Screening and Characterization of Marine Bacillus atrophaeus G4 Protease and Its Application in the Enzymatic Hydrolysis of Sheep (Ovis aries) Placenta for the Preparation of Antioxidant Peptides
Abstract
1. Introduction
2. Results and Discussion
2.1. Strain Screening and Identification
2.1.1. Strain Screening
2.1.2. Molecular Identification of Strain G4
2.2. Optimization of Protease Production by Fermentation of Strain G4
2.2.1. Effect of Cultivation Time
2.2.2. Effect of Temperature and pH on Protease Production
2.2.3. Optimization of the Fermentation Medium for Protease Production
2.2.4. Optimization of Enzyme Production Using Response Surface Methodology
2.3. Characterization of the Purified Protease
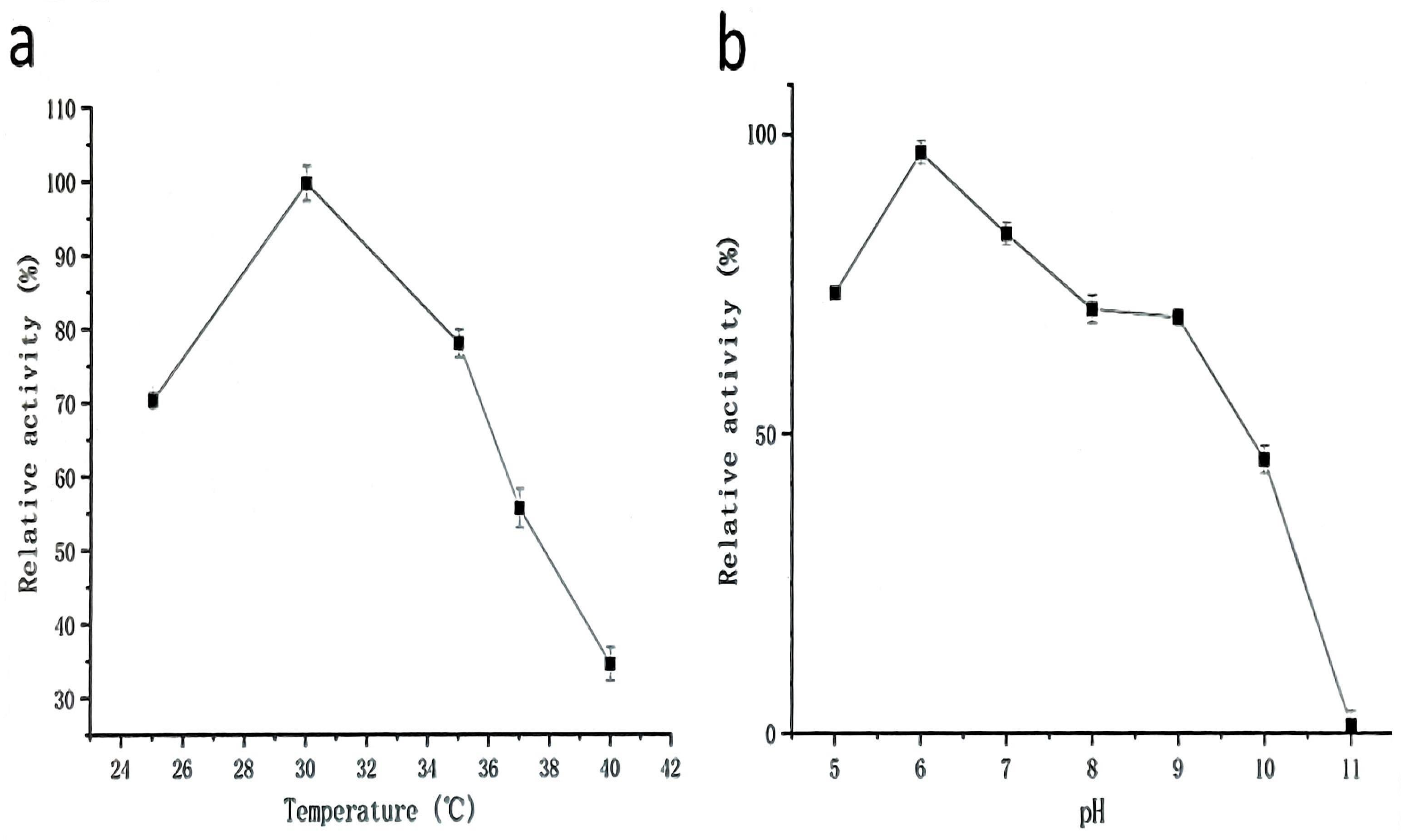
2.3.1. Effect of Temperature on the Activity and Stability of Protease from Strain G4
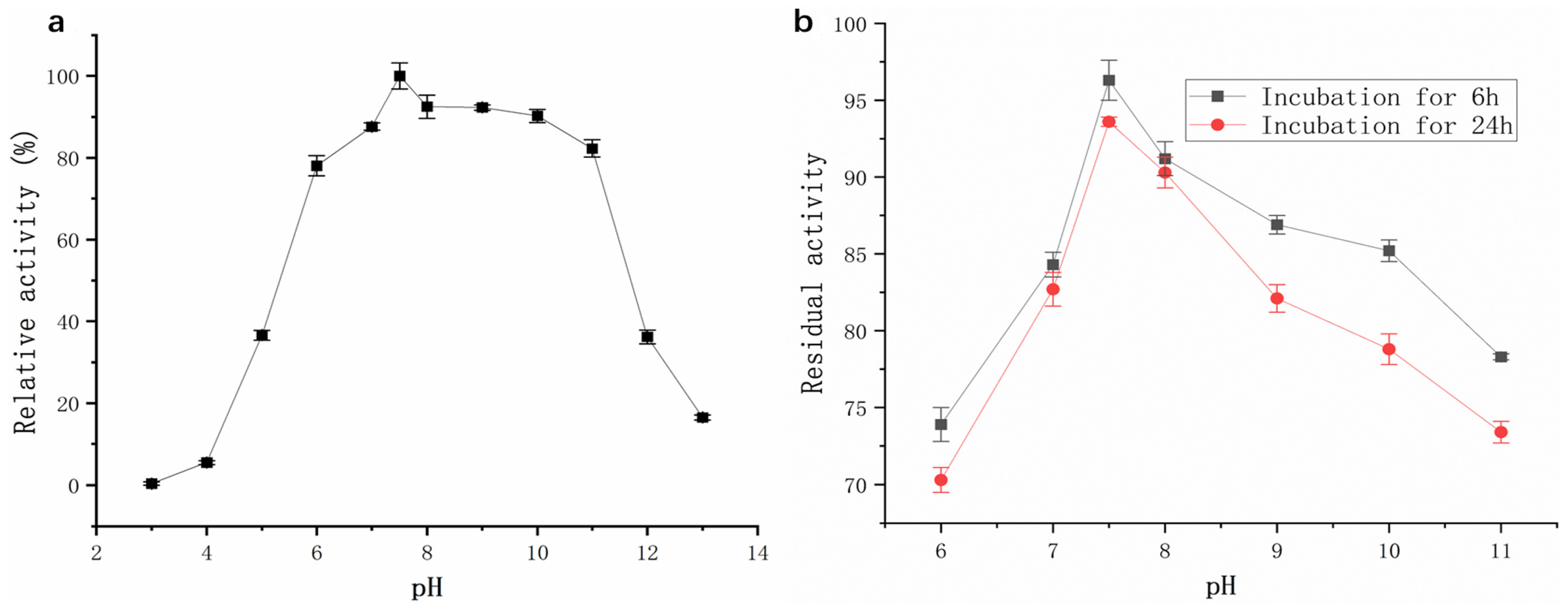
2.3.2. Effects of pH on Activity and Stability of Protease from Strain G4
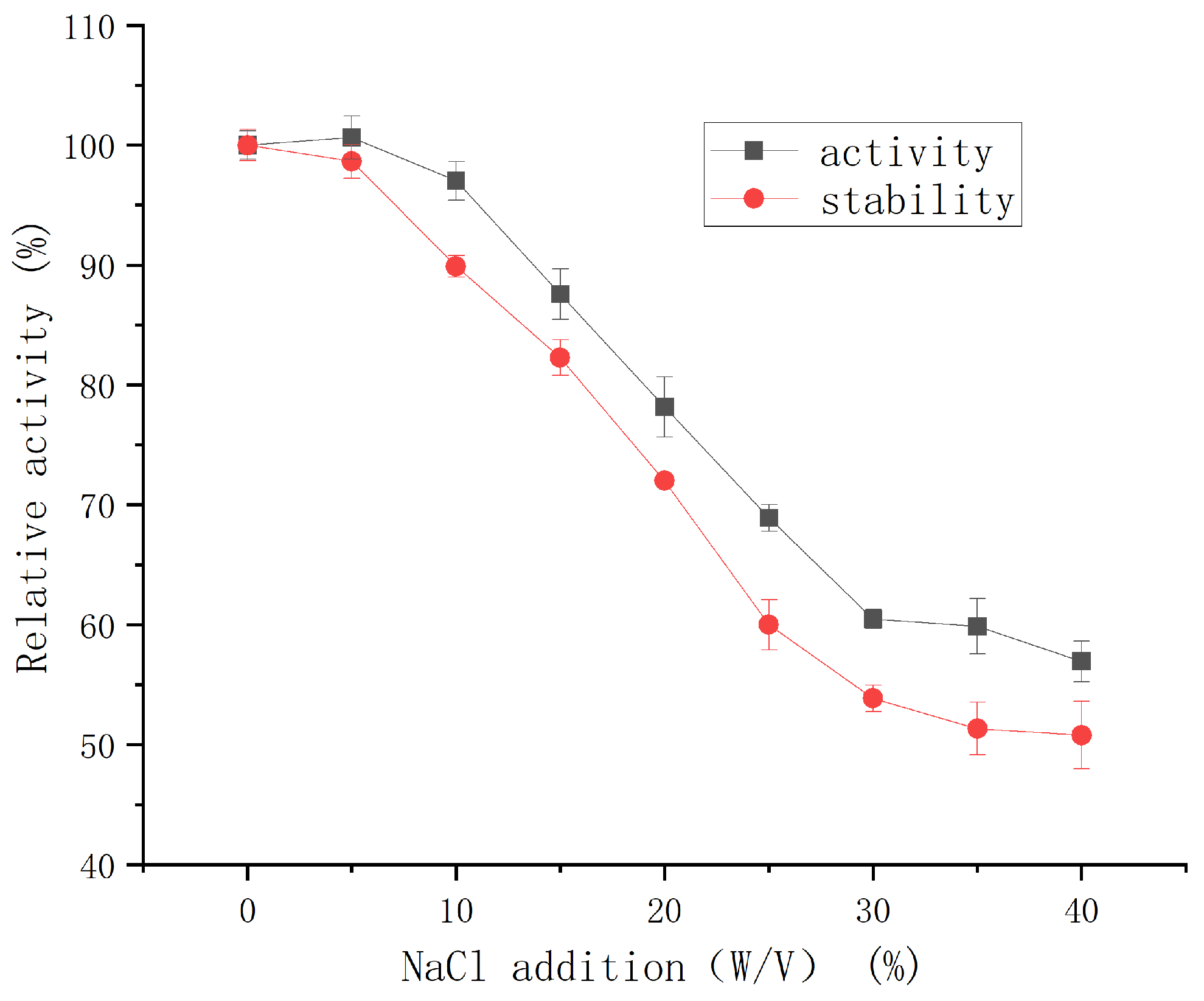
2.3.3. Effect of NaCl on the Activity and Salt Stability of the Protease
2.3.4. Effect of Metal Ions on the Activity of Protease from Strain G4
2.3.5. Effect of Inhibitors, Denaturants, and Surfactants on the Activity of the G4 Protease
2.4. Antioxidant Activity of Sheep Placenta Hydrolysates
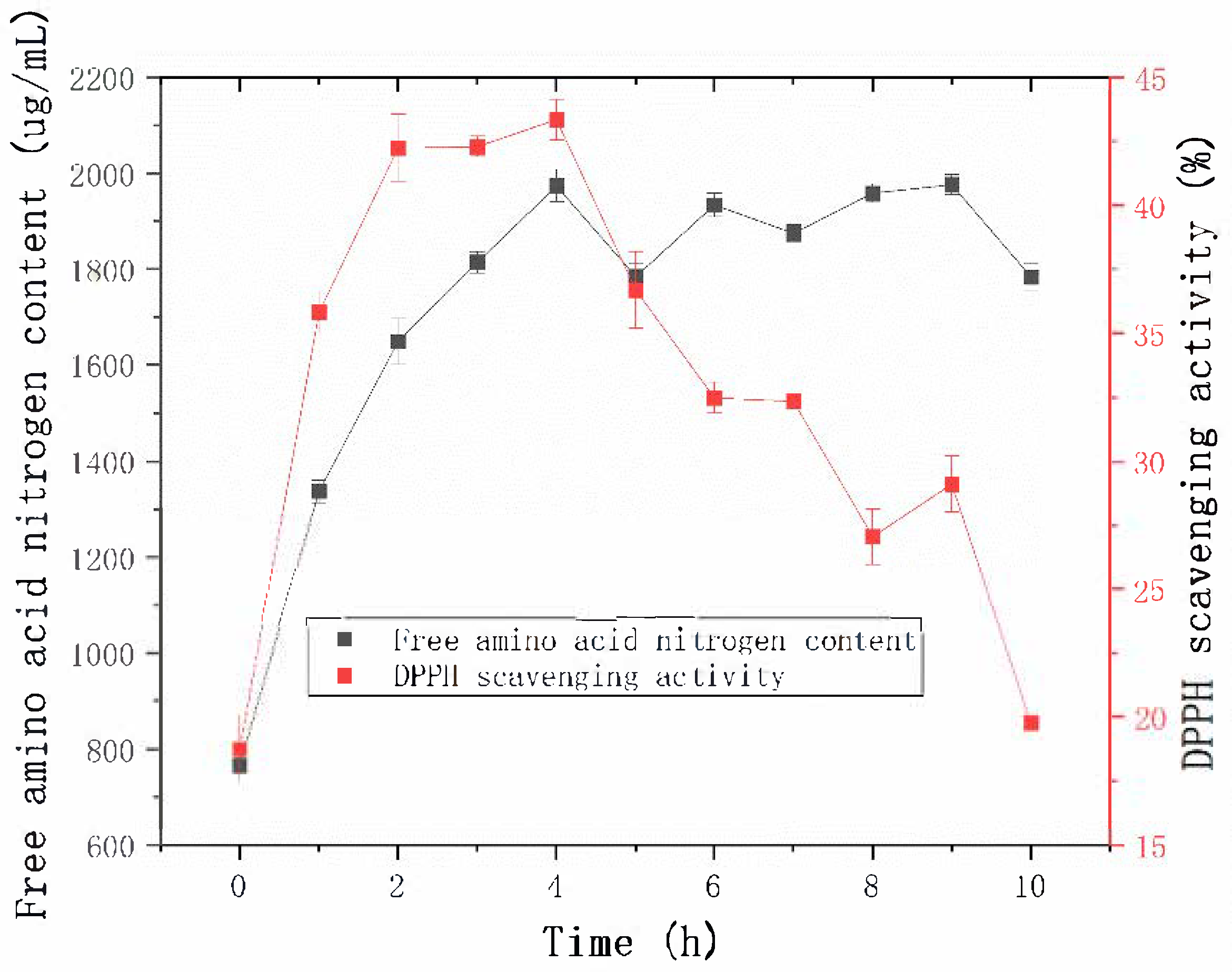
2.4.1. Effect of Hydrolysis Time on the Antioxidant Activity of Sheep Placenta Hydrolysates
2.4.2. Evaluation of the Antioxidant Activity of the Hydrolysate
3. Materials and Methods
3.1. Screening of Protease-Producing Strains
3.2. Protease Activity Assay
3.3. Molecular Identification of Bacillus atrophaeus G4
3.4. Optimization of Protease Production by Bacillus atrophaeus G4 Fermentation
3.5. Purification of Protease from Strain G4
3.6. Protein Analysis
3.7. Enzymatic Characterization of G4 Protease
3.8. Preparation of Sheep Placenta Hydrolysates Using G4 Protease
3.9. Evaluation of Antioxidant Activity of G4 Protease Hydrolysates
