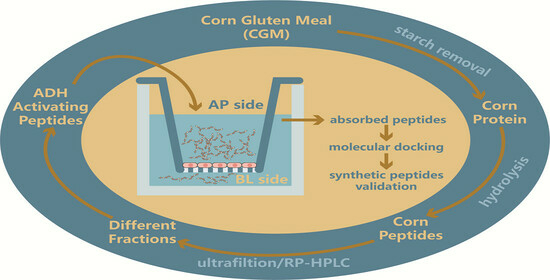
Identification of Corn Peptides with Alcohol Dehydrogenase Activating Activity Absorbed by Caco-2 Cell Monolayers
Abstract
1. Introduction
2. Results and Discussion
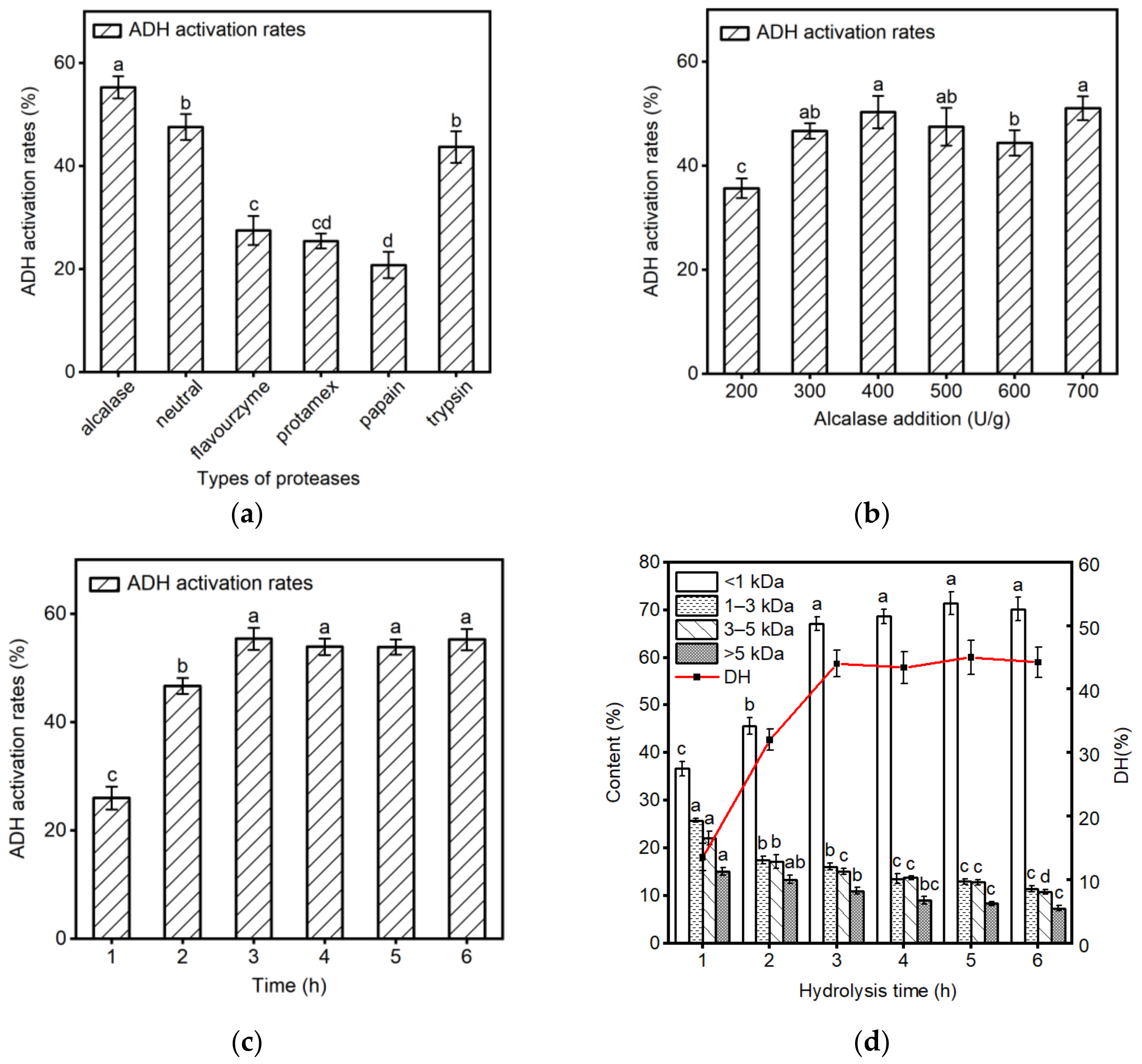
2.1. Screening for Optimal Enzymatic Conditions
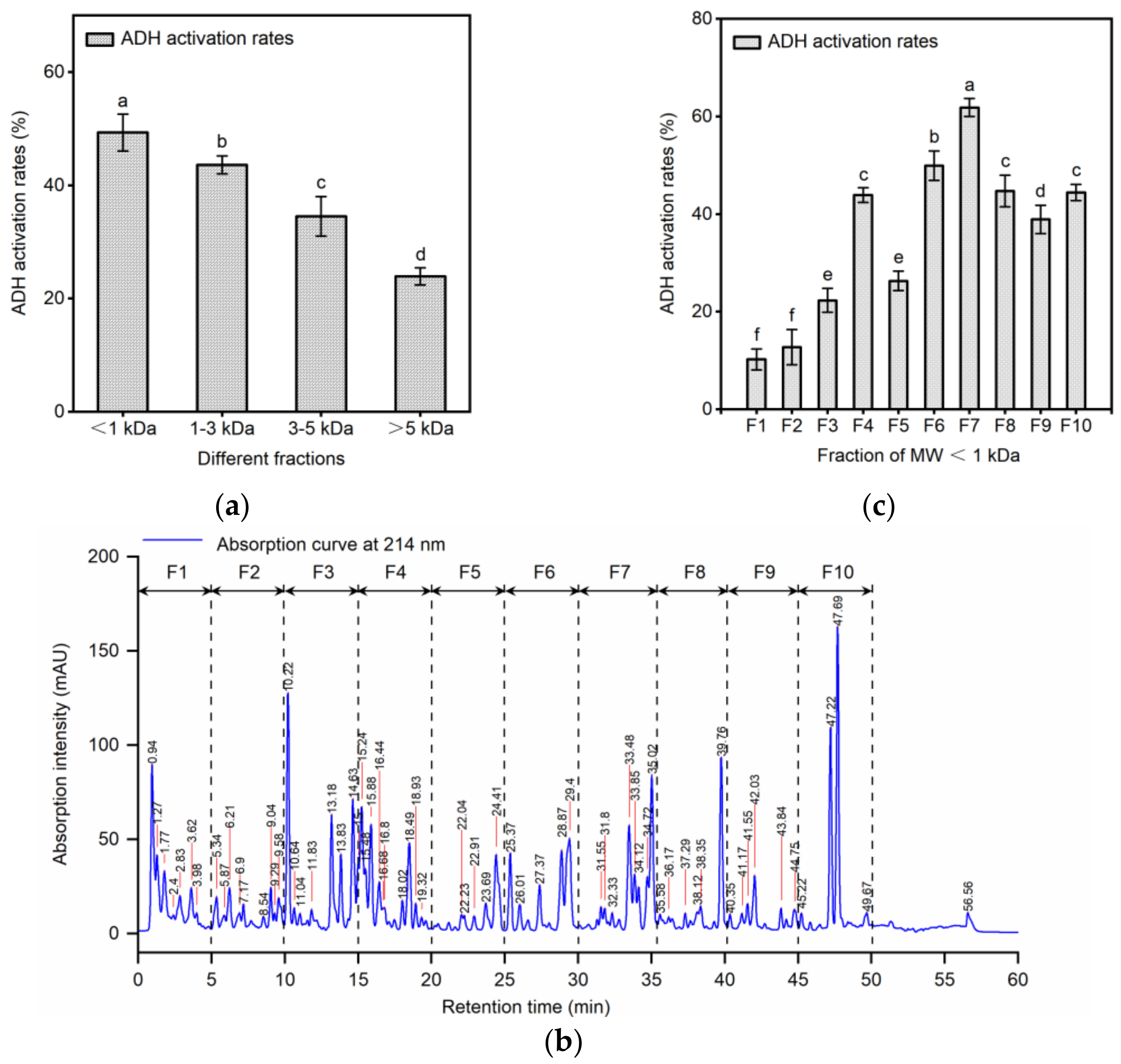
2.2. Separation of ADH Activating Peptides by Ultrafiltration and RP-HPLC
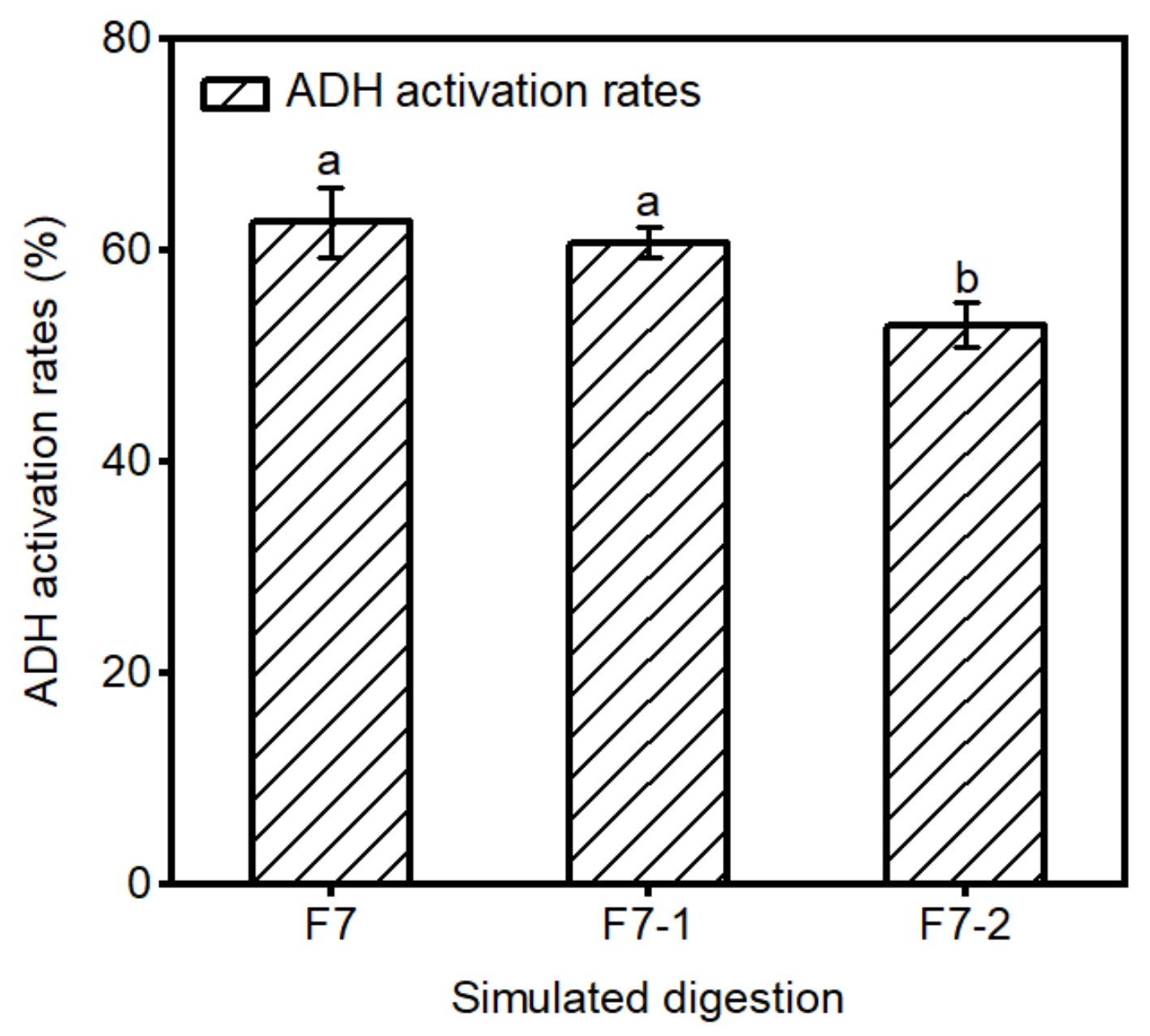
2.3. Stability of F7 in Gastrointestinal Digestion
2.4. Identification of Peptides Absorbed by Caco-2 Cells and Molecular Docking
2.5. In Vitro Activity Verification of Peptides
3. Materials and Methods
3.1. Materials and Chemicals
3.2. Removal of Starch from CGM
3.3. Preparation of Maize Protein Hydrolysates with ADH-Activating Activity
3.4. Separation and Purification of ADH-Activating Peptides
3.4.1. Ultrafiltration
3.4.2. High-Performance Liquid Chromatography in Reverse Phase (RP-HPLC)
3.5. Determination of ADH Activation Activity In Vitro
3.6. Simulated Gastrointestinal Digestion In Vitro
3.7. Transport Experiment
3.7.1. Cell Culture
3.7.2. Transport Experiments
3.8. Peptide Identification
3.9. Molecular Docking
3.10. Synthesis of Peptides
3.11. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- National Bureau of Statistics of China. Grain Yield Statistics Reports for the China. 2023 [EB/OL]. Available online: https://www.stats.gov.cn/sj/zxfb/202312/t20231211_1945417.html (accessed on 25 December 2023).
