Biological Valorization of Lignin-Derived Aromatics in Hydrolysate to Protocatechuic Acid by Engineered Pseudomonas putida KT2440
Abstract
1. Introduction
2. Results and Discussion
2.1. Evaluation of the Degradation Abilities of P. putida KT2440 towards Phenolic Compounds
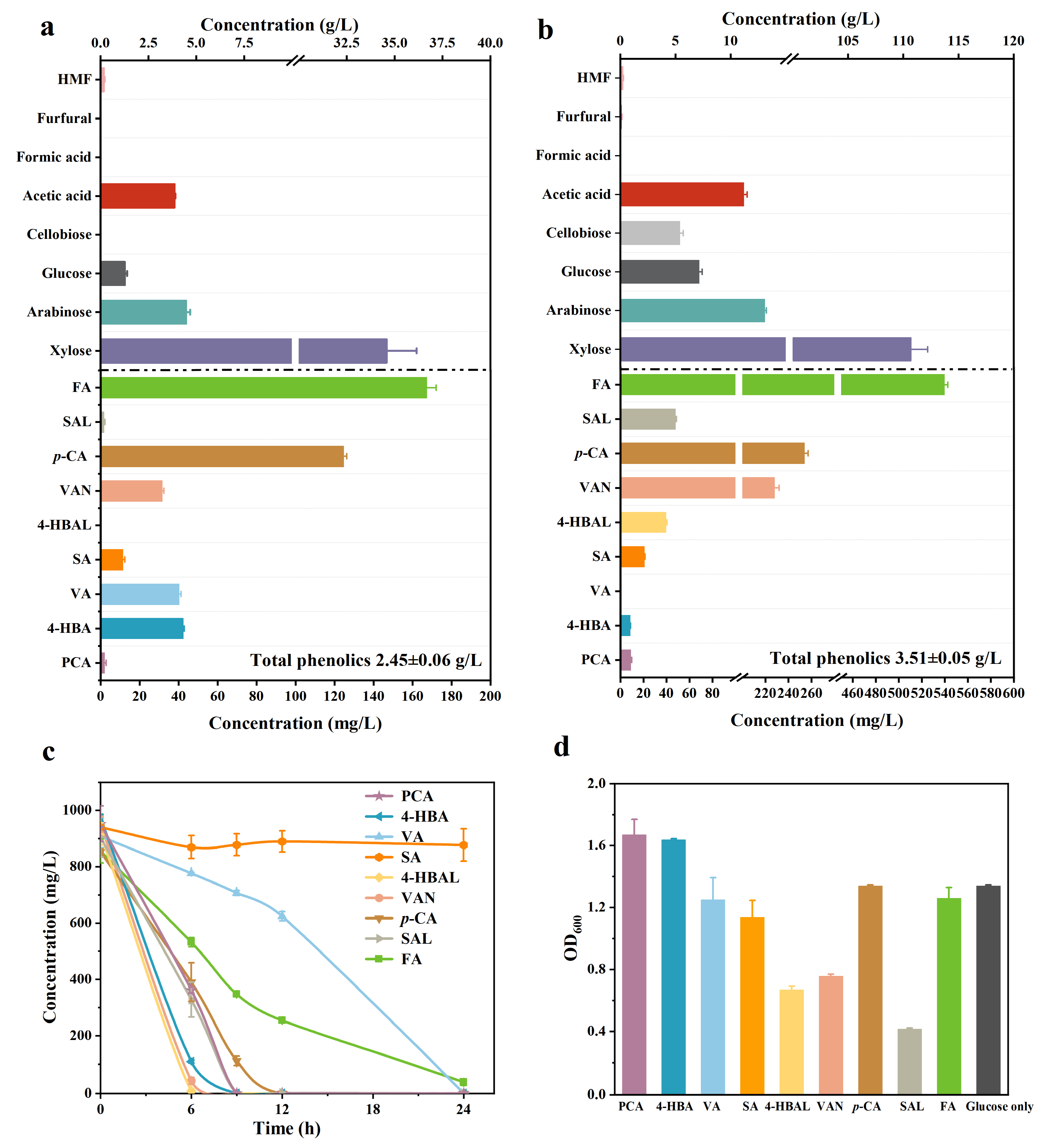
2.2. Effects of Sugar and Weak Acid in Hydrolysates on Phenolic Compound Metabolism
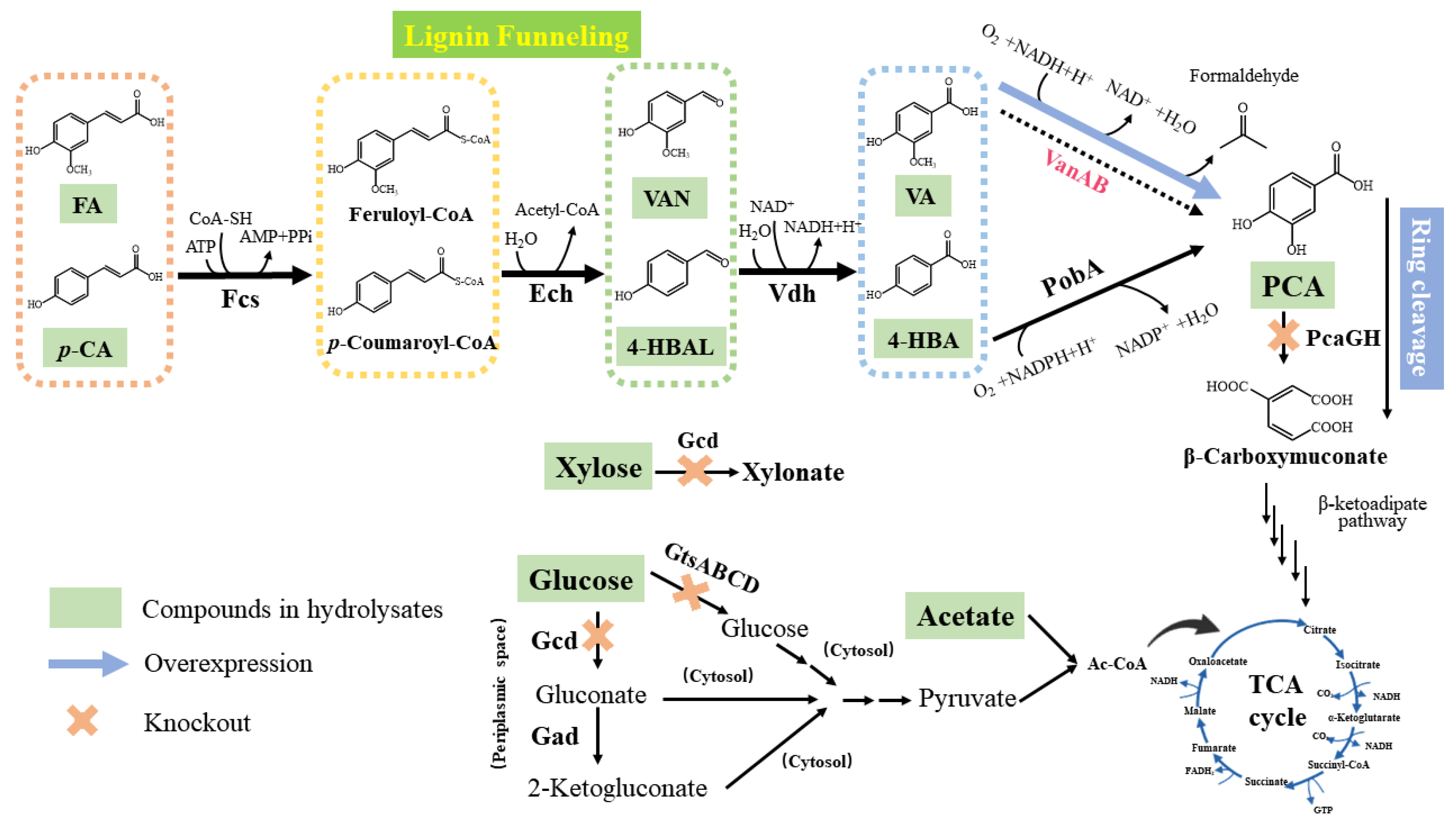
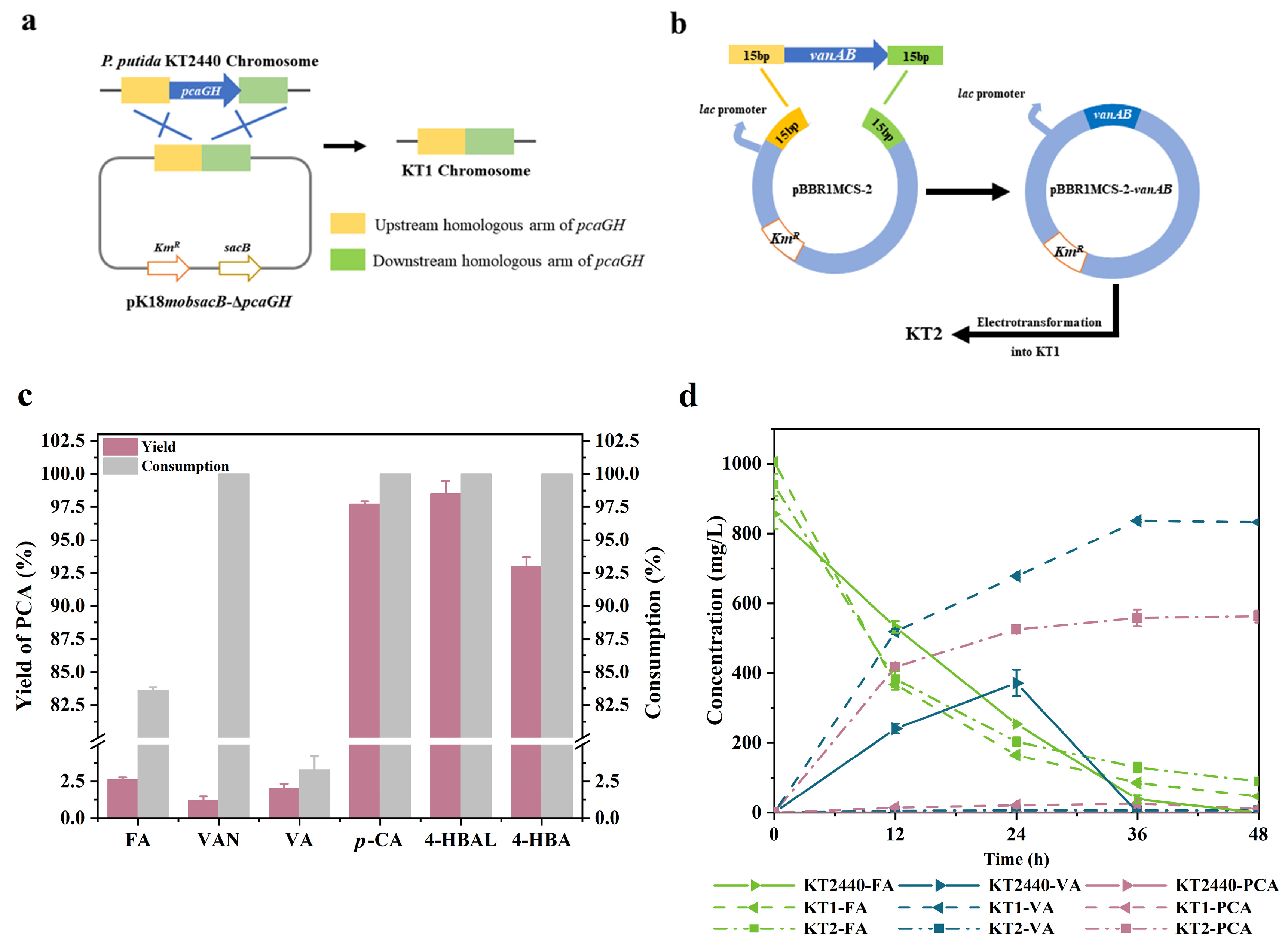
2.3. Construction of an Engineered P. putida Strain for Protocatechuic Acid Production
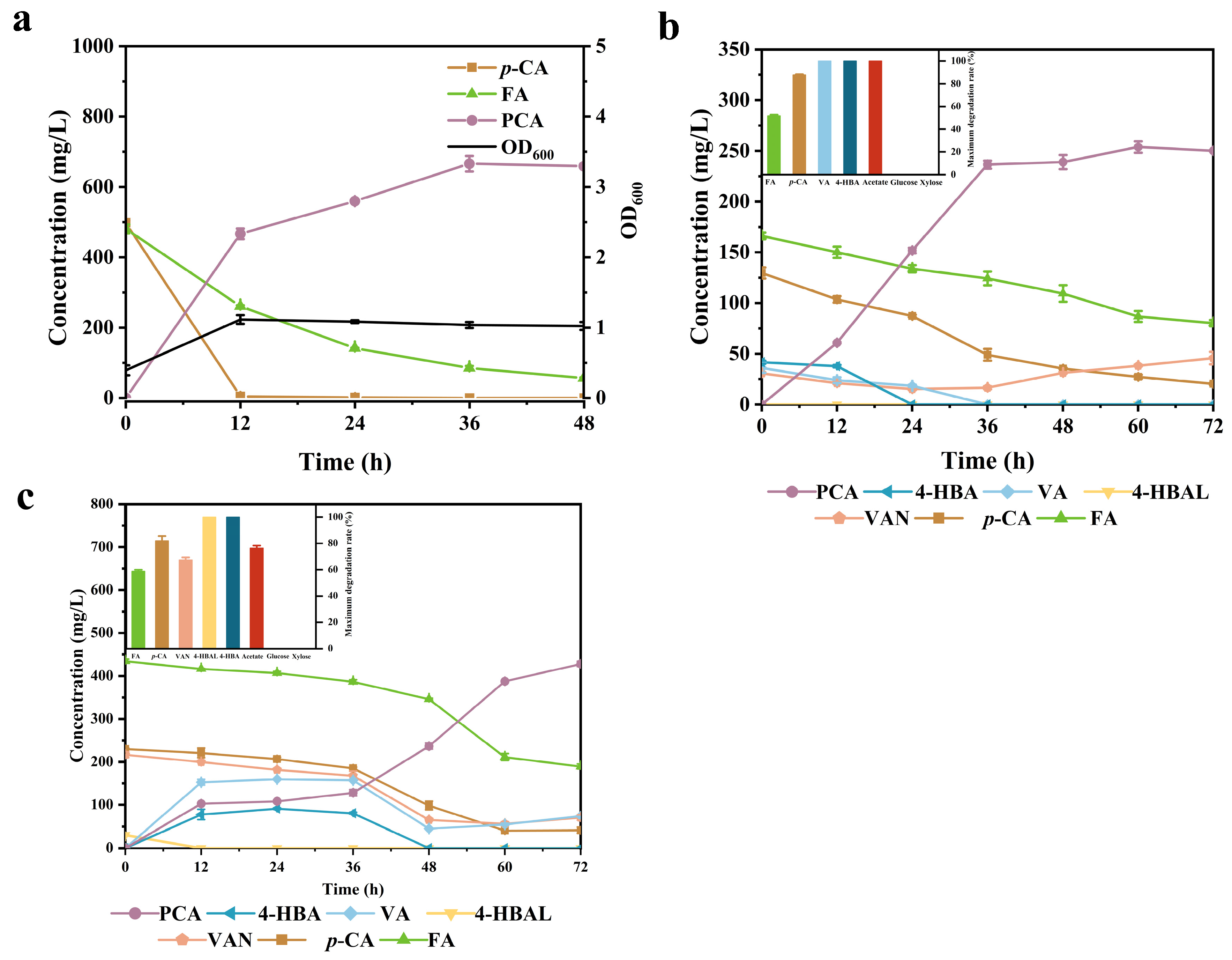
2.4. PCA Production from Single Phenolic Acid in the Engineered P. putida
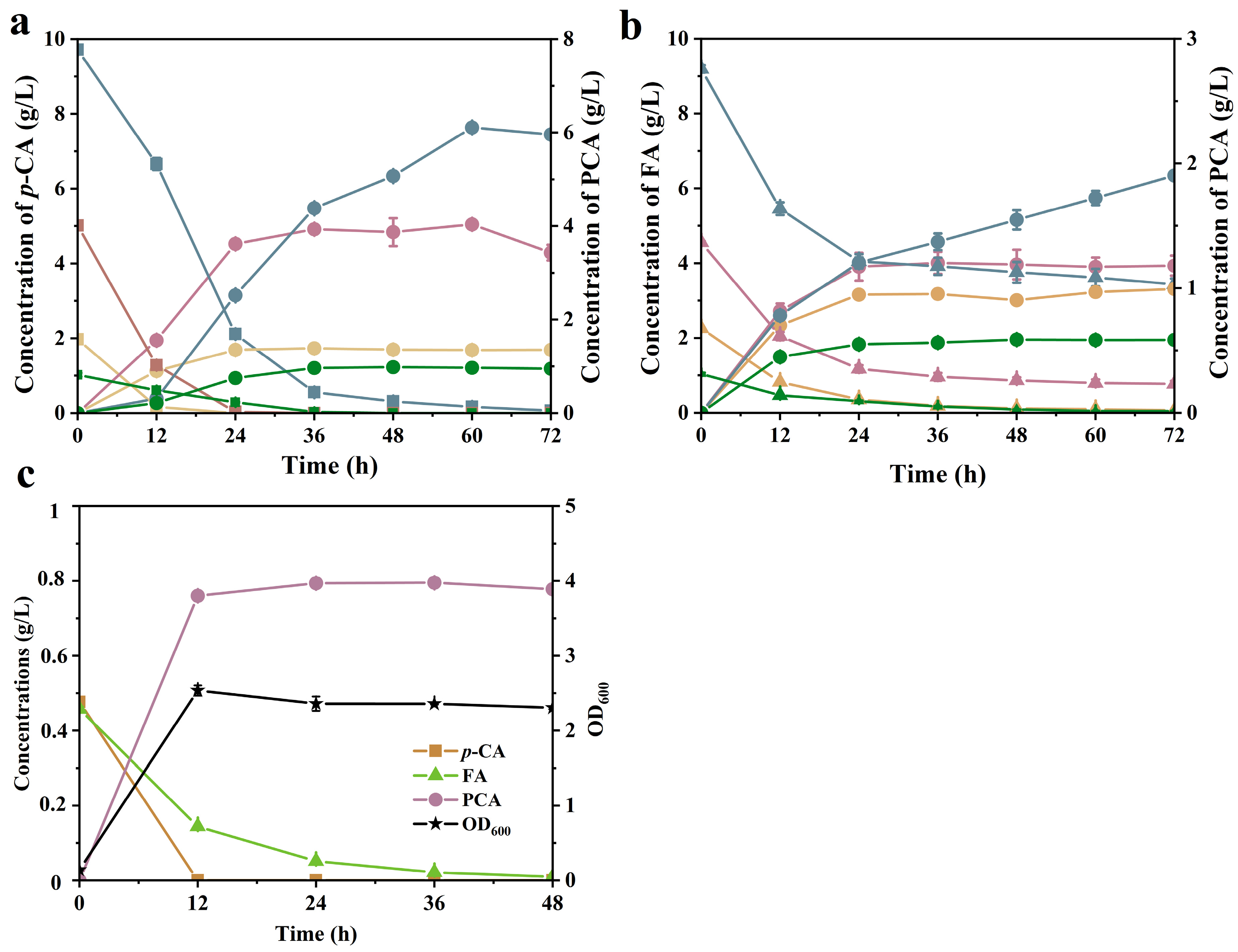
2.5. PCA Production from Hydrolysates
3. Materials and Methods
3.1. Materials
3.2. Strains and Plasmids
3.3. Genetic Manipulation and Plasmid Construction
3.4. P. putida Cultivations
3.5. Analytical Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.; Nie, Y.; Lu, X.; Zhang, X.; He, H.; Pan, F.; Zhou, L.; Liu, X.; Ji, X.; Zhang, S. Cascade Utilization of Lignocellulosic Biomass to High-Value Products. Green Chem. 2019, 21, 3499–3535. [Google Scholar] [CrossRef]
- Kant, S.; Sadashiv, S.; Ashok, A.; Kant, R. Bioresource Technology Recent Developments in Pretreatment Technologies on Lignocellulosic Biomass: Effect of Key Parameters, Technological Improvements, and Challenges. Bioresour. Technol. 2020, 300, 122724. [Google Scholar] [CrossRef]
- Hassan, S.S.; Williams, G.A.; Jaiswal, A.K. Lignocellulosic Biorefineries in Europe: Current State and Prospects. Trends Biotechnol. 2019, 37, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.S.; Maneerung, T.; Tsang, D.C.W.; Ok, Y.S.; Wang, C.H. Valorization of Biomass to Hydroxymethylfurfural, Levulinic Acid, and Fatty Acid Methyl Ester by Heterogeneous Catalysts. Chem. Eng. J. 2017, 328, 246–273. [Google Scholar] [CrossRef]
