Chemical and Enzymatic Synthesis of DisialylGb5 and Other Sialosides for Glycan Array Assembly and Evaluation of Siglec-Mediated Immune Checkpoint Inhibition
Abstract
1. Introduction
2. Results and Discussion
2.1. The mRNA Level of ST6GALNAC6 in Brain, Breast, Colon, Lung, Pancreas, and Prostate Cancers Decreased Compared to Normal Cells
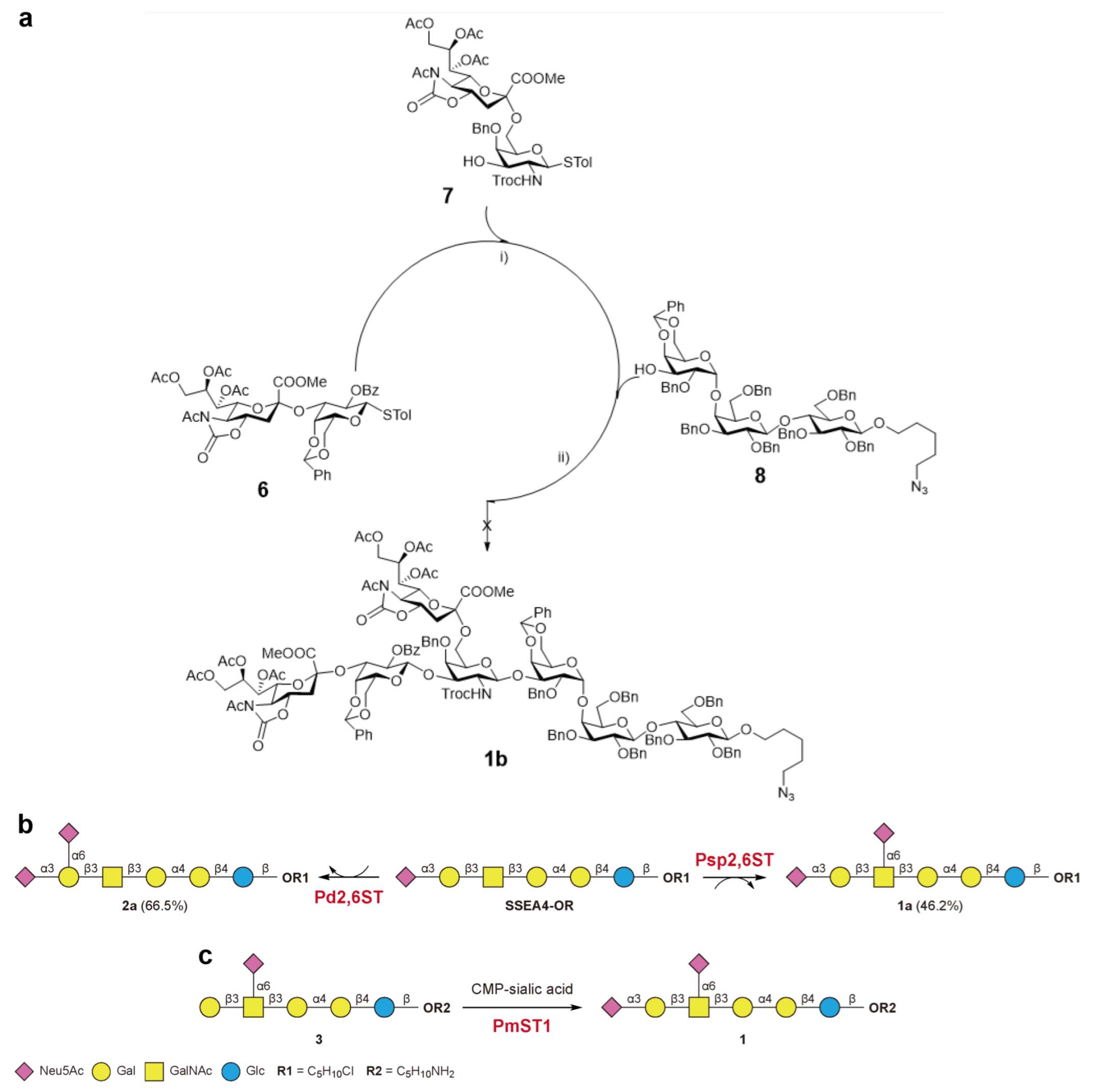
2.2. Synthesis of DSGb5 and Other Sialylated Derivatives
2.3. Glycan Array Analysis of Siglec-7, Siglec-9, Siglec-10, and Siglec-15 Binding to DSGb5 Glycan and Other Cancer-Associated Carbohydrate Antigens
2.4. Binding of Siglecs Toward Sialylated N-Glycans
2.5. Inhibition of Siglec-10 Binding to CD24 on Breast Cancer Cells with Synthetic Glycans
3. Materials and Methods
3.1. General Chemical Synthesis
3.2. Enzyme Preparations
3.3. Enzymatic Synthesis of DSGb5 via Human ST6GalNAc6
3.4. Glycan Microarray
3.5. Cell Culture
3.6. Flow Cytometry
3.7. Commercial Glycan Array
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hakomori, S. Tumor-associated carbohydrate antigens. Annu. Rev. Immunol. 1984, 2, 103–126. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Paulson, J.C. Siglecs as Immune Cell Checkpoints in Disease. Annu. Rev. Immunol. 2020, 38, 365–395. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.A.H.; Bertozzi, C.R. The clinical impact of glycobiology: Targeting selectins, Siglecs and mammalian glycans. Nat. Rev. Drug Discov. 2021, 20, 217–243. [Google Scholar] [CrossRef]
- Tsai, T.I.; Lee, H.Y.; Chang, S.H.; Wang, C.H.; Tu, Y.C.; Lin, Y.C.; Hwang, D.R.; Wu, C.Y.; Wong, C.H. Effective sugar nucleotide regeneration for the large-scale enzymatic synthesis of Globo H and SSEA4. J. Am. Chem. Soc. 2013, 135, 14831–14839. [Google Scholar] [CrossRef]
- Kung, C.C.; Lo, J.M.; Liao, K.S.; Wu, C.Y.; Cheng, L.C.; Chung, C.; Hsu, T.L.; Ma, C.; Wong, C.H. Expression of Human beta3GalT5-1 in Insect Cells as Active Glycoforms for the Efficient Synthesis of Cancer-Associated Globo-Series Glycans. J. Am. Chem. Soc. 2025, 147, 10864–10874. [Google Scholar] [CrossRef]
- Kannagi, R.; Cochran, N.A.; Ishigami, F.; Hakomori, S.; Andrews, P.W.; Knowles, B.B.; Solter, D. Stage-specific embryonic antigens (SSEA-3 and -4) are epitopes of a unique globo-series ganglioside isolated from human teratocarcinoma cells. EMBO J. 1983, 2, 2355–2361. [Google Scholar] [CrossRef]
- Krupnick, J.G.; Damjanov, I.; Damjanov, A.; Zhu, Z.M.; Fenderson, B.A. Globo-series carbohydrate antigens are expressed in different forms on human and murine teratocarcinoma-derived cells. Int. J. Cancer 1994, 59, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Li, Y.; Hu, Y.; Zhou, C.; Hu, Y.; Chen, H. Stage-specific embryonic antigen 4 expression in epithelial ovarian carcinoma. Int. J. Gynecol. Cancer 2010, 20, 958–964. [Google Scholar] [CrossRef]
