Design and Evaluation of NSAID Derivatives as AKR1C3 Inhibitors for Breast Cancer Treatment through Computer-Aided Drug Design and In Vitro Analysis
Abstract
1. Introduction
2. Results and Discussion
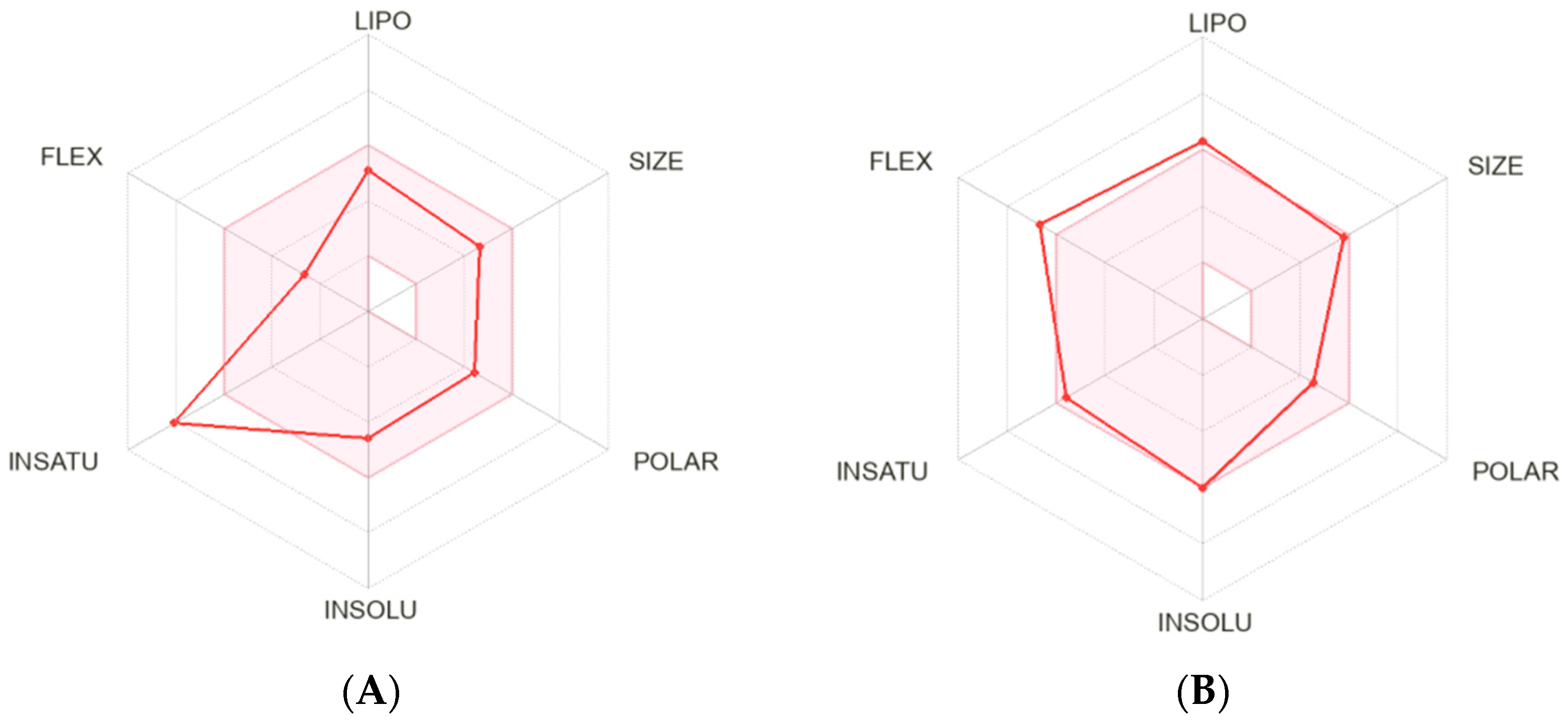
2.1. Bioinformatic
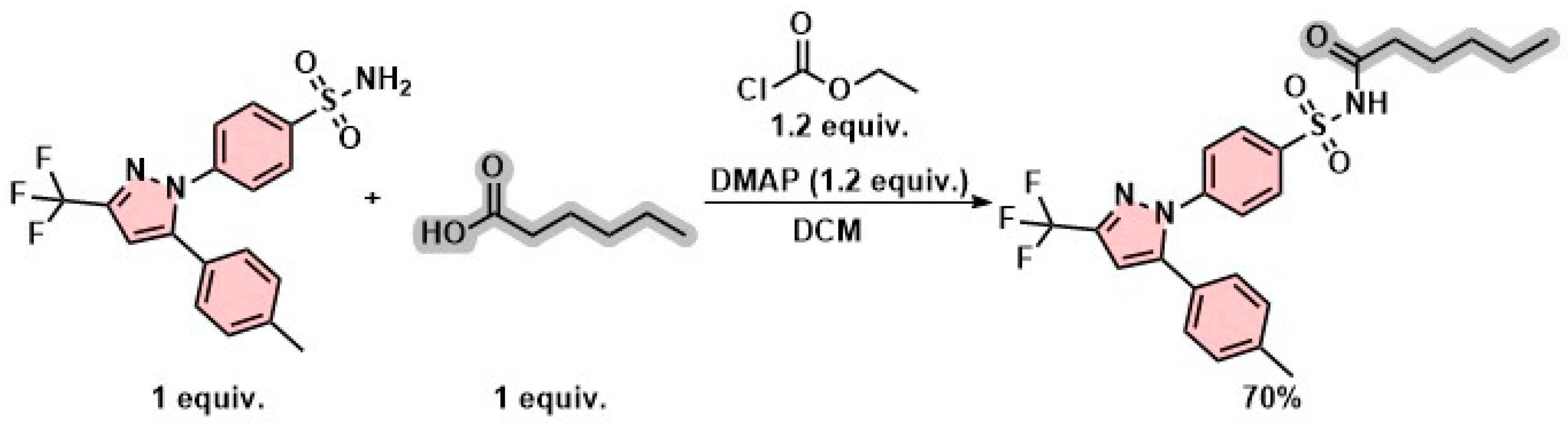
2.2. Synthesis and Characterization
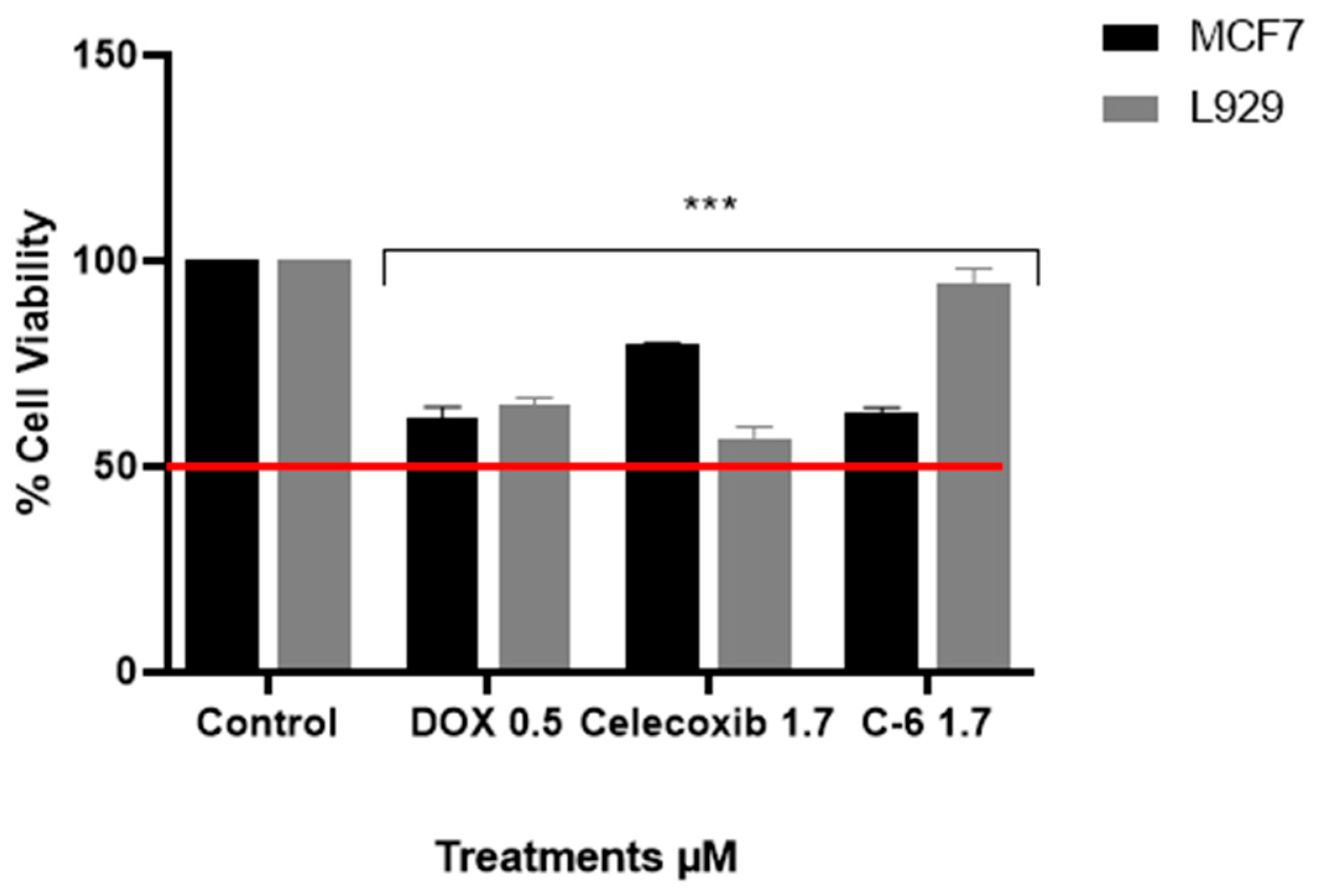
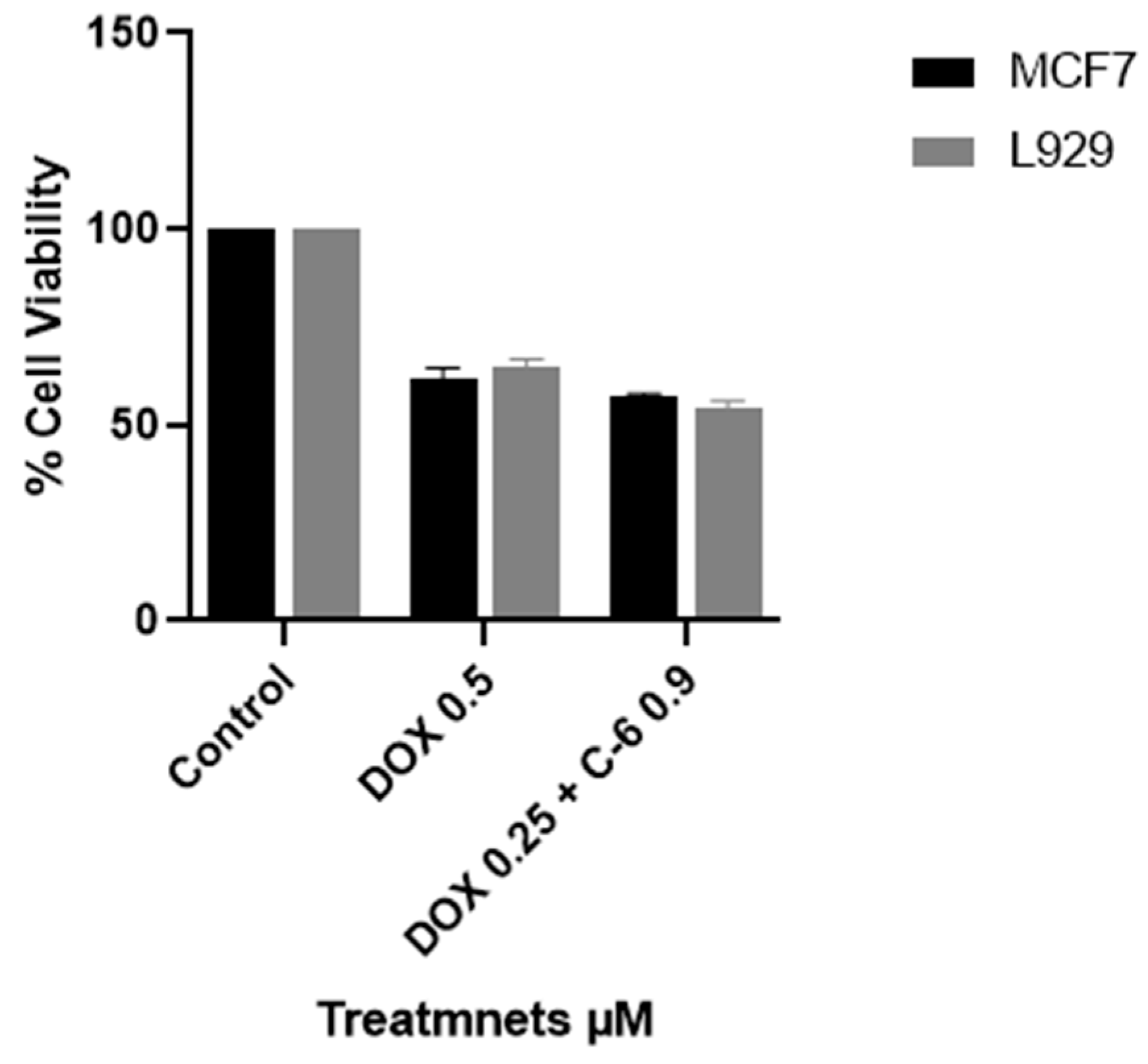
2.3. Viability Assay
3. Materials and Methods
3.1. Bioinformatics
3.1.1. Ligand-Based Virtual Screening (LBVS)
3.1.2. Structure-Based Virtual Screening (SBVS)
3.1.3. Toxicity and LogP Profile
3.2. Chemistry
Solubility and HPLC Method to C-6 and Celecoxib
3.3. Cell Lines and Culture Conditions
3.3.1. Treatments
3.3.2. Cytotoxicity Screening
3.3.3. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Ji, X.; Lu, Y.; Tian, H.; Meng, X.; Wei, M.; Cho, W.C. Chemoresistance mechanisms of breast cancer and their countermeasures. Biomed. Pharmacother. 2019, 114, 108800–108810. [Google Scholar] [CrossRef]
- David, R.B.; Stephen, I.R.; Lawrence, M.S.; Yi, J.; Sridhar, G.; Trevor, M.P. Development of Nonsteroidal Anti-Inflammatory Drug Analogs and Steroid Carboxylates Selective for Human Aldo-Keto Reductase Isoforms: Potential Antineoplastic Agents That Work Independently of Cyclooxygenase Isozymes. Mol. Pharmacol. 2005, 67, 60–68. [Google Scholar]
- Liu, Y.; Chen, Y.; Jiang, J.; Chu, X.; Guo, Q.; Zhao, L.; Feng, F.; Liu, W.; Zhang, X.; He, S.; et al. Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer. Eur. J. Med. Chem. 2023, 247, 115013–115020. [Google Scholar] [CrossRef]