- Toor, M.; Kumar, S.S.; Malyan, S.K.; Bishnoi, N.R.; Mathimani, T.; Rajendran, K.; Pugazhendhi, A. An Overview on Bioethanol Production from Lignocellulosic Feedstocks. Chemosphere 2020, 242, 125080. [Google Scholar] [CrossRef] [PubMed]
- Gogoi, G.; Hazarika, S. Coupling of Ionic Liquid Treatment and Membrane Filtration for Recovery of Lignin from Lignocellulosic Biomass. Sep. Purif. Technol. 2017, 173, 113–120. [Google Scholar] [CrossRef]
- Zhou, M.; Fakayode, O.A.; Ahmed Yagoub, A.E.G.; Ji, Q.; Zhou, C. Lignin Fractionation from Lignocellulosic Biomass Using Deep Eutectic Solvents and Its Valorization. Renew. Sustain. Energy Rev. 2022, 156, 111986. [Google Scholar] [CrossRef]
- Wang, Z.; Deuss, P.J. The Isolation of Lignin with Native-like Structure. Biotechnol. Adv. 2023, 68, 108230. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, R.; Jastrzebski, R.; Clough, M.T.; Ralph, J.; Kennema, M.; Bruijnincx, P.C.A.; Weckhuysen, B.M. Paving the Way for Lignin Valorisation: Recent Advances in Bioengineering, Biorefining and Catalysis. Angew. Chemie -Int. Ed. 2016, 55, 8164–8215. [Google Scholar] [CrossRef]
- Gillet, S.; Aguedo, M.; Petitjean, L.; Morais, A.R.C.; Da Costa Lopes, A.M.; Łukasik, R.M.; Anastas, P.T. Lignin Transformations for High Value Applications: Towards Targeted Modifications Using Green Chemistry. Green Chem. 2017, 19, 4200–4233. [Google Scholar] [CrossRef]
