved detection of β-amyloid aggregates by fluorescence. Nanomaterials 2020, 10, 690. [Google Scholar] [CrossRef]
- Moreira, F.T.C.; Correia, B.P.; Sousa, M.P.; Sales, G.F. Colorimetric cellulose-based test-strip for rapid detection of amyloid β-42. Microchim. Acta 2021, 188, 334–343. [Google Scholar] [CrossRef]
- Špringer, T.; Hemmerová, E.; Finocchiaro, G.; Krištofiková, Z.; Vyhnálek, M.; Homola, J. Surface plasmon resonance biosensor for the detection of tau-amyloid β complex. Sens. Actuators B Chem. 2020, 316, 128146. [Google Scholar] [CrossRef]
- Yu, X.K.; Hayden, E.Y.; Xia, M.; Liang, O.; Cheah, L.; Teplow, D.B.; Xie, Y.H. Surface enhanced Raman spectroscopy distinguishes amyloid Β-protein isoforms and conformational states. Protein Sci. 2018, 27, 1427–1438. [Google Scholar] [CrossRef] [PubMed]
- Bruggink, K.A.; Jongbloed, W.; Biemans, E.A.L.M.; Veerhuis, R.; Claassen, J.A.H.R.; Kuiperij, H.B.; Verbeek, M.M. Amyloid-β oligomer detection by ELISA in cerebrospinal fluid and brain tissue. Anal. Biochem. 2013, 433, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, M.; Zhang, D.; Li, P.; Wang, D.; Sun, S.; Wei, W. Detection of β-amyloid peptide aggregates by quartz crystal microbalance based on dual-aptamer assisted signal amplification. Anal. Chim. Acta. 2023, 1244, 340857. [Google Scholar] [CrossRef] [PubMed]
- Chiou, B.; Connor, J.R. Emerging and Dynamic Biomedical Uses of Ferritin. Pharmaceuticals 2018, 11, 124. [Google Scholar] [CrossRef]
- Gombos, J.; Balejcikova, L.; Kopcansky, P.; Batkova, M.; Siposova, K.; Kovac, J.; Zolochevska, K.; Safarik, I.; Lokajova, A.; Garamus, V.M.; et al. Destruction of Lysozyme Amyloid Fibrils Induced by Magnetoferritin and Reconstructed Ferritin. Int. J. Mol. Sci. 2022, 23, 13926. [Google Scholar] [CrossRef]
- Rodrigues, M.; Bhattacharjee, P.; Brinkmalm, A.; Do, D.T.; Pearson, C.M.; De, S.; Ponjavic, A.; Varela, J.A.; Kulenkampff, K.; Baudrexel, I.; et al. Structure-specific amyloid precipitation in biofluids. Nat. Chem. 2022, 14, 1045–1053. [Google Scholar] [CrossRef]
- Pansieri, J.; Gerstenmayer, M.; Lux, F.; Mériaux, S.; Tillement, O.; Forge, V.; Larrat, B.; Marquette, C. Magnetic Nanoparticles Applications for Am-yloidosis Study and Detection: A Review. Nanomaterials 2018, 8, 740. [Google Scholar] [CrossRef]
- Balistreri, A.; Goetzler, E.; Chapman, M. Functional Amyloids Are the Rule Rather Than the Exception in Cellular Biology. Microorganisms 2020, 8, 1951. [Google Scholar] [CrossRef]
- Abdelrahman, S.; Alghrably, M.; Lachowicz, J.I.; Emwas, A.H.; Hauser, C.A.E.; Jaremko, M. “What Doesn’t Kill You Makes You Stronger”: Future Applications of Amyloid Aggregates in Biomedicine. Molecules 2020, 25, 5245. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.J.; Mezzenga, R. Amyloid Fibrils as Building Blocks for Natural and Artificial Functional Materials. Adv. Mater. 2016, 28, 6546–6561. [Google Scholar] [CrossRef]
- Rufo, C.M.; Moroz, Y.S.; Moroz, O.V.; Stöhr, J.; Smith, T.A.; Hu, X.; DeGrado, W.F.; Korendovych, I.V. Short peptides self-assemble to produce catalytic amyloids. Nat. Chem. 2014, 6, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Espinoza, R. Catalytically Active Amyloids as Future Bionanomaterials. Nanomaterials 2022, 12, 3802. [Google Scholar] [CrossRef]
- Cherny, I.; Gazit, E. Amyloids: Not Only Pathological Agents but Also Ordered Nanomaterials. Angew. Chem. Int. Ed. Engl. 2008, 47, 4062–4069. [Google Scholar] [CrossRef] [PubMed]
- Lendel, C.; Solin, N. Protein nanofibrils and their use as building blocks of sustainable materials. RSC Adv. 2021, 11, 39188–39215. [Google Scholar] [CrossRef]