- Tołoczko-Iwaniuk, N.; Dziemiańczyk-Pakieła, D.; Nowaszewska, B.K.; Celińska-Janowicz, K.; Miltyk, W. Celecoxib in Cancer Therapy and Prevention—Review. Curr. Drug Targets 2019, 20, 302–315. [Google Scholar] [CrossRef]
- Yang, J.C.; Xu, P.; Ning, S.; Wasielewski, L.J.; Adomat, H.; Hwang, S.H.; Morisseau, C.; Gleave, M.; Corey, E.; Gao, A.C.; et al. Novel inhibition of AKR1C3 and androgen receptor axis by PTUPB synergizes enzalutamide treatment in advanced prostate cancer. Oncogene 2023, 42, 693–707. [Google Scholar] [CrossRef]
- Huang, C.; Chen, Y.; Hang Liu1, J.Y.; Song, X.; Zhao, J.; He, N.; Zhou, C.J.; Wang, Y.; Huang, C.; Dong, Q. Celecoxib targets breast cancer stem cells by inhibiting the synthesis of prostaglandin E2 and down-regulating the Wnt pathway activity. Oncotarget 2017, 8, 115254–115269. [Google Scholar] [CrossRef]
- Khafaga, A.F.; Shamma, R.N.; Abdeen, A.; Barakat, A.M.; Noreldin, A.E.; Elzoghby, A.O.; Sallam, M.A. Celecoxib repurposing in cancer therapy: Molecular mechanisms and nanomedicine-based delivery technologies. Nanomedicine 2021, 16, 1691–1712. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.E.; Schwartzbaum, J.A. Celecoxib may be a viable treatment option for breast cancer patients not treated with chemotherapy. Front. Oncol. 2022, 12, 958308. [Google Scholar] [CrossRef] [PubMed]
- El-Haj, B.M.; Ahmed, S.B.M.; Garawi, M.A.; Ali, H.S. Linking Aromatic Hydroxy Metabolic Functionalization of Drug Molecules to Structure and Pharmacologic Activity. Molecules 2018, 23, 2119. [Google Scholar] [CrossRef]
- El-Shahat, M.; Salama, M.A.M.; El-Farargyj, A.F.; Ali, M.M.; Ahmed, D.M. Effective Pharmacophore for CDC25 Phosphatases Enzyme Inhibitors: Newly Synthesized Bromothiazolopyrimidine Derivatives. Bentham Sci. 2022, 21, 118–131. [Google Scholar]