- Bajwa, D.S.; Pourhashem, G.; Ullah, A.H.; Bajwa, S.G. A Concise Review of Current Lignin Production, Applications, Products and Their Environment Impact. Ind. Crops Prod. 2019, 139, 111526. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, S.Y.; Lee, J.H.; Choi, I.G.; Choi, J.W. Sequential Solvent Fractionation of Lignin for Selective Production of Monoaromatics by Ru Catalyzed Ethanolysis. RSC Adv. 2017, 7, 53117–53125. [Google Scholar] [CrossRef]
- Liu, H.; Liu, Z.H.; Zhang, R.K.; Yuan, J.S.; Li, B.Z.; Yuan, Y.J. Bacterial Conversion Routes for Lignin Valorization. Biotechnol. Adv. 2022, 60, 108000. [Google Scholar] [CrossRef] [PubMed]
- Reshmy, R.; Athiyaman Balakumaran, P.; Divakar, K.; Philip, E.; Madhavan, A.; Pugazhendhi, A.; Sirohi, R.; Binod, P.; Kumar Awasthi, M.; Sindhu, R. Microbial Valorization of Lignin: Prospects and Challenges. Bioresour. Technol. 2022, 344, 126240. [Google Scholar] [CrossRef] [PubMed]
- Linger, J.G.; Vardon, D.R.; Guarnieri, M.T.; Karp, E.M.; Hunsinger, G.B.; Franden, M.A.; Johnson, C.W.; Chupka, G.; Strathmann, T.J.; Pienkos, P.T.; et al. Lignin Valorization through Integrated Biological Funneling and Chemical Catalysis. Proc. Natl. Acad. Sci. USA 2014, 111, 12013–12018. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.; Kuhl, M.; Kohlstedt, M.; Starck, S.; Wittmann, C. Metabolic Engineering of Corynebacterium Glutamicum for the Production of Cis, Cis-Muconic Acid from Lignin. Microb. Cell Fact. 2018, 17, 115. [Google Scholar] [CrossRef] [PubMed]
- Antony, F.M.; Wasewar, K. Effect of Temperature on Equilibria for Physical and Reactive Extraction of Protocatechuic Acid. Heliyon 2020, 6, e03664. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; He, Y.; Luo, C.; Feng, B.; Ran, F.; Xu, H.; Ci, Z.; Xu, R.; Han, L.; Zhang, D. New Progress in the Pharmacology of Protocatechuic Acid: A Compound Ingested in Daily Foods and Herbs Frequently and Heavily. Pharmacol. Res. 2020, 161, 105109. [Google Scholar] [CrossRef] [PubMed]
