Chemical Fingerprints of Honey Fermented by Conventional and Non-Conventional Yeasts
Abstract
1. Introduction
2. Results and Discussion
2.1. Yeast Growth at Different Concentrations of Glucose
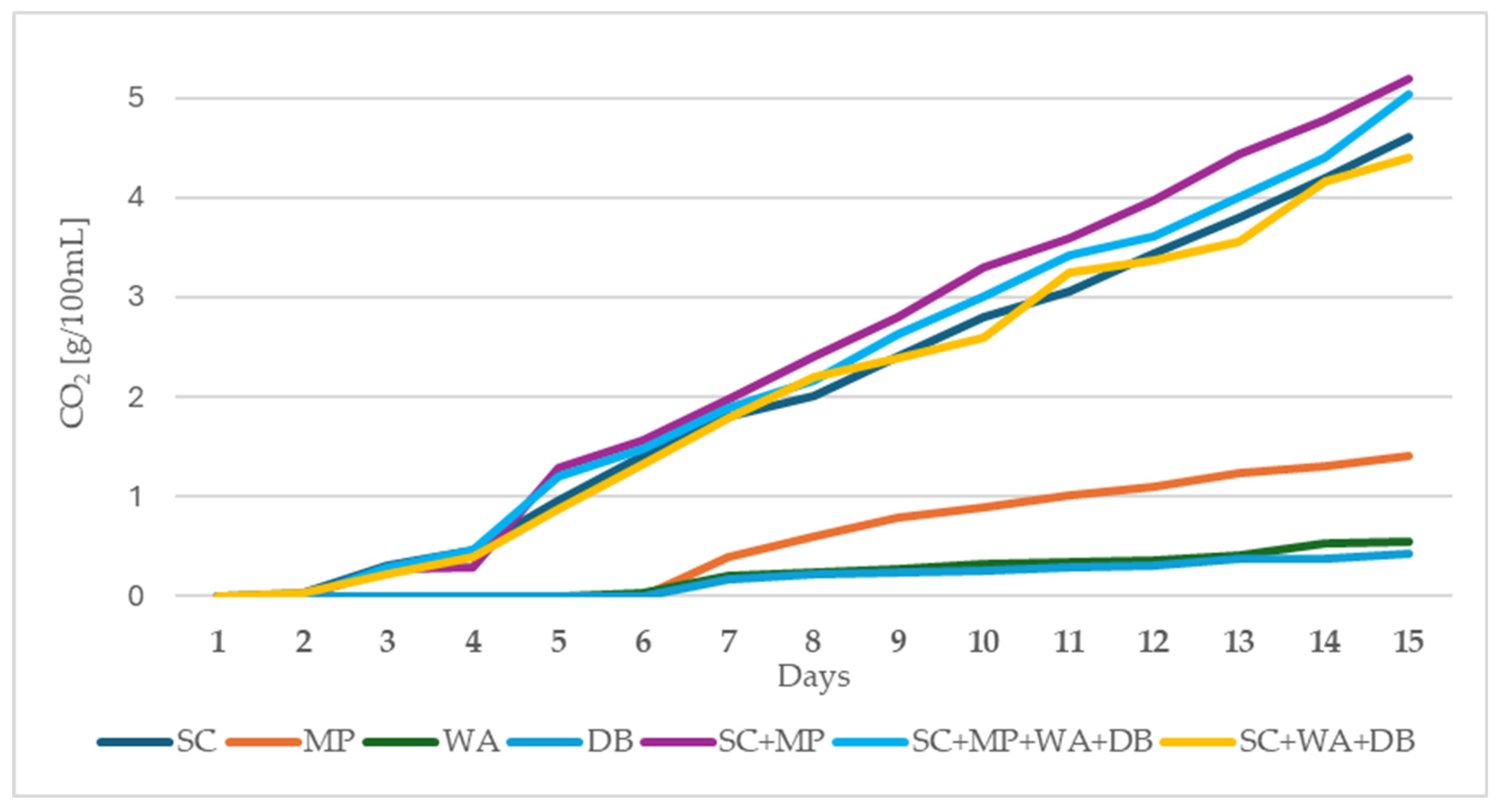
2.2. Fermentation Performance of Tested Yeast Strains
2.3. HPLC Analysis
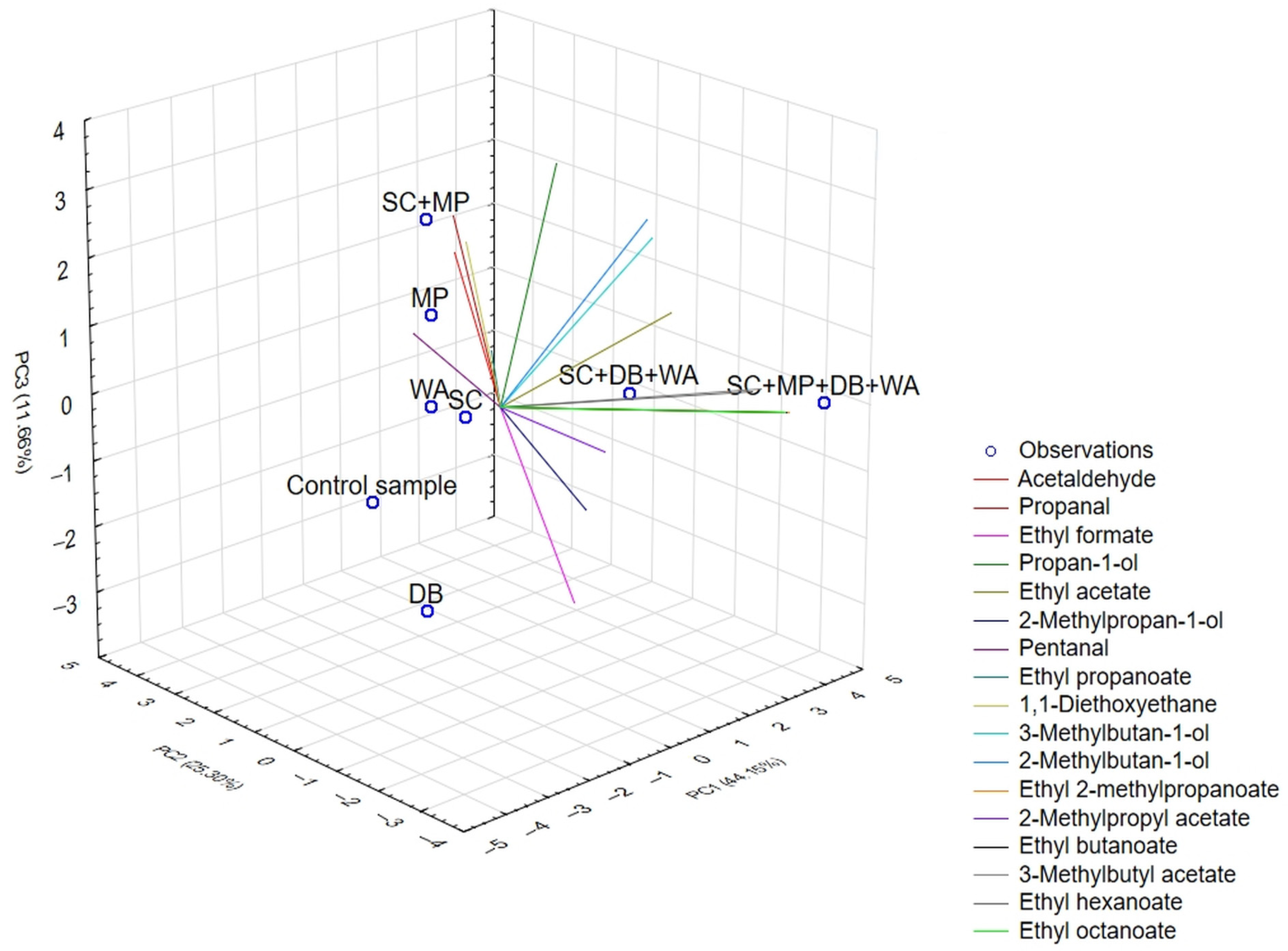
2.4. GC Analysis
3. Materials and Methods
3.1. Yeast Cultures
3.2. Yeast Osmotolerance
3.3. Honey Wort Preparation
3.4. Fermentation Trials
3.5. HPLC Analysis
3.6. GC-MS Analysis
3.7. Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bisson, L.F. Stuck and sluggish fermentations. AJEV 1999, 50, 107–119. [Google Scholar] [CrossRef]
- Binati, R.L.; Lemos Junior, W.J.F.; Luzzini, G.; Slaghenaufi, D.; Ugliano, M.; Torriani, S. Contribution of non-Saccharomyces yeasts to wine volatile and sensory diversity: A study on Lachancea thermotolerans, Metschnikowia spp. and Starmerella bacillaris strains isolated in Italy. Int. J. Food Microbiol. 2020, 318, 108470. [Google Scholar] [CrossRef]
- Vicente, J.; Ruiz, J.; Belda, I.; Benito-Vázquez, I.; Marquina, D.; Calderón, F.; Santos, A.; Benito, S. The genus Metschnikowia in enology. Microorganisms 2020, 8, 1038. [Google Scholar] [CrossRef]
- Zhang, B.; Tang, C.; Yang, D.; Liu, H.; Xue, J.; Duan, C.; Yan, G. Effects of three indigenous non-Saccharomyces yeasts and their pairwise combinations in co-fermentation with Saccharomyces cerevisiae on volatile compounds of Petit Manseng wines. Food Chem. 2022, 368, 130807. [Google Scholar] [CrossRef] [PubMed]
- Sipiczki, M. Overwintering of vineyard yeasts: Survival of interacting yeast communities in grapes mummified on vines. Front. Microbiol. 2016, 7, 212. [Google Scholar] [CrossRef]
- Morata, A.; Escott, C.; Bañuelos, M.A.; Loira, I.; Fresno, J.M.D.; González, C.; Suárez-Lepe, J.A. Contribution of non-Saccharomyces yeasts to wine freshness. A review. Biomolecules 2019, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Sipiczki, M. Metschnikowia pulcherrima and related pulcherrimin-producing yeasts: Fuzzy species boundaries and complex antimicrobial antagonism. Microorganisms 2020, 8, 1029. [Google Scholar] [CrossRef] [PubMed]
- Tufariello, M.; Fragasso, M.; Pico, J.; Panighel, A.; Castellarin, S.D.; Flamini, R.; Grieco, F. Influence of non-Saccharomyces on wine chemistry: A focus on aroma-related compounds. Molecules 2021, 26, 644. [Google Scholar] [CrossRef]