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, C.; Sun, C.; Wu, S. Screening and characterization of proteases produced by deep-sea cold seep bacteria. J. Oceanol. Limnol. 2021, 40, 678–689. [Google Scholar] [CrossRef]
- Tavano, O.L.; Berenguer-Murcia, A.; Secundo, F.; Fernandez-Lafuente, R. Biotechnological applications of proteases in food technology. Compr. Rev. Food Sci. Food Saf. 2018, 17, 412–436. [Google Scholar] [CrossRef]
- Gohel, S.D.; Singh, S.P. Thermodynamics of a Ca2+ dependent, highly thermostable and detergent compatible purified alkaline serine protease from Nocardiopsis xinjiangensis strain OM-6. Int. J. Biol. Macromol. 2018, 113, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Q. Genome-based studies of marine microorganisms to maximize the diversity of natural products discovery for medical treatments. Evid. Based Complement Alternat. Med. 2011, 2011, 384572. [Google Scholar] [CrossRef] [PubMed]
- Imada, C. Enzyme inhibitors of marine microbial origin with pharmaceutical importance. Mar. Biotechnol. 2004, 6, 193–198. [Google Scholar] [CrossRef]
- Barzkar, N. Marine microbial alkaline protease: An efficient and essential tool for various industrial applications. Int. J. Biol. Macromol. 2020, 161, 1216–1229. [Google Scholar] [CrossRef]
- Sun, C.; Groom, K.M.; Oyston, C.; Chamley, L.W.; Clark, A.R.; James, J.L. The placenta in fetal growth restriction: What is going wrong? Placenta 2020, 96, 10–18. [Google Scholar] [CrossRef]
- Chou, M.-Y.; Ou Yang, C.-P.; Li, W.-C.; Yang, Y.-M.; Huang, Y.-J.; Wang, M.-F.; Lin, W.-T. Evaluation of antiaging effect of sheep placenta extract using SAMP8 mice. Processes 2022, 10, 2242. [Google Scholar] [CrossRef]
- Ren, H.; Qin, T.; Zhou, Q.; Pan, L.; Yu, C.; Wang, Y.; Fan, W.; Li, Z.; Zheng, Y. Preparation, purification and identification of antioxidant peptides from the placenta of Tibetan sheep. Food Biosci. 2025, 63, 105560. [Google Scholar] [CrossRef]