- Muthukumaran, J.; Srinivasan, S.; Venkatesan, R.S.; Ramachandran, V.; Muruganathan, U. Syringic Acid, a Novel Natural Phenolic Acid, Normalizes Hyperglycemia with Special Reference to Glycoprotein Components in Experimental Diabetic Rats. J. Acute Dis. 2013, 2, 304–309. [Google Scholar] [CrossRef]
- Semaming, Y.; Pannengpetch, P.; Chattipakorn, S.C.; Chattipakorn, N. Pharmacological Properties of Protocatechuic Acid and Its Potential Roles as Complementary Medicine. Evid. -Based Complement. Altern. Med. 2015, 2015, 593902. [Google Scholar] [CrossRef]
- Yang, Y.C.; Wei, M.C.; Huang, T.C.; Lee, S.Z. Extraction of Protocatechuic Acid from Scutellaria Barbata D. Don Using Supercritical Carbon Dioxide. J. Supercrit. Fluids 2013, 81, 55–66. [Google Scholar] [CrossRef]
- Liu, Q.; Tang, G.Y.; Zhao, C.N.; Feng, X.L.; Xu, X.Y.; Cao, S.Y.; Meng, X.; Li, S.; Gan, R.Y.; Li, H. Bin Comparison of Antioxidant Activities of Different Grape Varieties. Molecules 2018, 23, 2432. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, X.; Chen, T.; Lu, Y.; Zhang, H. Comparing E. Coli Mono-Cultures and Co-Cultures for Biosynthesis of Protocatechuic Acid and Hydroquinone. Biochem. Eng. J. 2020, 156, 107518. [Google Scholar] [CrossRef]
- Kogure, T.; Suda, M.; Hiraga, K.; Inui, M. Protocatechuate Overproduction by Corynebacterium Glutamicum via Simultaneous Engineering of Native and Heterologous Biosynthetic Pathways. Metab. Eng. 2021, 65, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yue, C.; Wei, W.; Shang, Y.; Zhang, P.; Ye, B.C. Construction of a P-Coumaric and Ferulic Acid Auto-Regulatory System in Pseudomonas Putida KT2440 for Protocatechuate Production from Lignin-Derived Aromatics. Bioresour. Technol. 2022, 344, 126221. [Google Scholar] [CrossRef]