- Contreras, A.; Curtin, C.; Varela, C. Yeast population dynamics reveal a potential ‘collaboration’ between Metschnikowia pulcherrima and Saccharomyces uvarum for the production of reduced alcohol wines during Shiraz fermentation. Appl. Microbiol. Biotechnol. 2015, 99, 1885–1895. [Google Scholar] [CrossRef]
- Morales, P.; Rojas, V.; Quirós, M.; Gonzalez, R. The impact of oxygen on the final alcohol content of wine fermented by a mixed starter culture. Appl. Microbiol. Biotechnol. 2015, 99, 3993–4003. [Google Scholar] [CrossRef]
- Mencher, A.; Morales, P.; Valero, E.; Tronchoni, J.; Patil, K.R.; Gonzalez, R. Proteomic characterization of extracellular vesicles produced by several wine yeast species. Microb. Biotechnol. 2020, 13, 1581–1596. [Google Scholar] [CrossRef] [PubMed]
- Castrillo, D.; Blanco, P. Characterization of indigenous non-Saccharomyces yeast strains with potential use in winemaking. Front. Biosci. 2023, 15, 1. [Google Scholar] [CrossRef]
- Kregiel, D.; Pawlikowska, E.; Antolak, H.; Dziekonska-Kubczak, U.; Pielech-Przybylska, K. Exploring use of the Metschnikowia pulcherrima clade to improve properties of fruit wines. Fermentation 2022, 8, 247. [Google Scholar] [CrossRef]
- Puyo, M.; Simonin, S.; Bach, B.; Klein, G.; Alexandre, H.; Tourdot-Maréchal, R. Bio-protection in oenology by Metschnikowia pulcherrima: From field results to scientific inquiry. Front. Microbiol. 2023, 14, 1252973. [Google Scholar] [CrossRef] [PubMed]
- Altieri, V.; Rossi, V.; Fedele, G. Efficacy of preharvest application of biocontrol agents against gray mold in grapevine. Front. Plant Sci. 2023, 14, 1154370. [Google Scholar] [CrossRef]
- Ramalhosa, E.; Gomes, T.; Pereira, A.P.; Dias, T.; Estevinho, L.M. Mead production: Tradition versus modernity. Adv. Food Nutr. Res. 2011, 63, 101–118. [Google Scholar] [CrossRef]
- Iglesias, A.; Pascoal, A.; Choupina, A.B.; Carvalho, C.A.; Feás, X.; Estevinho, L.M. Developments in the fermentation process and quality improvement strategies for mead production. Molecules 2014, 19, 12577–12590. [Google Scholar] [CrossRef]
