The Effect of Salvianolic Acid A on Tumor-Associated Macrophage Polarization and Its Mechanisms in the Tumor Microenvironment of Triple-Negative Breast Cancer
Abstract
1. Introduction
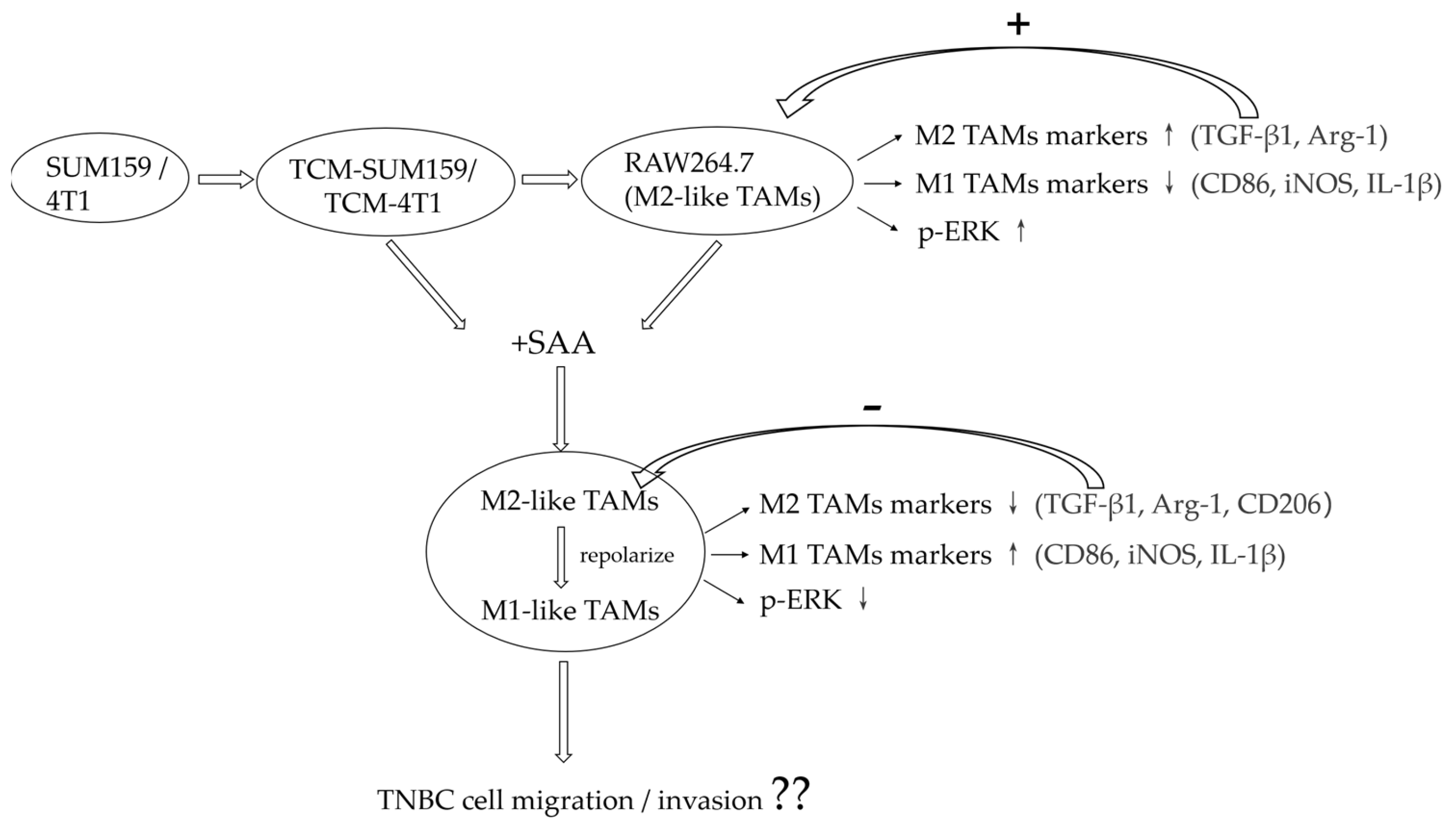
2. Results
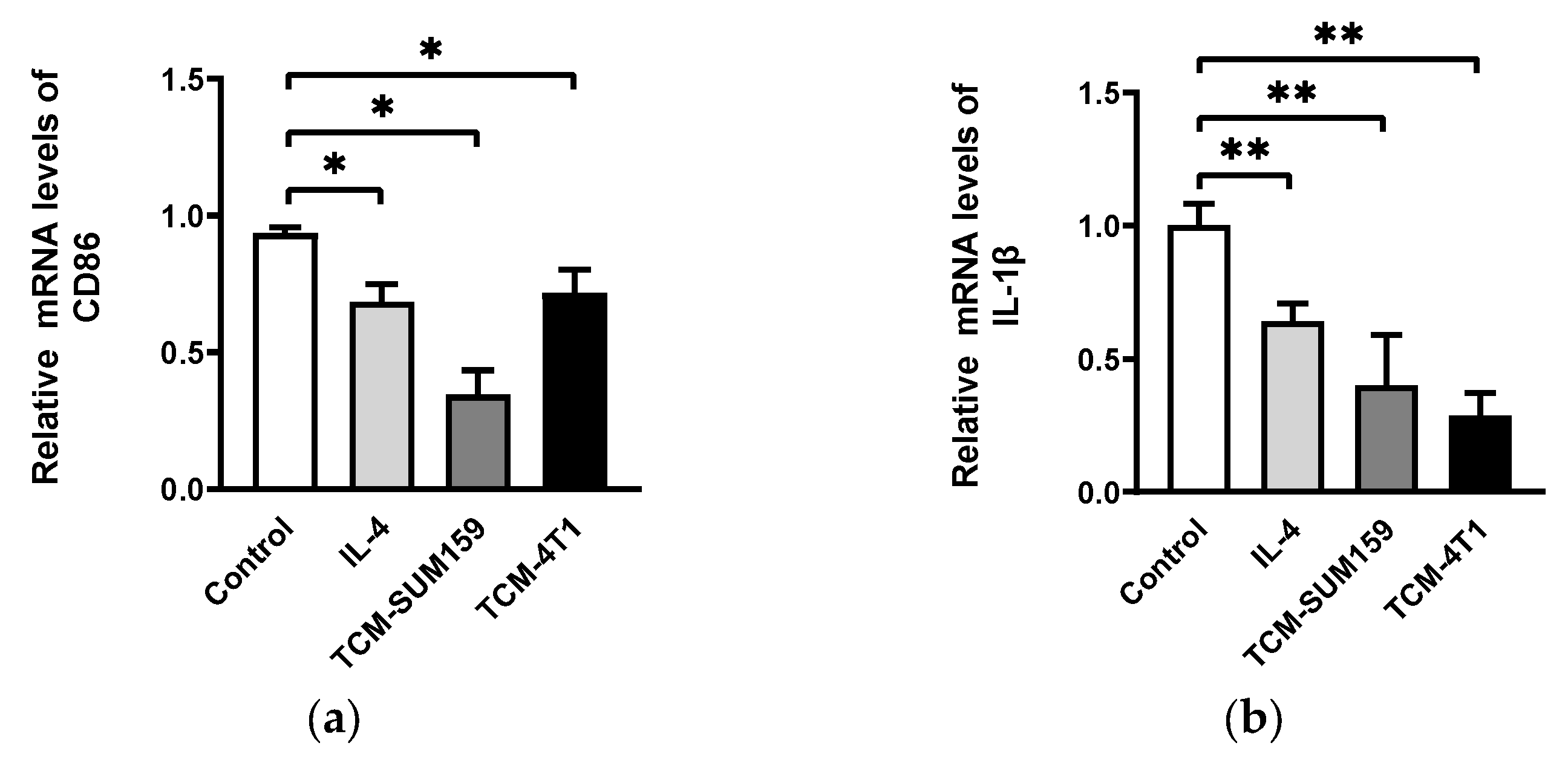