- Vignali, E.; Pollegioni, L.; Di Nardo, G.; Valetti, F.; Gazzola, S.; Gilardi, G.; Rosini, E. Multi-Enzymatic Cascade Reactions for the Synthesis of Cis,Cis-Muconic Acid. Adv. Synth. Catal. 2022, 364, 114–123. [Google Scholar] [CrossRef]
- Zou, L.; Ouyang, S.; Hu, Y.; Zheng, Z.; Ouyang, J. Efficient Lactic Acid Production from Dilute Acid-Pretreated Lignocellulosic Biomass by a Synthetic Consortium of Engineered Pseudomonas Putida and Bacillus Coagulans. Biotechnol. Biofuels 2021, 14, 227. [Google Scholar] [CrossRef]
- Fu, J.; Wang, Z.; Miao, H.; Yu, C.; Zheng, Z.; Ouyang, J. Rapid Adaptive Evolution of Bacillus Coagulans to Undetoxified Corncob Hydrolysates for Lactic Acid Production and New Insights into Its High Phenolic Degradation. Bioresour. Technol. 2023, 383, 129246. [Google Scholar] [CrossRef]
- Overhage, J.; Priefert, H.; Rabenhorst, J.; Steinbüchel, A. Biotransformation of Eugenol to Vanillin by a Mutant of Pseudomonas Sp. Strain HR199 Constructed by Disruption of the Vanillin Dehydrogenase (Vdh) Gene. Appl. Microbiol. Biotechnol. 1999, 52, 820–828. [Google Scholar] [CrossRef]
- Ravi, K.; García-hidalgo, J.; Gorwa-grauslund, M.F.; Lidén, G. Conversion of Lignin Model Compounds by Pseudomonas Putida KT2440 and Isolates from Compost. Appl. Microb. Cell Physiol. 2017, 101, 5059–5070. [Google Scholar] [CrossRef]
- Nikel, P.I.; Chavarría, M.; Fuhrer, T.; Sauer, U.; De Lorenzo, V. Pseudomonas Putida KT2440 Strain Metabolizes Glucose through a Cycle Formed by Enzymes of the Entner-Doudoroff, Embden-Meyerhof-Parnas, and Pentose Phosphate Pathways. J. Biol. Chem. 2015, 290, 25920–25932. [Google Scholar] [CrossRef]