- Wang, L.; Song, X.; Cui, H.; Man, S.; Li, W.; Muluye, R.A.; Bian, Y.; Chu, X.; Yan, D.; Cai, Y. Antifatigue effects of peptide isolated from sheep placenta. Chin. Herb. Med. 2018, 10, 279–284. [Google Scholar] [CrossRef]
- Haddar, A.; Agrebi, R.; Bougatef, A.; Hmidet, N.; Sellami-Kamoun, A.; Nasri, M. Two detergent stable alkaline serine-proteases from Bacillus mojavensis A21: Purification, characterization and potential application as a laundry detergent additive. Bioresour. Technol. 2009, 100, 3366–3373. [Google Scholar] [CrossRef]
- Jellouli, K.; Bougatef, A.; Manni, L.; Agrebi, R.; Siala, R.; Younes, I.; Nasri, M. Molecular and biochemical characterization of an extracellular serine-protease from Vibrio metschnikovii J1. J. Ind. Microbiol. Biotechnol. 2009, 36, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Gudzenko, O.V.; Varbanets, L.D.; Chernyshenko, V.O.; Stohnii, Y.M.; Ostapchuk, A.M.; Ivanytsia, V.O. Purification and physico-chemical properties of Bacillus atrophaeus protease with elastolytic and fibrinogenolytic activity. Ukr. Biochem. J. 2024, 96, 36–46. [Google Scholar] [CrossRef]
- Rahem, F.Z.; Badis, A.; Zenati, B.; Mechri, S.; Hadjidj, R.; Rekik, H.; Eddouaouda, K.; Annane, R.; Jaouadi, B. Characterization of a novel serine alkaline protease from Bacillus atrophaeus NIJ as a thermophilic hydrocarbonoclastic strain and its application in laundry detergent formulations. Algerian J. Environ. Sci. Technol. 2021, 7, 1707–1724. [Google Scholar]
- Rathod, B.G.; Poosarla, V.G.; Kuppili, S.K.; Chouhan, K.S.Y.R.; Shivshetty, N. Production and characterization of a halotolerant protease from Bacillus siamensis F2 for chicken byproducts valorization. Biomass Conv. Bioref. 2024, 14, 1343–1358. [Google Scholar] [CrossRef]
- Zhu, X.; Hua, Y.; Li, X.; Kong, X.; Zhang, C.; Chen, Y. Isolation and characterization of an activator-dependent protease from Aspergillus ochraceus screened from low denatured defatted soybean meal and the proteolysis of soy proteins. LWT 2021, 150, 112026. [Google Scholar] [CrossRef]