- Webster, C.E.; Barker, D.; Deed, R.C.; Pilkington, L.I. Mead production and quality: A review of chemical and sensory mead quality evaluation with a focus on analytical methods. Food Res. Int. 2025, 202, 115655. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.P.; Dias, T.; Andrade, J.; Ramalhosa, E.; Estevinho, L.M. Mead production: Selection and characterization assays of Saccharomyces cerevisiae strains. Food Chem. Toxicol. 2009, 47, 2057–2063. [Google Scholar] [CrossRef]
- Pawlikowska, E.; James, S.A.; Breierova, E.; Antolak, H.; Kregiel, D. Biocontrol capability of local Metschnikowia sp. isolates. Antonie Van Leeuwenhoek 2019, 112, 1425–1445. [Google Scholar] [CrossRef]
- Imura, M.; Nitta, K.; Iwakiri, R.; Matsuda, F.; Shimizu, H.; Fukusaki, E. Comparison of metabolic profiles of yeasts based on the difference of the Crabtree positive and negative. J. Biosci. Bioeng. 2020, 129, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Blomqvist, J.; Passoth, V. Dekkera bruxellensis-spoilage yeast with biotechnological potential, and a model for yeast evolution, physiology and competitiveness. FEMS Yeast Res. 2015, 15, fov021. [Google Scholar] [CrossRef]
- Němcová, A.; Szotkowski, M.; Samek, O.; Cagáňová, L.; Sipiczki, M.; Márová, I. Use of waste substrates for the lipid production by yeasts of the genus Metschnikowia-screening study. Microorganisms 2021, 9, 2295. [Google Scholar] [CrossRef] [PubMed]
- Breuer, U.; Harms, H. Debaryomyces hansenii-an extremophilic yeast with biotechnological potential. Yeast 2006, 23, 415–437. [Google Scholar] [CrossRef]
- Niu, C.; Yuan, Y.; Hu, Z.; Wang, Z.; Liu, B.; Wang, H.; Yue, T. Accessing spoilage features of osmotolerant yeasts identified from kiwifruit plantation and processing environment in Shaanxi, China. Int. J. Food Microbiol. 2016, 232, 126–133. [Google Scholar] [CrossRef]
- Thammaket, J.; Srimongkol, P.; Ekkaphan, P.; Thitiprasert, S.; Niyomsin, S.; Chaisuwan, T.; Chirachanchai, S.; Thongchul, N. Isolation, screening, and characterization of the newly isolated osmotolerant yeast Wickerhamomyces anomalus BKK11-4 for the coproduction of glycerol and arabitol. Braz. J. Microbiol. 2024, 55, 2149–2167. [Google Scholar] [CrossRef]
- Stratford, M.; Steels, H.; Novodvorska, M.; Archer, D.B.; Avery, S.V. Extreme osmotolerance and halotolerance in food-relevant yeasts and the role of glycerol-dependent cell individuality. Front. Microbiol. 2019, 9, 3238. [Google Scholar] [CrossRef]