- Lou, Y.W.; Wang, P.Y.; Yeh, S.C.; Chuang, P.K.; Li, S.T.; Wu, C.Y.; Khoo, K.H.; Hsiao, M.; Hsu, T.L.; Wong, C.H. Stage-specific embryonic antigen-4 as a potential therapeutic target in glioblastoma multiforme and other cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 2482–2487. [Google Scholar] [CrossRef]
- Lin, C.W.; Wang, Y.J.; Lai, T.Y.; Hsu, T.L.; Han, S.Y.; Wu, H.C.; Shen, C.N.; Dang, V.; Chen, M.W.; Chen, L.B.; et al. Homogeneous antibody and CAR-T cells with improved effector functions targeting SSEA-4 glycan on pancreatic cancer. Proc. Natl. Acad. Sci. USA 2021, 118, e2114774118. [Google Scholar] [CrossRef]
- Senda, M.; Ito, A.; Tsuchida, A.; Hagiwara, T.; Kaneda, T.; Nakamura, Y.; Kasama, K.; Kiso, M.; Yoshikawa, K.; Katagiri, Y.; et al. Identification and expression of a sialyltransferase responsible for the synthesis of disialylgalactosylgloboside in normal and malignant kidney cells: Downregulation of ST6GalNAc VI in renal cancers. Biochem. J. 2007, 402, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Hugonnet, M.; Singh, P.; Haas, Q.; von Gunten, S. The Distinct Roles of Sialyltransferases in Cancer Biology and Onco-Immunology. Front. Immunol. 2021, 12, 799861. [Google Scholar] [CrossRef] [PubMed]
- Freitas, D.P.; Teixeira, C.A.; Santos-Silva, F.; Vasconcelos, M.H.; Almeida, G.M. Therapy-induced enrichment of putative lung cancer stem-like cells. Int. J. Cancer 2014, 134, 1270–1278. [Google Scholar] [CrossRef]
- Brune, J.C.; Tormin, A.; Johansson, M.C.; Rissler, P.; Brosjo, O.; Lofvenberg, R.; von Steyern, F.V.; Mertens, F.; Rydholm, A.; Scheding, S. Mesenchymal stromal cells from primary osteosarcoma are non-malignant and strikingly similar to their bone marrow counterparts. Int. J. Cancer 2011, 129, 319–330. [Google Scholar] [CrossRef]
- Fonsato, V.; De Lena, M.; Tritta, S.; Brossa, A.; Calvetti, R.; Tetta, C.; Camussi, G.; Bussolati, B. Human liver stem cell-derived extracellular vesicles enhance cancer stem cell sensitivity to tyrosine kinase inhibitors through Akt/mTOR/PTEN combined modulation. Oncotarget 2018, 9, 36151–36165. [Google Scholar] [CrossRef][Green Version]
- Sourisseau, T.; Hassan, K.A.; Wistuba, I.; Penault-Llorca, F.; Adam, J.; Deutsch, E.; Soria, J.C. Lung cancer stem cell: Fancy conceptual model of tumor biology or cornerstone of a forthcoming therapeutic breakthrough? J. Thorac. Oncol. 2014, 9, 7–17. [Google Scholar] [CrossRef]