- Hu, X.; Tian, J.; Li, C.; Su, H.; Qin, R.; Wang, Y.; Cao, X.; Yang, P. Amyloid-Like Protein Aggregates: A New Class of Bioinspired Materials Merging an Interfacial Anchor with Antifouling. Adv. Mater. 2020, 32, 2000128. [Google Scholar] [CrossRef]
- Peydayesh, M.; Mezzenga, R. Protein nanofibrils for next generation sustainable water purification. Nat. Commun. 2021, 12, 3248. [Google Scholar] [CrossRef]
- Vinayagam, V.; Murugan, S.; Kumaresan, R.; Narayanan, M.; Sillanpää, M.; Vo, D.V.N.; Kushwaha, O.S. Protein nanofibrils as versatile and sustainable adsorbents for an effective removal of heavy metals from wastewater: A review. Chemosphere 2022, 301, 134635. [Google Scholar] [CrossRef]
- Li, D.; Jones, E.M.; Sawaya, M.R. Structure-Based Design of Functional Amyloid Materials. J. Am. Chem. Soc. 2014, 136, 18044–18051. [Google Scholar] [CrossRef]
- Wei, G.; Su, Z.; Reynolds, N.P.; Arosio, P.; Hamley, I.W.; Gazit, E.; Mezzenga, R. Self-assembling peptide and protein amyloids: From structure to tailored function in nanotechnology. Chem. Soc. Rev. 2017, 46, 4661–4708. [Google Scholar] [CrossRef] [PubMed]
- Bolisetty, S.; Arcari, M.; Adamcik, J.; Mezzenga, R. Hybrid Amyloid Membranes for Continuous Flow Catalysis. Langmuir 2015, 31, 13867–13873. [Google Scholar] [CrossRef]
- Knowles, T.P.; Oppenheim, T.W.; Buell, A.K.; Chirgadze, D.Y.; Welland, M.E. Nanostructured films from hierarchical self-assembly of amyloidogenic proteins. Nat. Nanotechnol. 2010, 5, 204–207. [Google Scholar] [CrossRef]
- Jordens, S.; Schwenke, K.; Usov, I.; Del Gado, E.; Mezzenga, R. Nematic field transfer in a two-dimensional protein fibril assembly. Soft Matter. 2016, 12, 1830–1835. [Google Scholar] [CrossRef]
- Qin, R.; Guo, Y.; Ren, H.; Liu, Y.; Su, H.; Chu, X.; Jin, Y.; Lu, F.; Wang, B.; Yang, P. Instant Adhesion of Amyloid-like Nanofilms with Wet Surfaces. ACS Cent. Sci. 2022, 8, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tao, F.; Miao, S.; Yang, P. Controlling the Structure and Function of Protein Thin Films through Amyloid-like Aggregation. Acc. Chem. Res. 2021, 54, 3016–3027. [Google Scholar] [CrossRef] [PubMed]
- Pechkova, E.; Nicolini, C. Langmuir-Blodgett nanotemplates for protein crystallography. Nat. Protoc. 2017, 12, 2570–2589. [Google Scholar] [CrossRef]
- Pechkova, E.; Nicolini, C.; Burghammer, M.; Riekel, C. Emergence of amyloidic fibrillation in 2D-ordered Langmuir-Blodgett protein multilayers upon heating. Appl. Phys. Lett. 2020, 117, 053701. [Google Scholar] [CrossRef]
- Pechkova, E.; Burghammer, M.; Nicolini, C.; Riekel, C. New structural features appear in thermally treated Lan-muir-Blodgett protein multilayers. NanoWorld J. 2020, 6, 66–67. [Google Scholar] [CrossRef]
- Pechkova, E.; Nicolini, C.; Fiordoro, S.; Riekel, C. Mesoscale ordering of Phycocyanin molecules in Langmuir-Blodgett multilayers. Langmuir 2022, 38, 86–91. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pechkova, E.; Fiordoro, S.; Izzotti, A.; Riekel, C. Recent Advances in Amyloids Structural Studies and Thin Film Applications. Molecules 2025, 30, 2908. https://doi.org/10.3390/molecules30142908
Pechkova E, Fiordoro S, Izzotti A, Riekel C. Recent Advances in Amyloids Structural Studies and Thin Film Applications. Molecules. 2025; 30(14):2908. https://doi.org/10.3390/molecules30142908
Chicago/Turabian StylePechkova, Eugenia, Stefano Fiordoro, Alberto Izzotti, and Christian Riekel. 2025. "Recent Advances in Amyloids Structural Studies and Thin Film Applications" Molecules 30, no. 14: 2908. https://doi.org/10.3390/molecules30142908
APA StylePechkova, E., Fiordoro, S., Izzotti, A., & Riekel, C. (2025). Recent Advances in Amyloids Structural Studies and Thin Film Applications. Molecules, 30(14), 2908. https://doi.org/10.3390/molecules30142908