- Dvořák, P.; de Lorenzo, V. Refactoring the Upper Sugar Metabolism of Pseudomonas Putida for Co-Utilization of Cellobiose, Xylose, and Glucose. Metab. Eng. 2018, 48, 94–108. [Google Scholar] [CrossRef]
- Erickson, E.; Bleem, A.; Kuatsjah, E.; Werner, A.Z.; DuBois, J.L.; McGeehan, J.E.; Eltis, L.D.; Beckham, G.T. Critical Enzyme Reactions in Aromatic Catabolism for Microbial Lignin Conversion. Nat. Catal. 2022, 5, 86–98. [Google Scholar] [CrossRef]
- Li, J.; Ye, B.C. Metabolic Engineering of Pseudomonas Putida KT2440 for High-Yield Production of Protocatechuic Acid. Bioresour. Technol. 2021, 319, 124239. [Google Scholar] [CrossRef]
- Upadhyay, P.; Lali, A. Protocatechuic Acid Production from Lignin-Associated Phenolics. Prep. Biochem. Biotechnol. 2021, 51, 979–984. [Google Scholar] [CrossRef]
- Graf, N.; Altenbuchner, J. Genetic Engineering of Pseudomonas Putida KT2440 for Rapid and High-Yield Production of Vanillin from Ferulic Acid. Appl. Microbiol. Biotechnol. 2014, 98, 137–149. [Google Scholar] [CrossRef]
- Johnson, C.W.; Salvachúa, D.; Khanna, P.; Smith, H.; Peterson, D.J.; Beckham, G.T. Enhancing Muconic Acid Production from Glucose and Lignin-Derived Aromatic Compounds via Increased Protocatechuate Decarboxylase Activity. Metab. Eng. Commun. 2016, 3, 111–119. [Google Scholar] [CrossRef]
- Zhang, R.K.; Tan, Y.S.; Cui, Y.Z.; Xin, X.; Liu, Z.H.; Li, B.Z.; Yuan, Y.J. Lignin Valorization for Protocatechuic Acid Production in Engineered: Saccharomyces Cerevisiae. Green Chem. 2021, 23, 6515–6526. [Google Scholar] [CrossRef]