- Keshapaga, U.R.; Jathoth, K.; Singh, S.S.; Gogada, R.; Burgula, S. Characterization of high-yield Bacillus subtilis cysteine protease for diverse industrial applications. Braz. J. Microbiol. 2023, 54, 739–752. [Google Scholar] [CrossRef]
- Lu, J.; Zhao, Y.; Hu, R.; Cheng, Y.; Qin, J.; Yang, J.; Fang, Y.; Lyu, M.; Wang, S. Screening and characteristics of marine Bacillus velezensis Z-1 protease and its application of enzymatic hydrolysis of mussels to prepare antioxidant active substances. Molecules 2022, 27, 6570. [Google Scholar] [CrossRef]
- Thomas, N.N.; Archana, V.; Shibina, S.; Edwin, B.T. Isolation and characterization of a protease from Bacillus sps. Mater. Today Proc. 2021, 41, 685–691. [Google Scholar] [CrossRef]
- Gupta, R.; Beg, Q.K.; Lorenz, P. Bacterial alkaline proteases: Molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 2002, 59, 15–32. [Google Scholar] [CrossRef]
- Zhang, H.; Li, H.; Lang, D.A.; Xu, H.; Zhu, H. Purification, characterization and potential applications of a novel halotolerant metalloprotease from marine bacterium Vibrio sp. LA-05. J. Chem. Technol. Biotechnol. 2018, 93, 3627–3637. [Google Scholar] [CrossRef]
- Liu, X.; Lu, Q.; Xiao, H.; Feng, Y.; Su, G.; Zhao, M.; Huang, M. Expression of a salt-tolerant pseudolysin in yeast for efficient protein hydrolysis under high-salt conditions. Biomolecules 2023, 13, 83. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Yin, Y.; Yan, J.; Zhang, J.; Ma, H.; Zhou, C. Separation, biochemical characterization and salt-tolerant mechanisms of alkaline protease from Aspergillus oryzae. J. Sci. Food Agric. 2019, 99, 3359–3366. [Google Scholar] [CrossRef]
- Gao, X.; Yin, Y.; Zhou, C. Purification, characterisation and salt-tolerance molecular mechanisms of aspartyl aminopeptidase from Aspergillus oryzae 3.042. Food Chem. 2018, 240, 377–385. [Google Scholar] [CrossRef]
- Suwannaphan, S.; Fufeungsombut, E.; Promboon, A.; Chim-anage, P. A serine protease from newly isolated Bacillus sp. for efficient silk degumming, sericin degrading and colour bleaching activities. Int. Biodeterior. Biodegrad. 2017, 117, 141–149. [Google Scholar] [CrossRef]