- Cronwright, G.R.; Rohwer, J.M.; Prior, B.A. Metabolic control analysis of glycerol synthesis in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2002, 68, 4448–4456. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Ren, L.; Wang, H.; Xu, D.; Zeng, X.; Li, F. Glycerol uptake and synthesis systems contribute to the osmotic tolerance of Kluyveromyces marxianus. Enzyme Microb. Technol. 2020, 140, 109641. [Google Scholar] [CrossRef]
- Dušková, M.; Ferreira, C.; Lucas, C.; Sychrová, H. Two glycerol uptake systems contribute to the high osmotolerance of Zygosaccharomyces rouxii. Mol. Microbiol. 2015, 97, 541–559. [Google Scholar] [CrossRef]
- Chen, A.; Qu, T.; Smith, J.R.; Li, J.; Du, G.; Chen, J. Osmotic tolerance in Saccharomyces cerevisiae: Implications for food and bioethanol industries. Food Biosci. 2024, 60, 104451. [Google Scholar] [CrossRef]
- Siavoshi, F.; Sahraee, M.; Heydari, S.; Sarrafnejad, A.; Saniee, P.; Tavakolian, A.; Heidarian, S. Sugar-rich foods carry osmotolerant yeasts with intracellular Helicobacter pylori and Staphylococcus spp. Middle East J. Dig. Dis. 2020, 12, 182–193. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Jadhav, R.; Avchar, R.; Lanjekar, V.; Datar, M.; Baghela, A. Nectar yeast community of tropical flowering plants and assessment of their osmotolerance and xylitol-producing potential. Curr. Microbiol. 2021, 79, 28. [Google Scholar] [CrossRef] [PubMed]
- Maicas, S.; Mateo, J.J. The life of Saccharomyces and non-Saccharomyces yeasts in drinking wine. Microorganisms 2023, 11, 1178. [Google Scholar] [CrossRef]
- Romano, A.; Perello, M.C.; de Revel, G.; Lonvaud-Funel, A. Growth and volatile compound production by Brettanomyces/Dekkera bruxellensis in red wine. J. Appl. Microbiol. 2008, 104, 1577–1585. [Google Scholar] [CrossRef]
- Aplin, J.J.; White, K.P.; Edwards, C.G. Growth and metabolism of non-Saccharomyces yeasts isolated from Washington state vineyards in media and high sugar grape musts. Food Microbiol. 2019, 77, 158–165. [Google Scholar] [CrossRef]
- Mencher, A.; Morales, P.; Curiel, J.A.; Gonzalez, R.; Tronchoni, J. Metschnikowia pulcherrima represses aerobic respiration in Saccharomyces cerevisiae suggesting a direct response to co-cultivation. Food Microbiol. 2021, 94, 103670. [Google Scholar] [CrossRef]