- Ranum, P.; Pena-Rosas, J.P.; Garcia-Casal, M.N. Global maize production, utilization, and consumption. Ann. N. Y. Acad. Sci. 2014, 1312, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Li, X.L.; Wu, K.N.; Hao, S.H.; Yue, Z.; Ran, Z.; Ma, J.L. Mapping cropland suitability in China using optimized MaxEnt model. Field Crop. Res. 2023, 302, 109064. [Google Scholar] [CrossRef]
- Silva, W.R.; Carvalho, F.R.; Silva, R.B.; Pereira, R.A.N.; Avila, C.L.S.; Devries, T.J.; Pereira, M.N. Fibrous coproducts of corn and citrus as forage and concentrate sources for dairy cows. J. Dairy Sci. 2022, 105, 8099–8114. [Google Scholar] [CrossRef]
- Lobos, N.E.; Wattiaux, M.A.; Broderick, G.A. Effect of rumen-protected lysine supplementation of diets based on corn protein fed to lactating dairy cows. J. Dairy Sci. 2021, 104, 6620–6632. [Google Scholar] [CrossRef]
- Landry, J.; Delhaye, S.; Di Gioia, L. Protein distribution in gluten products isolated during and after wet-milling of maize grains. Cereal Chem. 1999, 76, 503–505. [Google Scholar] [CrossRef]
- Cederbaum, A.I. Alcohol Metabolism. Clin. Liver Dis. 2012, 16, 667–685. [Google Scholar] [CrossRef] [PubMed]
- Jelski, W.; Szmitkowski, M. Alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) in the cancer diseases. Clin. Chim. Acta 2008, 395, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Hyun, J.; Han, J.; Lee, C.; Yoon, M.; Jung, Y. Pathophysiological Aspects of Alcohol Metabolism in the Liver. Int. J. Mol. Sci. 2021, 22, 5717. [Google Scholar] [CrossRef] [PubMed]
- Plapp, B.V.; Charlier, H.A.; Ramaswamy, S. Mechanistic implications from structures of yeast alcohol dehydrogenase complexed with coenzyme and an alcohol. Arch. Biochem. Biophys. 2016, 591, 35–42. [Google Scholar] [CrossRef]
- WHO. Global Status Report on Alcohol and Health. 2018. Available online: https://www.who.int/substance_abuse/publications/global_alcohol_report/en/ (accessed on 23 December 2023).