- Bedel, A.; Pasquet, J.M.; Lippert, E.; Taillepierre, M.; Lagarde, V.; Dabernat, S.; Dubus, P.; Charaf, L.; Beliveau, F.; de Verneuil, H.; et al. Variable behavior of iPSCs derived from CML patients for response to TKI and hematopoietic differentiation. PLoS ONE 2013, 8, e71596. [Google Scholar] [CrossRef]
- Miyauchi, M.; Koya, J.; Arai, S.; Yamazaki, S.; Honda, A.; Kataoka, K.; Yoshimi, A.; Taoka, K.; Kumano, K.; Kurokawa, M. ADAM8 Is an Antigen of Tyrosine Kinase Inhibitor-Resistant Chronic Myeloid Leukemia Cells Identified by Patient-Derived Induced Pluripotent Stem Cells. Stem Cell Rep. 2018, 10, 1115–1130. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, G.; Lian, X.; Kron, S.J.; Palecek, S.P. Properties of resistant cells generated from lung cancer cell lines treated with EGFR inhibitors. BMC Cancer 2012, 12, 95. [Google Scholar] [CrossRef]
- Chen, N.Y.; Lin, C.W.; Lai, T.Y.; Wu, C.Y.; Liao, P.C.; Hsu, T.L.; Wong, C.H. Increased expression of SSEA-4 on TKI-resistant non-small cell lung cancer with EGFR-T790M mutation. Proc. Natl. Acad. Sci. USA 2024, 121, e2313397121. [Google Scholar] [CrossRef]
- Huang, Y.L.; Hung, J.T.; Cheung, S.K.; Lee, H.Y.; Chu, K.C.; Li, S.T.; Lin, Y.C.; Ren, C.T.; Cheng, T.J.; Hsu, T.L.; et al. Carbohydrate-based vaccines with a glycolipid adjuvant for breast cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 2517–2522. [Google Scholar] [CrossRef] [PubMed]
- Danishefsky, S.J.; Shue, Y.K.; Chang, M.N.; Wong, C.H. Development of Globo-H cancer vaccine. Acc. Chem. Res. 2015, 48, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.W.; Ko, Y.A.; Chen, C.Y.; Liao, K.S.; Chang, Y.H.; Lee, H.Y.; Yu, Y.H.; Lih, Y.H.; Cheng, Y.Y.; Lin, H.H.; et al. Mechanism of Antigen Presentation and Specificity of Antibody Cross-Reactivity Elicited by an Oligosaccharide-Conjugate Cancer Vaccine. J. Am. Chem. Soc. 2023, 145, 9840–9849. [Google Scholar] [CrossRef]
- Chuang, P.K.; Hsiao, M.; Hsu, T.L.; Chang, C.F.; Wu, C.Y.; Chen, B.R.; Huang, H.W.; Liao, K.S.; Chen, C.C.; Chen, C.L.; et al. Signaling pathway of globo-series glycosphingolipids and beta1,3-galactosyltransferase V (beta3GalT5) in breast cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 3518–3523. [Google Scholar] [CrossRef]
- Kawasaki, Y.; Ito, A.; Withers, D.A.; Taima, T.; Kakoi, N.; Saito, S.; Arai, Y. Ganglioside DSGb5, preferred ligand for Siglec-7, inhibits NK cell cytotoxicity against renal cell carcinoma cells. Glycobiology 2010, 20, 1373–1379. [Google Scholar] [CrossRef]