- Mejias-Ortiz, M.; Mencher, A.; Morales, P.; Tronchoni, J.; Gonzalez, R. Saccharomyces cerevisiae responds similarly to co-culture or to a fraction enriched in Metschnikowia pulcherrima extracellular vesicles. Microb. Biotechnol. 2023, 16, 1027–1040. [Google Scholar] [CrossRef]
- Carlson, M. Glucose repression in yeast. Curr. Opin. Microbiol. 1999, 2, 202–207. [Google Scholar] [CrossRef]
- Blumenthal, P.; Steger, M.C.; Einfalt, D.; Rieke-Zapp, J.; Quintanilla Bellucci, A.; Sommerfeld, K.; Schwarz, S.; Lachenmeier, D.W. Methanol mitigation during manufacturing of fruit spirits with special consideration of novel coffee cherry spirits. Molecules 2021, 26, 2585. [Google Scholar] [CrossRef]
- Shen, J.; Huang, W.; You, Y.; Zhan, J. Controlling strategies of methanol generation in fermented fruit wine: Pathways, advances, and applications. Compr. Rev. Food Sci. Food Saf. 2024, 23, e70048. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, Y.; Cui, Z.; Wang, T.; Liu, T.; Liu, G. Analysis of free amino acid composition and honey plant species in seven honey species in China. Foods 2024, 13, 1065. [Google Scholar] [CrossRef] [PubMed]
- Oro, L.; Ciani, M.; Comitini, F. Antimicrobial activity of Metschnikowia pulcherrima on wine yeasts. J. Appl. Microbiol. 2014, 116, 1209–1217. [Google Scholar] [CrossRef]
- Canonico, L.; Comitini, F.; Ciani, M. Metschnikowia pulcherrima selected strain for ethanol reduction in wine: Influence of cell immobilization and aeration condition. Foods 2019, 8, 378. [Google Scholar] [CrossRef]
- Binati, R.L.; Maule, M.; Luzzini, G.; Martelli, F.; Felis, G.E.; Ugliano, M.; Torriani, S. From bioprotective effects to diversification of wine aroma: Expanding the knowledge on Metschnikowia pulcherrima oenological potential. Food Res. Int. 2023, 74 Pt 1, 113550. [Google Scholar] [CrossRef]
- Tapia, S.M.; Cuevas, M.; Abarca, V.; Delgado, V.; Rojas, V.; García, V.; Brice, C.; Martínez, C.; Salinas, F.; Larrondo, L.F.; et al. GPD1 and ADH3 natural variants underlie glycerol yield differences in wine fermentation. Front. Microbiol. 2018, 9, 1460. [Google Scholar] [CrossRef]
- Carrau, F.; Boido, E.; Dellacassa, E. Yeast diversity and flavor compounds. In Fungal Metabolities; Merillon, J.M., Ramawat, K.G., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 569–598. [Google Scholar]