- El-Sofany, W.I.; El-sayed, W.A.; Abd-Rabou, A.A.; El-Shahat, M. Synthesis of new imidazole-triazole-glycoside hybrids as anti-breast cancer candidates. J. Mol. Struct. 2022, 1270, 133942–133954. [Google Scholar] [CrossRef]
- Flefel, E.M.; El-Sofany, W.I.; Al-Harbi, R.A.K.; El-Shahat, M. Development of a Novel Series of Anticancer and Antidiabetic: Spirothiazolidines Analogs. Molecules 2019, 24, 2511. [Google Scholar] [CrossRef] [PubMed]
- Shamroukh, A.H.; El-Shahat, M.; Drabowicz, J.; Ali, M.M.; Rashad, A.E.; Ali, H.S. Anticancer evaluation of some newly synthesized N-nicotinonitrile derivative. Eur. J. Med. Chem. 2013, 69, 521–526. [Google Scholar] [CrossRef] [PubMed]
- Berdigaliyev, N.; Aljofan, M. An overview of drug discovery and development. Future Med. Chem. 2020, 12, 939–947. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Moudgil, M.n.; Mandal, S.K. Rational drug design. Eur. J. Pharmacol. 2009, 625, 90–100. [Google Scholar] [CrossRef]
- Rizzuti, B.; Grande, F. Chapter 14—Virtual screening in drug discovery: A precious tool for a still-demanding challenge. In Protein Homeostasis Diseases; Pey, A.L., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 309–327. [Google Scholar]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Guedes, I.A.; de Magalhães, C.S.; Dardenne, L.E. Receptor-ligand molecular docking. Biophys. Rev. 2014, 6, 75–87. [Google Scholar] [CrossRef]
- Lawless, M.S.; Waldman, M.; Fraczkiewicz, R.; Clark, R.D. Using Cheminformatics in Drug Discovery. In New Approaches to Drug Discovery; Nielsch, U., Fuhrmann, U., Jaroch, S., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 139–168. [Google Scholar]