- Okai, N.; Masuda, T.; Takeshima, Y.; Tanaka, K.; Yoshida, K.-I.; Miyamoto, M.; Ogino, C.; Kondo, A. Biotransformation of Ferulic Acid to Protocatechuic Acid by Corynebacterium Glutamicum ATCC 21420 Engineered to Express Vanillate O-Demethylase. AMB Express 2017, 7, 130. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Qiao, H.; Zheng, Z.; Chu, Q.; Li, X.; Yong, Q.; Ouyang, J. Lactic Acid Production from Pretreated Hydrolysates of Corn Stover by a Newly Developed Bacillus Coagulans Strain. PLoS ONE 2016, 11, e0149101. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Jiang, W.; Jia, R.; Zhou, X.; Xu, Y. Directional Enhancement of 2-Keto-Gluconic Acid Production from Enzymatic Hydrolysate by Acetic Acid-Mediated Bio-Oxidation with Gluconobacter Oxydans. Bioresour. Technol. 2022, 348, 126811. [Google Scholar] [CrossRef] [PubMed]

| Strain | Substrate | Fermentation Duration (h) | Titer (g/L) | Yield | Reference |
|---|---|---|---|---|---|
| Engineered P. putida KT2440 | 1 g/L VA | n. a. | 0.51 | 0.51 | [35] |
| P. putida KT14 | 3.28 g/L p-CA | 48 | 3.1 | 1.00 | [25] |
| S. Cerevisiae yPCA12 | 1.6 g/L p-CA | 96 | 0.72 | 0.48 | [38] |
| C. glutamicum ATCC 21420 FVan | 0.89 g/L FA | 48 | 0.44 | 0.63 | [39] |
| P. putida KT2440 KT16 | 3.88 g/L FA | 48 | 2.1 | 0.68 | [25] |
| P. putida KT2 | 1.02 g/L p-CA | 72 | 0.95 | 0.99 | This work |
| P. putida KT2 | 1 g/L FA | 72 | 0.76 | 0.76 | This work |
| P. putida KT2 | p-CA:FA = 1:1 (mol/mol) | 48 | 0.8 | 0.98 | This work |
| S. cerevisiae yPCA12 | 0.5×Corn stover APL | 96 | 0.81 | 0.74 a | [38] |
| P. putida KT3 | Corncob hydrolysate 1 | 72 | 0.25 | 0.70 b | This work |
| P. putida KT3 | Corncob hydrolysate 2 | 72 | 0.43 | 0.57 b | This work |
| Strains/Plasmids | Characteristics | Source |
|---|---|---|
| Escherichia coli | ||
| Tans1-T1 | Cloning host | TransGen Biotech |
| Pseudomonas putida | ||
| KT2440 | Wild-type strain | Lab stock |
| KT1 | KT2440 with scarless deletion of pcaGH (PP_4655-4656) encoding protocatechuate 3,4-dioxygenase dehydrogenase | This study |