- Li, P.J.; Huang, S.Y.; Chiang, P.Y.; Fan, C.Y.; Guo, L.J.; Wu, D.Y.; Angata, T.; Lin, C.C. Chemoenzymatic Synthesis of DSGb5 and Sialylated Globo-series Glycans. Angew. Chem. Int. Ed. Engl. 2019, 58, 11273–11278. [Google Scholar] [CrossRef]
- Chuang, H.Y.; Ren, C.T.; Chao, C.A.; Wu, C.Y.; Shivatare, S.S.; Cheng, T.J.; Wu, C.Y.; Wong, C.H. Synthesis and vaccine evaluation of the tumor-associated carbohydrate antigen RM2 from prostate cancer. J. Am. Chem. Soc. 2013, 135, 11140–11150. [Google Scholar] [CrossRef] [PubMed]
- Shivatare, S.S.; Chang, S.H.; Tsai, T.I.; Ren, C.T.; Chuang, H.Y.; Hsu, L.; Lin, C.W.; Li, S.T.; Wu, C.Y.; Wong, C.H. Efficient convergent synthesis of bi-, tri-, and tetra-antennary complex type N-glycans and their HIV-1 antigenicity. J. Am. Chem. Soc. 2013, 135, 15382–15391. [Google Scholar] [CrossRef]
- Shivatare, S.S.; Chang, S.H.; Tsai, T.I.; Tseng, S.Y.; Shivatare, V.S.; Lin, Y.S.; Cheng, Y.Y.; Ren, C.T.; Lee, C.C.; Pawar, S.; et al. Modular synthesis of N-glycans and arrays for the hetero-ligand binding analysis of HIV antibodies. Nat. Chem. 2016, 8, 338–346. [Google Scholar] [CrossRef]
- Chu, K.C.; Ren, C.T.; Lu, C.P.; Hsu, C.H.; Sun, T.H.; Han, J.L.; Pal, B.; Chao, T.A.; Lin, Y.F.; Wu, S.H.; et al. Efficient and stereoselective synthesis of alpha(2-->9) oligosialic acids: From monomers to dodecamers. Angew. Chem. Int. Ed. Engl. 2011, 50, 9391–9395. [Google Scholar] [CrossRef]
- Hsu, C.H.; Hung, S.C.; Wu, C.Y.; Wong, C.H. Toward automated oligosaccharide synthesis. Angew. Chem. Int. Ed. Engl. 2011, 50, 11872–11923. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.H.; Chu, K.C.; Lin, Y.S.; Han, J.L.; Peng, Y.S.; Ren, C.T.; Wu, C.Y.; Wong, C.H. Highly alpha-selective sialyl phosphate donors for efficient preparation of natural sialosides. Chemistry 2010, 16, 1754–1760. [Google Scholar] [CrossRef]
- Yu, H.; Huang, S.; Chokhawala, H.; Sun, M.; Zheng, H.; Chen, X. Highly efficient chemoenzymatic synthesis of naturally occurring and non-natural alpha-2,6-linked sialosides: A P. damsela alpha-2,6-sialyltransferase with extremely flexible donor-substrate specificity. Angew. Chem. Int. Ed. Engl. 2006, 45, 3938–3944. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Zhao, C.; Qu, J.; Li, Y.; Sugiarto, G.; Yu, H.; Wang, J.; Chen, X. A Photobacterium sp. alpha2-6-sialyltransferase (Psp2,6ST) mutant with an increased expression level and improved activities in sialylating Tn antigens. Carbohydr. Res. 2015, 408, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Yu, H.; Lau, K.; Li, Y.; Muthana, S.; Wang, J.; Chen, X. Efficient chemoenzymatic synthesis of sialyl Tn-antigens and derivatives. Chem. Commun. 2011, 47, 8691–8693. [Google Scholar] [CrossRef]