- Jaramillo, D.N.; Millán, D.; Guevara-Pulido, J. Design, synthesis and cytotoxic evaluation of a selective serotonin reuptake inhibitor (SSRI) by virtual screening. Eur. J. Pharm. Sci. 2023, 183, 106403–106412. [Google Scholar] [CrossRef]
- Guevara-Pulido, J.; Jiménez, R.A.; Morantes, S.J.; Jaramillo, D.N.; Acosta-Guzmán, P. Design, Synthesis, and Development of 4-[(7-Chloroquinoline-4-yl)amino]phenol as a Potential SARS-CoV-2 Mpro Inhibitor. ChemistrySelect 2022, 7, e202200125. [Google Scholar] [CrossRef]
- Zambrano, D.; Millán, D.; Guevara-Pulido, J. In silico design, synthesis and evaluation of a less toxic octinoxate alternative with suitable photoprotection properties. Eur. J. Pharm. Sci. 2023, 180, 106332–106340. [Google Scholar] [CrossRef] [PubMed]
- Byrns, M.C.; Jin, Y.; Penning, T.M. Inhibitors of type 5 17β-hydroxysteroid dehydrogenase (AKR1C3): Overview and structural insights. J. Steroid Biochem. Mol. Biol. 2011, 125, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Penning, T.M. Aldo-Keto Reductase (AKR) 1C3 inhibitors: A patent review. Expert Opin. Ther. Pat. 2017, 27, 1329–1340. [Google Scholar] [CrossRef] [PubMed]
- Prieto, M.; Niño, A.; Acosta-Guzmán, P.; Guevara-Pulido, J. Design and synthesis of a potential selective JAK-3 inhibitor for the treatment of rheumatoid arthritis using predictive QSAR models. Inform. Med. Unlocked 2024, 45, 101464–101476. [Google Scholar] [CrossRef]
- Pirela-Ocando, S.; Romero-Cabezas, A.; Guevara-Pulido, J. Construction of a predictive model for the design of triptamin analogues with potential activity in Parkinson’s and Alzheimer’s diseases. Inform. Med. Unlocked 2023, 43, 101413–101423. [Google Scholar] [CrossRef]