- Arias-Pérez, I.; Sáenz-Navajas, M.P.; De-La-Fuente-Blanco, A.; Ferreira, V.; Escudero, A. Insights on the role of acetaldehyde and other aldehydes in the odour and tactile nasal perception of red wine. Food Chem. 2021, 361, 130081. [Google Scholar] [CrossRef]
- Yu, H.; Xie, T.; Xie, J.; Ai, L.; Tian, H. Characterization of key aroma compounds in Chinese rice wine using gas chromatography-mass spectrometry and gas chromatography-olfactometry. Food Chem. 2019, 293, 8–14. [Google Scholar] [CrossRef]
- Felipe, A.L.D.; Souza, C.O.; Santos, L.F.; Cestari, A. Synthesis and characterization of mead: From the past to the future and development of a new fermentative route. J. Food Sci. Technol. 2019, 56, 4966–4971. [Google Scholar] [CrossRef] [PubMed]
- De-La-Fuente-Blanco, A.; Saenz-Navajas, M.P.; Ferreira, V. On the effects of higher alcohols on red wine aroma. Food Chem. 2016, 210, 107–114. [Google Scholar] [CrossRef]
- Qureshi, N.; Tamhane, D.V. Production of mead by immobilized cells of Hansenula anomala. Appl. Microbiol. Biotechnol. 1987, 27, 27–30. [Google Scholar] [CrossRef]
- Barry, J.P.; Metz, M.S.; Hughey, J.; Quirk, A.; Bochman, M.L. Two novel strains of Torulaspora delbrueckii isolated from the honey bee microbiome and their use in honey fermentation. Fermentation 2018, 4, 22. [Google Scholar] [CrossRef]
- Prestianni, R.; Matraxia, M.; Naselli, V.; Pirrone, A.; Badalamenti, N.; Ingrassia, M.; Gaglio, R.; Settanni, L.; Columba, P.; Maggio, A.; et al. Use of sequentially inoculation of Saccharomyces cerevisiae and Hanseniaspora uvarum strains isolated from honey by-products to improve and stabilize the quality of mead produced in Sicily. Food Microbiol. 2022, 107, 104064. [Google Scholar] [CrossRef]
- Liu, Y.; Jiang, B.; Wang, K. A review of fermented bee products: Sources, nutritional values, and health benefits. Food Res. Int. 2023, 174 Pt 1, 113506. [Google Scholar] [CrossRef] [PubMed]
- Czabaj, S.; Kawa-Rygielska, J.; Kucharska, A.Z.; Kliks, J. Effects of mead wort heat treatment on the mead fermentation process and antioxidant activity. Molecules 2017, 22, 803. [Google Scholar] [CrossRef]
- García, M.; Greetham, D.; Wimalasena, T.T.; Phister, T.G.; Cabellos, J.M.; Arroyo, T. The phenotypic characterization of yeast strains to stresses inherent to wine fermentation in warm climates. J. Appl. Microbiol. 2016, 121, 215–233. [Google Scholar] [CrossRef]