- Pereira, J.Q.; Ambrosini, A.; Passaglia, L.M.P.; Brandelli, A. A new cold-adapted serine peptidase from Antarctic Lysobacter sp. A03: Insights about enzyme activity at low temperatures. Int. J. Biol. Macromol. 2017, 103, 854–862. [Google Scholar] [CrossRef]
- Siddiqui, K.S.; Ertan, H.; Poljak, A.; Bridge, W.J. Evaluating enzymatic productivity—The missing link to enzyme utility. Int. J. Mol. Sci. 2022, 23, 6908. [Google Scholar] [CrossRef]
- Nikhita, R.; Sachindra, N.M. Optimization of chemical and enzymatic hydrolysis for production of chicken blood protein hydrolysate rich in angiotensin-I converting enzyme inhibitory and antioxidant activity. Poult. Sci. 2021, 100, 101047. [Google Scholar] [CrossRef] [PubMed]
- Babini, E.; Tagliazucchi, D.; Martini, S.; Più, L.D.; Gianotti, A. LC-ESI-QTOF-MS identification of novel antioxidant peptides obtained by enzymatic and microbial hydrolysis of vegetable proteins. Food Chem. 2017, 228, 186–196. [Google Scholar] [CrossRef]
- Chen, X.; Liang, L.; Han, C. Borate suppresses the scavenging activity of gallic acid and plant polyphenol extracts on DPPH radical: A potential interference to DPPH assay. LWT 2020, 131, 109769. [Google Scholar] [CrossRef]
- Guo, X.; Dong, Z.; Li, Q.; Wan, D.; Zhong, J.; Dongzhi, D.; Huang, M. Flavonoids from Rhododendron nivale Hook. f delay aging via modulation of gut microbiota and glutathione metabolism. Phytomedicine 2022, 104, 154270. [Google Scholar] [CrossRef] [PubMed]
- Karunakaran, U.; Elumalai, S.; Moon, J.S.; Won, K.C. c-Abl tyrosine kinase inhibition attenuate oxidative stress-induced pancreatic β-Cell dysfunction via glutathione antioxidant system. Transl. Res. 2022, 249, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Jiang, H.; Cao, G.; Yang, C.; Cao, Y. Enzymolysis of by-product derived from sheep placenta to production of highly active antioxidant peptide. Synth. Catal. 2017, 2, 3. [Google Scholar] [CrossRef]