- Yap, C.W. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011, 32, 1466–1474. [Google Scholar] [CrossRef] [PubMed]
- Maunz, A.; Gütlein, M.; Rautenberg, M.; Vorgrimmler, D.; Gebele, D.; Helma, C. lazar: A modular predictive toxicology framework. Front. Pharmacol. 2013, 4, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Xing, E.; Wu, S.; Zhuang, T.; Li, P.-K.; Li, C.; Cheng, X. Computational modeling studies reveal the origin of the binding preference of 3-(3,4-di hydroisoquinolin-2(1H)-ylsulfonyl)benzoic acids for AKR1C3 over its isoforms. Protein Sci. 2022, 31, e4499. [Google Scholar] [CrossRef] [PubMed]
- Pippione, A.C.; Kilic-Kurt, Z.; Kovachka, S.; Sainas, S.; Rolando, B.; Denasio, E.; Pors, K.; Adinolfi, S.; Zonari, D.; Bagnati, R.; et al. New aldo-keto reductase 1C3 (AKR1C3) inhibitors based on the hydroxytriazole scaffold. Eur. J. Med. Chem. 2022, 237, 114366–114372. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. iLOGP: A Simple, Robust, and Efficient Description of n-Octanol/Water Partition Coefficient for Drug Design Using the GB/SA Approach. J. Chem. Inf. Model. 2014, 54, 3284–3301. [Google Scholar] [CrossRef]
- Byrns, M.C.; Duan, L.; Lee, S.H.; Blair, I.A.; Penning, T.M. Aldo-keto reductase 1C3 expression in MCF-7 cells reveals roles in steroid hormone and prostaglandin metabolism that may explain its over-expression in breast cancer. J. Steroid Biochem. Mol. Biol. 2010, 118, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Zhong, T.; Xu, F.; Xu, J.; Liu, L.; Chen, Y. Aldo-keto reductase 1C3 (AKR1C3) is associated with the doxorubicin resistance in human breast cancer via PTEN Loss. Biomed. Pharmacother. 2015, 69, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17–42. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions, and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions, or products referred to in the content. |