- Holt, S.; Mukherjee, V.; Lievens, B.; Verstrepen, K.J.; Thevelein, J.M. Bioflavoring by non-conventional yeasts in sequential beer fermentations. Food Microbiol. 2018, 72, 55–66. [Google Scholar] [CrossRef]
- Cioch-Skoneczny, M.; Satora, P.; Skoneczny, S.; Skotniczny, M. Biodiversity of yeasts isolated during spontaneous fermentation of cool climate grape musts. Arch. Microbiol. 2021, 203, 153–162. [Google Scholar] [CrossRef]
- Dziekońska-Kubczak, U.; Berłowska, J.; Dziugan, P.; Patelski, P.; Pielech-Przybylska, K.; Balcerek, M. Nitric acid pretreatment of Jerusalem artichoke stalks for enzymatic saccharification and bioethanol production. Energies 2018, 11, 2153. [Google Scholar] [CrossRef]
- Hubaux, A.; Vos, G. Decision and detection limits for calibration curves. Anal. Chem. 1970, 42, 849–855. [Google Scholar] [CrossRef]
| Yeast Strain | Glucose Concentration in the Culture Medium [% w/v] | |||
|---|---|---|---|---|
| 1 | 10 | 20 | 30 | |
| S. cerevisiae | 8.967 ± 0.252 | 7.600 ± 0.458 | 4.867 ± 0.306 | 3.567 ± 0.321 |
| M. pulcherrima | 8.367 ± 0.208 | 7.933 ± 0.551 | 6.667 ± 0.321 A | 5.900 ± 0.100 B |
| D. bruxellensis | 8.067 ± 0.493 | 6.767 ± 0.321 | 3.833 ± 0.153 A | 2.867 ± 0.153 B |
| W. anomalus | 8.967 ± 0.115 | 7.867 ± 0.208 | 6.233 ± 0.416 | 4.167 ± 0.404 |
| p value | p > 0.05 | p > 0.05 | A 0.019 | B 0.013 |
| Yeasts | Sample | Glucose [g/L] | Fructose [g/L] |
|---|---|---|---|
| Control | 182.466 ± 6.513 | 189.375 ± 8.407 | |
| Monocultures | SC * | 65.555 ± 1.092 A | 110.476 ± 5.472 B |
| MP | 89.211 ± 2.321 | 159.559 ± 10.990 | |
| WA | 145.564 ± 4.127 | 173.857 ± 5.550 B | |
| DB | 152.324 ± 4.052 A | 166.734 ± 4.202 | |
| Mixed populations | SC + MP | 70.123 ± 3.830 | 128.480 ± 2.703 |
| SC + MP + WA + DB | 79.390 ± 2.683 | 130.322 ± 6.252 | |
| SC + WA + DB | 73.015 ± 2.804 | 126.511 ± 2.984 | |
| p-value | p = 0.013 | p = 0.013 | |
| Yeast Strains | Compound [g/L] | ||||
|---|---|---|---|---|---|
| Glycerol | Acetic Acid | Methanol | Ethanol | ||
| Control | 0.197 ± 0.006 | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.000 ± 0.000 | |
| Monocultures | SC * | 5.762 ± 0.345 | 0.858 ± 0.016 | 2.281 ± 0.103 C | 45.563 ± 0.903 |
| MP | 7.928 ± 0.178 A | 0.562 ± 0.011 | 1.247 ± 0.193 | 12.781 ± 0.547 | |
| WA | 1.873 ± 0.054 | 0.387 ± 0.007 | 0.153 ± 0.017 | 12.442 ± 0.586 | |