- Sun, M.; Li, Y.; Chokhawala, H.A.; Henning, R.; Chen, X. N-Terminal 112 amino acid residues are not required for the sialyltransferase activity of Photobacterium damsela alpha2,6-sialyltransferase. Biotechnol. Lett. 2008, 30, 671–676. [Google Scholar] [CrossRef]
- Teo, C.F.; Hwang, T.S.; Chen, P.H.; Hung, C.H.; Gao, H.S.; Chang, L.S.; Lin, C.H. Synthesis of sialyl T Glycopeptides -: Enzymatic sialylation by α2,6-sialyltransferase from. Adv. Synth. Catal. 2005, 347, 967–972. [Google Scholar] [CrossRef]
- Meng, X.; Yao, W.; Cheng, J.; Zhang, X.; Jin, L.; Yu, H.; Chen, X.; Wang, F.; Cao, H. Regioselective chemoenzymatic synthesis of ganglioside disialyl tetrasaccharide epitopes. J. Am. Chem. Soc. 2014, 136, 5205–5208. [Google Scholar] [CrossRef]
- Ito, A.; Handa, K.; Withers, D.A.; Satoh, M.; Hakomori, S. Binding specificity of siglec7 to disialogangliosides of renal cell carcinoma: Possible role of disialogangliosides in tumor progression. FEBS Lett. 2001, 498, 116–120. [Google Scholar] [CrossRef]
- Yamaji, T.; Teranishi, T.; Alphey, M.S.; Crocker, P.R.; Hashimoto, Y. A small region of the natural killer cell receptor, Siglec-7, is responsible for its preferred binding to alpha 2,8-disialyl and branched alpha 2,6-sialyl residues. A comparison with Siglec-9. J. Biol. Chem. 2002, 277, 6324–6332. [Google Scholar] [CrossRef]
- Büll, C.; den Brok, M.H.; Adema, G.J. Sweet escape: Sialic acids in tumor immune evasion. Biochim. Biophys. Acta 2014, 1846, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Ravindranath, M.H.; Yesowitch, P.; Sumobay, C.; Morton, D.L. Glycoimmunomics of human cancer: Current concepts and future perspectives. Future Oncol. 2007, 3, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Krengel, U.; Bousquet, P.A. Molecular recognition of gangliosides and their potential for cancer immunotherapies. Front. Immunol. 2014, 5, 325. [Google Scholar] [CrossRef]
- t’Hart, I.M.E.; Li, T.; Wolfert, M.A.; Wang, S.; Moremen, K.W.; Boons, G.J. Chemoenzymatic synthesis of the oligosaccharide moiety of the tumor-associated antigen disialosyl globopentaosylceramide. Org. Biomol. Chem. 2019, 17, 7304–7308. [Google Scholar] [CrossRef]
- Hashimoto, N.; Ito, S.; Tsuchida, A.; Bhuiyan, R.H.; Okajima, T.; Yamamoto, A.; Furukawa, K.; Ohmi, Y.; Furukawa, K. The ceramide moiety of disialoganglioside (GD3) is essential for GD3 recognition by the sialic acid-binding lectin SIGLEC7 on the cell surface. J. Biol. Chem. 2019, 294, 10833–10845. [Google Scholar] [CrossRef] [PubMed]
- Mereiter, S.; Balmaña, M.; Campos, D.; Gomes, J.; Reis, C.A. Glycosylation in the Era of Cancer-Targeted Therapy: Where Are We Heading? Cancer Cell 2019, 36, 6–16. [Google Scholar] [CrossRef]