- Xiao, J.; Wang, F.; Wong, N.-K.; He, J.; Zhang, R.; Sun, R.; Xu, Y.; Liu, Y.; Li, W.; Koike, K.; et al. Global liver disease burdens and research trends: Analysis from a Chinese perspective. J. Hepatol. 2019, 71, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yu, Z.; Zhao, W. Identification and action mechanism of novel antioxidative peptides from copra meal protein. LWT—Food Sci. Technol. 2023, 188, 115425. [Google Scholar] [CrossRef]
- Iram, D.; Sansi, M.S.; Zanab, S.; Vij, S.; Ashutosh; Meena, S. In silico identification of antidiabetic and hypotensive potential bioactive peptides from the sheep milk proteins—A molecular docking study. J. Food Biochem. 2022, 46, e14137. [Google Scholar] [CrossRef]
- Li, G.; Liu, X.; Miao, Z.; Hu, N.; Zheng, X. Preparation of corn peptides with anti-adhesive activity and its functionality to alleviate gastric injury induced by helicobacter pylori infection in vivo. Nutrients 2023, 15, 3467. [Google Scholar] [CrossRef]
- Wang, Z.; Xing, L.; Cai, J.; Toldrá, F.; Hao, Y.; Zhang, W. Identification of hepatoprotective peptides from porcine liver and its interaction with ethanol metabolizing enzymes in vitro. Food Biosci. 2023, 55, 115425. [Google Scholar] [CrossRef]
- Wang, X.J.; Liu, X.L.; Zheng, X.Q.; Qu, Y.; Shi, Y.G. Preparation of corn glycopeptides and evaluation of their antagonistic effects on alcohol-induced liver injury in rats. J. Funct. Foods 2020, 66, 103776. [Google Scholar] [CrossRef]
- Ma, Z.L.; Hou, T.; Shi, W.; Liu, W.W.; Ibrahim, S.A.; He, H. Purification and identification of corn peptides that facilitate alcohol metabolism by semi-preparative high-performance liquid chromatography and nano liquid chromatography with electrospray ionization tandem mass spectrometry. J. Sep. Sci. 2016, 39, 4234–4242. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Li, S.; Song, C.; Sun, X.; Liu, X. Black soybean-derived peptides exerted protective effect against alcohol-induced liver injury in mice. J. Funct. Foods 2021, 87, 104828. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, C.; Qin, X.; Cao, W.; Chen, J.; Li, Y.; Zheng, H.; Lin, H.; Chen, Z. Hepatoprotective effect of clam (Corbicula fluminea) protein hydrolysate on alcohol-induced liver injury in mice and partial identification of a hepatoprotective peptide from the hydrolysate. Food Sci. Technol. 2022, 42, e61522. [Google Scholar] [CrossRef]
- Zan, R.; Zhu, L.; Wu, G.; Zhang, H. Identification of Novel Peptides with Alcohol Dehydrogenase (ADH) Activating Ability in Chickpea Protein Hydrolysates. Foods 2023, 12, 1574. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.Q.; Zhou, F.B.; Zheng, L.; Cai, Y.J.; Su, G.W.; Luo, D.H.; Zhao, M.M. Chicken breast-derived alcohol dehydrogenase-activating peptides in response to physicochemical changes and digestion simulation: The vital role of hydrophobicity. Food Res. Int. 2020, 136, 109592. [Google Scholar] [CrossRef] [PubMed]
- Bröer, S. Intestinal Amino Acid Transport and Metabolic Health. Annu. Rev. Nutr. 2023, 43, 73–99. [Google Scholar] [CrossRef]
- Zheng, X.Q.; Liu, X.L.; Yu, S.F.; Wang, X.J.; Ma, Y.Q.; Yang, S.; Jing, S.S. Effects of Extrusion and Starch Removal Pretreatment on Zein Proteins Extracted from Corn Gluten Meal. Cereal. Chem. 2014, 91, 496–501. [Google Scholar] [CrossRef]