| Entry | Compound | LBVS Exp IC50 (µM) | Pred IC50 a (µM) | SBVS Affinity b (kcal/mol) | Lipophilicity Pred c |
|---|---|---|---|---|---|
| 1 | Naproxen | 0.5 | 0.6 | −8.6 | 2.76 |
| 2 | Diclofenac | 2.6 | 1.2 | −8.9 | 3.66 |
| 3 | Flurbiprofen | 7.8 | 4 | −9.3 | 3.59 |
| 4 | Lornoxicam | 0.7 | 0.6 | −8.7 | 1.50 |
| 5 | Mefenamic acid | 0.3 | 0.1 | −9.0 | 3.30 |
| 6 | Ibuprofen | 33.0 | 30 | −7.7 | 3.00 |
| 7 | Celecoxib | 5.2 | 2.3 | −10.4 | 3.40 d |
| 8 | Ketoprofen | 6.0 | 3.0 | −9.0 | 2.84 |
| 9 | Sulindac | 3.4 | 3.6 | −9.9 | 3.96 |
| 10 | Indomethacin | 2.3 | 0.4 | −9.4 | 3.63 |
| 11 | A1 | -- | 2.0 | −9.4 | 3.75 |
| 12 | A2 | -- | 2.2 | −9.3 | 4.03 |
| 13 | A3 | -- | 2.3 | −9.5 | 4.35 |
| 14 | A4 | -- | 2.2 | −9.6 | 4.74 |
| 15 | A5 | -- | 2.0 | −9.3 | 5.10 |
| 16 | A6 | -- | 2.1 | −9.4 | 5.42 |
| 17 | B1 | -- | 2.2 | −10.7 | 3.50 |
| 18 | B2 | -- | 2.3 | −10.9 | 3.82 |
| 19 | B3 | -- | 2.3 | −11.1 | 4.22 |
| 22 | B4 | -- | 2.5 | −11.5 | 4.55 |
| 23 | B5 | -- | 2.4 | −10.1 | 4.86 |
| 24 | B6 | -- | 2.4 | −10.1 | 5.14 |
| 25 | C2 | -- | 2.1 | −11.4 | 3.46 |
| 26 | C3 | -- | 1.9 | −11.3 | 3.76 |
| 27 | C4 | -- | 1.9 | −11.2 | 4.10 |
| 28 | C5 | -- | 1.8 | −11.1 | 4.46 |
| 29 | C6 | -- | 1.7 | −11.4 | 4.81 d |
| 30 | C7 | -- | 1.7 | −11.0 | 5.16 |
| Descriptor | Description |
|---|---|
| naAromAtom | Number of aromatic atoms |
| TopoPSA | Topological polar surface area |
| McGowan_Volume | Volume of a mole when the molecules are not in motion |