- Lv, J.Y.; Nawaz, M.A.H.; Liu, N.; Zhou, H.P.; Hussain, E.; Wen, X.; Gou, X.Y.; Jin, X.; Yu, C. A Nile red-based near-infrared fluorescent probe for the detection of superoxide radical anion in living cells. Chin. J. Anal. Chem. 2022, 50, 100140. [Google Scholar] [CrossRef]
- Lee, E.J.; Jang, Y.S.; Hong, I.K. Effects of lamb placenta peptide powder on skin moisturizing and wrinkle formation. J. Korean Soc. Food Sci. Nutr. 2023, 52, 1013–1034. [Google Scholar] [CrossRef]
- Lowry, O.; Rosebrough, N.; Farr, A.; Randall, R. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Chalita, M.; Kim, Y.O.; Park, S.; Oh, H.S.; Cho, J.H.; Moon, J.; Baek, N.; Moon, C.; Lee, K.; Yang, J.; et al. EzBioCloud: A genome-driven database and platform for microbiome identification and discovery. Int. J. Syst. Evol. Microbiol. 2024, 74, 006421. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Hassabo, A.A.; Selim, M.H.; Saad, M.M.; Abdelraof, M. Optimization of L-methioninase and L-arginase production by newly isolated marine yeast using response surface methodology. Biocatal. Agric. Biotechnol. 2022, 42, 102383. [Google Scholar] [CrossRef]
- Karray, A.; Alonazi, M.; Horchani, H.; Bacha, A.B. A novel thermostable and alkaline protease produced from Bacillus stearothermophilus isolated from olive oil mill sols suitable to industrial biotechnology. Molecules 2021, 26, 1139. [Google Scholar] [CrossRef]
- Weber, K.; Osborn, M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 1969, 244, 4406–4412. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Feng, Z. The measurement of the degree of hydrolysis of hydrolysed protein. Food Sci. 1994, 179, 65–67. (In Chinese) [Google Scholar]
- Skelton, G.S. Papaya proteinases. I. Temperature-and pH-stability curves. Enzymologia 1968, 35, 270–274. [Google Scholar] [PubMed]






| Run | A: Casein Content (%) | B: Yeast Extract Content (%) | C: NaCl Addition (%) | R: Protease Activity (U/mL) |
|---|---|---|---|---|
| 1 | 1 | 0 | 1 | 1191 |
| 2 | 3 | 0 | 1 | 1527 |