- Giansanti, P.; Tsiatsiani, L.; Low, T.Y.; Heck, A.J.R. Six alternative proteases for mass spectrometry-based proteomics beyond trypsin. Nat. Protoc. 2016, 11, 993–1006. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Zhao, M.; Zhou, F.; Gallego, M.; Gao, J.; Toldrá, F.; Mora, L. Isolation and identification of alcohol dehydrogenase stabilizing peptides from Alcalase digested chicken breast hydrolysates. J. Funct. Foods 2020, 64, 103617. [Google Scholar] [CrossRef]
- Zheng, X.Q.; Wang, J.T.; Liu, X.L.; Sun, Y.; Zheng, Y.J.; Wang, X.J.; Liu, Y. Effect of hydrolysis time on the physicochemical and functional properties of corn glutelin by Protamex hydrolysis. Food Chem. 2015, 172, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Sonklin, C.; Laohakunjit, N.; Kerdchoechuen, O. Assessment of antioxidant properties of membrane ultrafiltration peptides from mungbean meal protein hydrolysates. Peerj 2018, 6, e5337. [Google Scholar] [CrossRef]
- Gao, S.; Shi, J.; Wang, K.; Tan, Y.; Hong, H.; Luo, Y. Protective effects of oyster protein hydrolysates on alcohol-induced liver disease (ALD) in mice: Based on the mechanism of anti-oxidative metabolism. Food Funct. 2022, 13, 8411–8424. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Yu, F.; Wu, Y.; Dai, L.; Feng, Y.; Chen, S.; Wang, G.; Ma, H.; Li, X.; Dai, C. Identification of a novel peptide that activates alcohol dehydrogenase from crucian carp swim bladder and how it protects against acute alcohol-induced liver injury in mice. J. Pharmaceut. Biomed. 2022, 207, 114426. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.-J.; Huo, C.-Y.; Qian, Y.; Ren, D.-F.; Lu, J. Ultra-high-pressure processing improves proteolysis and release of bioactive peptides with activation activities on alcohol metabolic enzymes in vitro from mushroom foot protein. Food Chem. 2017, 231, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Acquah, C.; Chan, Y.W.; Pan, S.; Agyei, D.; Udenigwe, C.C. Structure-informed separation of bioactive peptides. J. Food Biochem. 2019, 43, e12765. [Google Scholar] [CrossRef]
- Ahmed, T.; Sun, X.; Udenigwe, C.C. Role of structural properties of bioactive peptides in their stability during simulated gastrointestinal digestion: A systematic review. Trends Food Sci. Technol. 2022, 120, 265–273. [Google Scholar] [CrossRef]
- Huang, P.; Zhao, W.; Cai, L.; Liu, Y.; Wu, J.; Cui, C. Enhancement of functional properties, digestive properties, and in vitro digestion product physiological activity of extruded corn gluten meal by enzymatic modification. J. Sci. Food Agric. 2024, 104, 3477–3486. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Liu, X.; Miao, Z.; Zheng, X. Purification and structural characterization of three novel anti-adhesive peptides against Helicobacter pylori from corn gluten meal. J. Funct. Foods 2024, 112, 105992. [Google Scholar] [CrossRef]
- Marchiando, A.M.; Graham, W.V.; Turner, J.R. Epithelial Barriers in Homeostasis and Disease. Annu. Rev. Pathol.-Mech. 2010, 5, 119–144. [Google Scholar] [CrossRef] [PubMed]
- Iftikhar, M.; Iftikhar, A.; Zhang, H.; Gong, L.; Wang, J. Transport, metabolism and remedial potential of functional food extracts (FFEs) in Caco-2 cells monolayer: A review. Food Res. Int. 2020, 136, 109240. [Google Scholar] [CrossRef] [PubMed]
- Stanzione, F.; Giangreco, I.; Cole, J.C. Use of molecular docking computational tools in drug discovery. Prog. Med. Chem. 2021, 60, 273–343. [Google Scholar]