| KT2 | KT01 harboring plasmid pBBR1MCS2-vanAB (PP_3736-3737) encoding vanillate O-demethylase oxygenase | This study |
| KT3 | KT02 with scarless deletion of glucose dehydrogenase encoding gcd (PP_1444) and glucose ABC transporter gtsABCD (PP_1015-1018) | This study |
| Plasmids | ||
| pBBR1MCS2 | Broad-host-range cloning vector; KmR | Lab stock |
| pK18mobsacB | The suicide vector containing the sacB gene; KmR | Lab stock |
| pBBR1MCS2-vanAB | pBBR1MCS2 with vanAB from P. putida KT2440 | This study |
| pK18mobsacB-ΔpcaGH | pK18mobsacB containing the homology arms of pcaGH from P. putida KT2440 | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, X.; Li, X.; Zou, L.; Zheng, Z.; Ouyang, J. Biological Valorization of Lignin-Derived Aromatics in Hydrolysate to Protocatechuic Acid by Engineered Pseudomonas putida KT2440. Molecules 2024, 29, 1555. https://doi.org/10.3390/molecules29071555
Jin X, Li X, Zou L, Zheng Z, Ouyang J. Biological Valorization of Lignin-Derived Aromatics in Hydrolysate to Protocatechuic Acid by Engineered Pseudomonas putida KT2440. Molecules. 2024; 29(7):1555. https://doi.org/10.3390/molecules29071555
Chicago/Turabian StyleJin, Xinzhu, Xiaoxia Li, Lihua Zou, Zhaojuan Zheng, and Jia Ouyang. 2024. "Biological Valorization of Lignin-Derived Aromatics in Hydrolysate to Protocatechuic Acid by Engineered Pseudomonas putida KT2440" Molecules 29, no. 7: 1555. https://doi.org/10.3390/molecules29071555
APA StyleJin, X., Li, X., Zou, L., Zheng, Z., & Ouyang, J. (2024). Biological Valorization of Lignin-Derived Aromatics in Hydrolysate to Protocatechuic Acid by Engineered Pseudomonas putida KT2440. Molecules, 29(7), 1555. https://doi.org/10.3390/molecules29071555