| Compound | Mutagenicity | Carcinogenicity in Rats | Carcinogenicity in Mice |
|---|---|---|---|
| Celecoxib | Non-mutagen | Negative | Positive |
| C-6 | Non-mutagen | Negative | Negative |
| C-7 | Non-mutagen | Negative | Negative |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fonseca-Benítez, V.; Acosta-Guzmán, P.; Sánchez, J.E.; Alarcón, Z.; Jiménez, R.A.; Guevara-Pulido, J. Design and Evaluation of NSAID Derivatives as AKR1C3 Inhibitors for Breast Cancer Treatment through Computer-Aided Drug Design and In Vitro Analysis. Molecules 2024, 29, 1802. https://doi.org/10.3390/molecules29081802
Fonseca-Benítez V, Acosta-Guzmán P, Sánchez JE, Alarcón Z, Jiménez RA, Guevara-Pulido J. Design and Evaluation of NSAID Derivatives as AKR1C3 Inhibitors for Breast Cancer Treatment through Computer-Aided Drug Design and In Vitro Analysis. Molecules. 2024; 29(8):1802. https://doi.org/10.3390/molecules29081802
Chicago/Turabian StyleFonseca-Benítez, Victoria, Paola Acosta-Guzmán, Juan Esteban Sánchez, Zaira Alarcón, Ronald Andrés Jiménez, and James Guevara-Pulido. 2024. "Design and Evaluation of NSAID Derivatives as AKR1C3 Inhibitors for Breast Cancer Treatment through Computer-Aided Drug Design and In Vitro Analysis" Molecules 29, no. 8: 1802. https://doi.org/10.3390/molecules29081802
APA StyleFonseca-Benítez, V., Acosta-Guzmán, P., Sánchez, J. E., Alarcón, Z., Jiménez, R. A., & Guevara-Pulido, J. (2024). Design and Evaluation of NSAID Derivatives as AKR1C3 Inhibitors for Breast Cancer Treatment through Computer-Aided Drug Design and In Vitro Analysis. Molecules, 29(8), 1802. https://doi.org/10.3390/molecules29081802