| 3 | 1 | 0.5 | 1 | 2590 |
| 4 | 3 | 0.5 | 1 | 2388 |
| 5 | 1 | 0.25 | 0.5 | 2294 |
| 6 | 3 | 0.25 | 0.5 | 2480 |
| 7 | 1 | 0.25 | 1.5 | 2154 |
| 8 | 3 | 0.25 | 1.5 | 2810 |
| 9 | 2 | 0 | 0.5 | 1493 |
| 10 | 2 | 0.5 | 0.5 | 2862 |
| 11 | 2 | 0 | 1.5 | 1658 |
| 12 | 2 | 0.5 | 1.5 | 2850 |
| 13 | 2 | 0.25 | 1 | 3144 |
| 14 | 2 | 0.25 | 1 | 2998 |
| 15 | 2 | 0.25 | 1 | 3195 |
| 16 | 2 | 0.25 | 1 | 3090 |
| 17 | 2 | 0.25 | 1 | 3278 |
| Source | Sum of Squares | df | Mean Square | F-Value | p-Value |
|---|---|---|---|---|---|
| Model | 6.880 × 106 | 9 | 7.645 × 105 | 44.89 | <0.0001 |
| A-casein | 1.195 × 105 | 1 | 1.195 × 105 | 7.02 | 0.0330 |
| B-Yeast extra | 2.907 × 106 | 1 | 2.907 × 106 | 170.70 | <0.0001 |
| C-NaCl | 14,644 | 1 | 14,644 | 0.86 | 0.3846 |
| AB | 72,395 | 1 | 72,395 | 4.25 | 0.0781 |
| AC | 55,117 | 1 | 55,117 | 3.24 | 0.1150 |
| BC | 7796 | 1 | 7796 | 0.46 | 0.5204 |
| A2 | 1.048 × 106 | 1 | 1.048 × 106 | 61.56 | 0.0001 |
| B2 | 2.168 × 106 | 1 | 2.168 × 106 | 127.30 | <0.0001 |
| C2 | 1.809 × 105 | 1 | 1.809 × 105 | 10.62 | 0.0139 |
| Residual | 1.192 × 105 | 7 | 17,028 | ||
| Lack of Fit | 74,324 | 3 | 24,775 | 2.21 | 0.2296 |
| Pure Error | 44,875 | 4 | 11,219 | ||
| Cor Total | 6.999 × 106 | 16 | |||
| C.V.% = 5.28 R2 = 0.9830 R2adj = 0.9611 R2pred = 0.8201 | |||||
| Purification Step | Total Protein (mg) | Total Activity (U) | Specific Activity (U/mg) | Purification Fold | Recovery (%) |
|---|---|---|---|---|---|
| Crude | 305.30 | 133,790 | 438 | 1.00 | 100.00% |
| 20–60% (NH4)2SO4 | 20.38 | 94,496 | 4637 | 10.58 | 70.63% |
| DEAE FF | 9.30 | 69,424 | 7467 | 17.04 | 51.89% |
| Ultra-15 | 0.50 | 5445 | 10,976 | 25.05 | 4.07% |
| Metal Ion | Relative Enzyme Activity (%) (1 mM) | Relative Enzyme Activity (%) (10 mM) |
|---|---|---|
| Blank | 100 ± 0.8 | 100 ± 2.1 |
| Cu2+ | 99.7 ± 1.3 | 62.8 ± 3.2 |
| Fe3+ | 90.3 ± 2.3 | 0.0 ± 0.1 |
| Mg2+ | 94.7 ± 1.7 | 94.0 ± 1.1 |
| K+ | 94.3 ± 2.9 | 97.3 ± 0.3 |
| Zn2+ | 93.1 ± 2.1 | 77.3 ± 1.5 |
| Ni2+ | 87.3 ± 1.5 | 86.5 ± 0.8 |