- Gonzalez-Gil, A.; Schnaar, R.L. Siglec Ligands. Cells 2021, 10, 1260. [Google Scholar] [CrossRef]
- Pirruccello, S.J.; Lebien, T.W. The Human B-Cell-Associated Antigen Cd24 Is a Single Chain Sialoglycoprotein. J. Immunol. 1986, 136, 3779–3784. [Google Scholar] [CrossRef]
- Fang, X.; Zheng, P.; Tang, J.; Liu, Y. CD24: From A to Z. Cell Mol. Immunol. 2010, 7, 100–103. [Google Scholar] [CrossRef]
- Kristiansen, G.; Sammar, M.; Altevogt, P. Tumour biological aspects of CD24, a mucin-like adhesion molecule. J. Mol. Histol. 2004, 35, 255–262. [Google Scholar] [CrossRef]
- Lim, S.C. CD24 and human carcinoma: Tumor biological aspects. Biomed. Pharmacother. 2005, 59 (Suppl. 2), S351–S354. [Google Scholar] [CrossRef] [PubMed]
- Munday, J.; Kerr, S.; Ni, J.; Cornish, A.L.; Zhang, J.Q.; Nicoll, G.; Floyd, H.; Mattei, M.G.; Moore, P.; Liu, D.; et al. Identification, characterization and leucocyte expression of Siglec-10, a novel human sialic acid-binding receptor. Biochem. J. 2001, 355 Pt 2, 489–497. [Google Scholar] [CrossRef]
- Barkal, A.A.; Brewer, R.E.; Markovic, M.; Kowarsky, M.; Barkal, S.A.; Zaro, B.W.; Krishnan, V.; Hatakeyama, J.; Dorigo, O.; Barkal, L.J.; et al. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature 2019, 572, 392–396. [Google Scholar] [CrossRef]
- Lim, J.; Sari-Ak, D.; Bagga, T. Siglecs as Therapeutic Targets in Cancer. Biology 2021, 10, 1178. [Google Scholar] [CrossRef]
- Chen, G.Y.; Tang, J.; Zheng, P.; Liu, Y. CD24 and Siglec-10 selectively repress tissue damage-induced immune responses. Science 2009, 323, 1722–1725. [Google Scholar] [CrossRef]
- Toubai, T.; Hou, G.; Mathewson, N.; Liu, C.; Wang, Y.; Oravecz-Wilson, K.; Cummings, E.; Rossi, C.; Evers, R.; Sun, Y.; et al. Siglec-G-CD24 axis controls the severity of graft-versus-host disease in mice. Blood 2014, 123, 3512–3523. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.S.; Gao, F.H. Molecular Mechanism of Tumor Cell Immune Escape Mediated by CD24/Siglec-10. Front. Immunol. 2020, 11, 1324. [Google Scholar] [CrossRef]
- Sammar, M.; Siwetz, M.; Meiri, H.; Fleming, V.; Altevogt, P.; Huppertz, B. Expression of CD24 and Siglec-10 in first trimester placenta: Implications for immune tolerance at the fetal-maternal interface. Histochem. Cell Biol. 2017, 147, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Forgione, R.E.; Di Carluccio, C.; Guzman-Caldentey, J.; Gaglione, R.; Battista, F.; Chiodo, F.; Manabe, Y.; Arciello, A.; Del Vecchio, P.; Fukase, K.; et al. Unveiling Molecular Recognition of Sialoglycans by Human Siglec-10. iScience 2020, 23, 101231. [Google Scholar] [CrossRef]