| DB | 1.362 ± 0.057 A | 0.227 ± 0.010 B | 0.126 ± 0.007 C | 10.495 ± 0.497 D | |
| Mixed populations | SC + MP | 7.155 ± 0.046 | 0.928 ± 0.026 | 1.242 ± 0.122 | 63.511 ± 1.948 D |
| SC + MP + WA + DB | 7.107 ± 0.097 | 1.087 ± 0.028 B | 2.081 ± 0.085 | 57.630 ± 1.377 | |
| SC + WA + DB | 7.019 ± 0.186 | 0.944 ± 0.043 | 1.241 ± 0.119 | 59.549 ± 1.899 | |
| p-value | 0.008 | 0.008 | 0.008 | 0.008 | |
| Principal Component | Eigenvalue | Variability [%] | Cumulative [%] |
|---|---|---|---|
| PC1 | 7.505 | 44.147 | 44.147 |
| PC2 | 4.301 | 25.298 | 69.445 |
| PC3 | 1.982 | 11.658 | 81.103 |
| Strain | Origin | Strain Abbreviation | References |
|---|---|---|---|
| Saccharomyces cerevisiae Tokay LOCK0203 | LOCK * | SC | [13,20] |
| Metschnikowia pulcherrima NCYC747 | NCYC ** | MP | [13,20] |
| Dekkera bruxellensis NCYC D5300 | NCYC | DB | [13,20] |
| Wickerhamomyces anomalus NCYC D5299 | NCYC | WA | [13,20] |
| Monocultures | Mixed Populations |
|---|---|
| Saccharomyces cerevisiae SC | S. cerevisiae SC + M. pulcherrima MP |
| Metschnikowia pulcherrima MP | S. cerevisiae SC + M. pulcherrima MP + W. anomalus WA + D. bruxellensis DB |
| Dekkera bruxellensis DB | |
| Wickerhamomyces anomalus WA | S. cerevisiae SC + W. anomalus WA + D. bruxellensis DB |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kregiel, D.; Dziekonska-Kubczak, U.; Czarnecka-Chrebelska, K.; Pielech-Przybylska, K. Chemical Fingerprints of Honey Fermented by Conventional and Non-Conventional Yeasts. Molecules 2025, 30, 2319. https://doi.org/10.3390/molecules30112319
Kregiel D, Dziekonska-Kubczak U, Czarnecka-Chrebelska K, Pielech-Przybylska K. Chemical Fingerprints of Honey Fermented by Conventional and Non-Conventional Yeasts. Molecules. 2025; 30(11):2319. https://doi.org/10.3390/molecules30112319
Chicago/Turabian StyleKregiel, Dorota, Urszula Dziekonska-Kubczak, Karolina Czarnecka-Chrebelska, and Katarzyna Pielech-Przybylska. 2025. "Chemical Fingerprints of Honey Fermented by Conventional and Non-Conventional Yeasts" Molecules 30, no. 11: 2319. https://doi.org/10.3390/molecules30112319
APA StyleKregiel, D., Dziekonska-Kubczak, U., Czarnecka-Chrebelska, K., & Pielech-Przybylska, K. (2025). Chemical Fingerprints of Honey Fermented by Conventional and Non-Conventional Yeasts. Molecules, 30(11), 2319. https://doi.org/10.3390/molecules30112319