- Igbokwe, C.J.; Feng, Y.; Louis, H.; Benjamin, I.; Quaisie, J.; Duan, Y.; Tuly, J.A.; Cai, M.; Zhang, H. Novel antioxidant peptides identified from coix seed by molecular docking, quantum chemical calculations and invitro study in HepG2 cells. Food Chem. 2024, 440, 138234. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Toldrá, F.; Zhou, F.; Mora, L.; Luo, L.; Zheng, L.; Luo, D.; Zhao, M. Chicken-derived tripeptide KPC (Lys-Pro-Cys) stabilizes alcohol dehydrogenase (ADH) through peptide-enzyme interaction. LWT 2022, 161, 113376. [Google Scholar] [CrossRef]


| No. | Peptide Sequence | Measured m/z (Da) | Charge | Calculated MW (Da) | Binding Energy (kcal/moL) |
|---|---|---|---|---|---|
| 1 | CENPILQ | 408.6982 | 2 | 815.3847 | −5.34 |
| 2 | TGCPVLQ | 717.3574 | 1 | 716.3527 | −8.20 |
| 3 | EVFEPF | 767.3594 | 1 | 766.3538 | 0.95 |
| 4 | SPFLGQ | 648.3331 | 1 | 647.3279 | −3.67 |
| 5 | SSNCQPF | 782.3066 | 1 | 781.3065 | −8.69 |
| 6 | DTPYSEF | 429.6775 | 2 | 857.3443 | −2.21 |
| 7 | EVGDGVFE | 426.1901 | 2 | 850.3709 | 1.26 |
| 8 | TPYSEF | 372.1648 | 2 | 742.3174 | −3.42 |
| 9 | FTPVLQ | 704.3978 | 1 | 703.3905 | −3.52 |
| 10 | SVCENPAL | 416.6947 | 2 | 831.3797 | 0.85 |
| 11 | IFPQC | 304.1461 | 2 | 606.2836 | −5.65 |
| 12 | TIFPQ | 303.1659 | 2 | 604.3221 | −3.49 |
| 13 | QPQQPW | 783.3767 | 1 | 782.3711 | −7.31 |
| Sequence | Binding Energy (kcal/moL) | Hydrogen Bonds | Hydrophobic Interaction | Salt Bridge | π-Stacking | π-Cation Interaction |
|---|---|---|---|---|---|---|
| SSNCQPF | −8.69 | His 44 (3.55 Å), Gly 181 (3.56 Å), Leu 182 (3.11 Å), Gly 183 (3.63 Å), Val 245 (3.20 Å), Ser 246 (2.96 Å), Val 247 (3.56 Å, 3.72 Å), Met 270 (3.82 Å), Glu 333 (3.12 Å), Gly 339 (4.04 Å), Arg 340 (3.64 Å) | Phe 221 (3.62 Å, 3.90 Å), Val 247 (3.52 Å), Ala 251 (3.67 Å) | - | Phe 221 (3.78 Å) | - |
| QPQQPW | −7.31 | His 44 (3.98 Å, 3.47 Å), Asp 53 (3.13 Å), Gly 181 (3.15 Å), Asp 201 (3.89 Å), Lys 206 (3.06 Å), Glu 333 (2.99 Å, 3.42 Å), Gly 335 (3.79 Å), Arg 340 (3.87 Å) | Leu 182 (3.84 Å, 3.58 Å, 3.58 Å), Phe 221 (3.27 Å), Val 247 (3.67 Å), Ile 337 (3.69 Å) | His 44 (4.31 Å, 5.14 Å) | - | His 44 (5.97 Å) |
| TGCPVLQ | −8.20 | His 44 (2.80 Å, 3.15 Å), Thr 45 (3.64 Å), His 66 (3.45 Å), Thr 157 (2.72 Å), Gly 181 (3.85 Å), Asp 201 (3.09 Å), Lys 206 (4.06 Å), Val 245 (2.79 Å), Gly 335 (4.06 Å), Arg 340 (3.92 Å) | Thr 45 (3.89 Å), Trp 54 (3.61 Å), Trp 92 (3.69 Å), Leu 182 (3.78 Å), Phe 221 (3.57 Å), Val 245 (3.93 Å) | Lys 206 (5.09 Å) | - | - |
| Sequence | ADH Activation EC50 (mM) |
|---|---|
| SSNCQPF | 1.35 ± 0.22 b |
| TGCPVLQ | 2.26 ± 0.16 a |
| QPQQPW | 2.73 ± 0.13 a |
| Enzyme | pH | Temperature (°C) |
|---|---|---|
| Alcalase | 8.5 | 60 |
| Neutral | 7.0 | 45 |
| Flavourzyme | 7.5 | 50 |
| Protamex | 7.0 | 55 |
| Papain | 8.0 | 50 |
| Trypsin | 8.0 | 37 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Li, G.; Liu, X. Identification of Corn Peptides with Alcohol Dehydrogenase Activating Activity Absorbed by Caco-2 Cell Monolayers. Molecules 2024, 29, 1523. https://doi.org/10.3390/molecules29071523
Wang Z, Li G, Liu X. Identification of Corn Peptides with Alcohol Dehydrogenase Activating Activity Absorbed by Caco-2 Cell Monolayers. Molecules. 2024; 29(7):1523. https://doi.org/10.3390/molecules29071523
Chicago/Turabian StyleWang, Zhe, Guanlong Li, and Xiaolan Liu. 2024. "Identification of Corn Peptides with Alcohol Dehydrogenase Activating Activity Absorbed by Caco-2 Cell Monolayers" Molecules 29, no. 7: 1523. https://doi.org/10.3390/molecules29071523
APA StyleWang, Z., Li, G., & Liu, X. (2024). Identification of Corn Peptides with Alcohol Dehydrogenase Activating Activity Absorbed by Caco-2 Cell Monolayers. Molecules, 29(7), 1523. https://doi.org/10.3390/molecules29071523