- Crocker, P.R.; Paulson, J.C.; Varki, A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007, 7, 255–266. [Google Scholar] [CrossRef]
- Liang, P.H.; Wang, S.K.; Wong, C.H. Quantitative analysis of carbohydrate-protein interactions using glycan microarrays: Determination of surface and solution dissociation constants. J. Am. Chem. Soc. 2007, 129, 11177–11184. [Google Scholar] [CrossRef] [PubMed]
- Shivatare, V.S.; Shivatare, S.S.; Lee, C.D.; Liang, C.H.; Liao, K.S.; Cheng, Y.Y.; Saidachary, G.; Wu, C.Y.; Lin, N.H.; Kwong, P.D.; et al. Unprecedented Role of Hybrid N-Glycans as Ligands for HIV-1 Broadly Neutralizing Antibodies. J. Am. Chem. Soc. 2018, 140, 5202–5210. [Google Scholar] [CrossRef] [PubMed]
- Cheung, S.K.; Chuang, P.K.; Huang, H.W.; Hwang-Verslues, W.W.; Cho, C.H.; Yang, W.B.; Shen, C.N.; Hsiao, M.; Hsu, T.L.; Chang, C.F.; et al. Stage-specific embryonic antigen-3 (SSEA-3) and beta3GalT5 are cancer specific and significant markers for breast cancer stem cells. Proc. Natl. Acad. Sci. USA 2016, 113, 960–965. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, K.-S.; Zhou, Y.; Chung, C.; Kung, C.-C.; Ren, C.-T.; Wu, C.-Y.; Lou, Y.-W.; Chuang, P.-K.; Imre, B.; Hsieh, Y.S.Y.; et al. Chemical and Enzymatic Synthesis of DisialylGb5 and Other Sialosides for Glycan Array Assembly and Evaluation of Siglec-Mediated Immune Checkpoint Inhibition. Molecules 2025, 30, 2264. https://doi.org/10.3390/molecules30112264
Liao K-S, Zhou Y, Chung C, Kung C-C, Ren C-T, Wu C-Y, Lou Y-W, Chuang P-K, Imre B, Hsieh YSY, et al. Chemical and Enzymatic Synthesis of DisialylGb5 and Other Sialosides for Glycan Array Assembly and Evaluation of Siglec-Mediated Immune Checkpoint Inhibition. Molecules. 2025; 30(11):2264. https://doi.org/10.3390/molecules30112264
Chicago/Turabian StyleLiao, Kuo-Shiang, Yixuan Zhou, Cinya Chung, Chih-Chuan Kung, Chien-Tai Ren, Chung-Yi Wu, Yi-Wei Lou, Po-Kai Chuang, Balázs Imre, Yves S. Y. Hsieh, and et al. 2025. "Chemical and Enzymatic Synthesis of DisialylGb5 and Other Sialosides for Glycan Array Assembly and Evaluation of Siglec-Mediated Immune Checkpoint Inhibition" Molecules 30, no. 11: 2264. https://doi.org/10.3390/molecules30112264
APA StyleLiao, K.-S., Zhou, Y., Chung, C., Kung, C.-C., Ren, C.-T., Wu, C.-Y., Lou, Y.-W., Chuang, P.-K., Imre, B., Hsieh, Y. S. Y., & Wong, C.-H. (2025). Chemical and Enzymatic Synthesis of DisialylGb5 and Other Sialosides for Glycan Array Assembly and Evaluation of Siglec-Mediated Immune Checkpoint Inhibition. Molecules, 30(11), 2264. https://doi.org/10.3390/molecules30112264