| Co2+ | 99.2 ± 3.1 | 95.6 ± 1.4 |
| Mn2+ | 110.3 ± 1.2 | 115.4 ± 2.7 |
| Al3+ | 91.3 ± 0.6 | 0.6 ± 0.2 |
| Ca2+ | 97.1 ± 1.4 | 98.1 ± 1.9 |
| Inhibitor/Denaturing/Surfactant Agent | Residual Activity (%) | |
|---|---|---|
| None | 100 ± 0.7 | 100 ± 1.1 |
| Inhibitor/Denaturing agents | 1 mmol/L | 10 mmol/L |
| PMSF | 20.1 ± 1.2 | 2.7 ± 0.4 |
| EDTA | 62.5 ± 0.6 | 48.9 ± 1.2 |
| 1, 10-Phenanthroline | 99.0 ± 1.7 | 98.5 ± 1.0 |
| Dithiothreitol | 92.3 ± 1.3 | 90.5 ± 1.5 |
| Surfactant agents | 1% (v/v) | 10% (v/v) |
| Tween 80 | 102.5 ± 0.9 | 105.6 ± 0.5 |
| OP-10 | 94.6 ± 2.1 | 101.3 ± 1.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Peng, G.; Sun, J.; Jiang, C.; Hao, J.; Zhang, X. Screening and Characterization of Marine Bacillus atrophaeus G4 Protease and Its Application in the Enzymatic Hydrolysis of Sheep (Ovis aries) Placenta for the Preparation of Antioxidant Peptides. Molecules 2025, 30, 2217. https://doi.org/10.3390/molecules30102217
Wang W, Peng G, Sun J, Jiang C, Hao J, Zhang X. Screening and Characterization of Marine Bacillus atrophaeus G4 Protease and Its Application in the Enzymatic Hydrolysis of Sheep (Ovis aries) Placenta for the Preparation of Antioxidant Peptides. Molecules. 2025; 30(10):2217. https://doi.org/10.3390/molecules30102217
Chicago/Turabian StyleWang, Wei, Guoqing Peng, Jingjing Sun, Chengcheng Jiang, Jianhua Hao, and Xiu Zhang. 2025. "Screening and Characterization of Marine Bacillus atrophaeus G4 Protease and Its Application in the Enzymatic Hydrolysis of Sheep (Ovis aries) Placenta for the Preparation of Antioxidant Peptides" Molecules 30, no. 10: 2217. https://doi.org/10.3390/molecules30102217
APA StyleWang, W., Peng, G., Sun, J., Jiang, C., Hao, J., & Zhang, X. (2025). Screening and Characterization of Marine Bacillus atrophaeus G4 Protease and Its Application in the Enzymatic Hydrolysis of Sheep (Ovis aries) Placenta for the Preparation of Antioxidant Peptides. Molecules, 30(10), 2217. https://doi.org/10.3390/molecules30102217