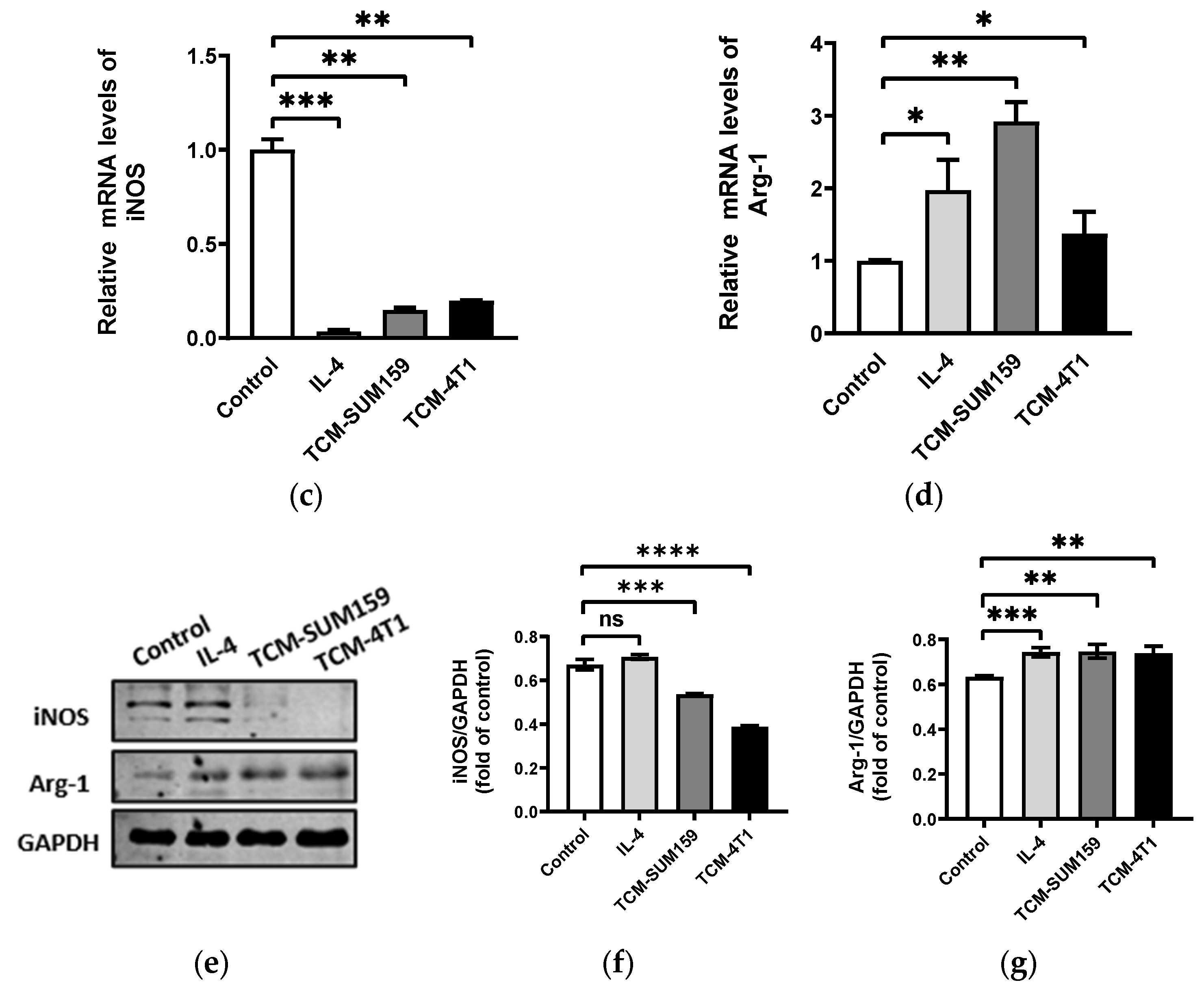
2.1. TNBC Cell-Conditioned Medium Induced the Polarization of M2-like TAMs
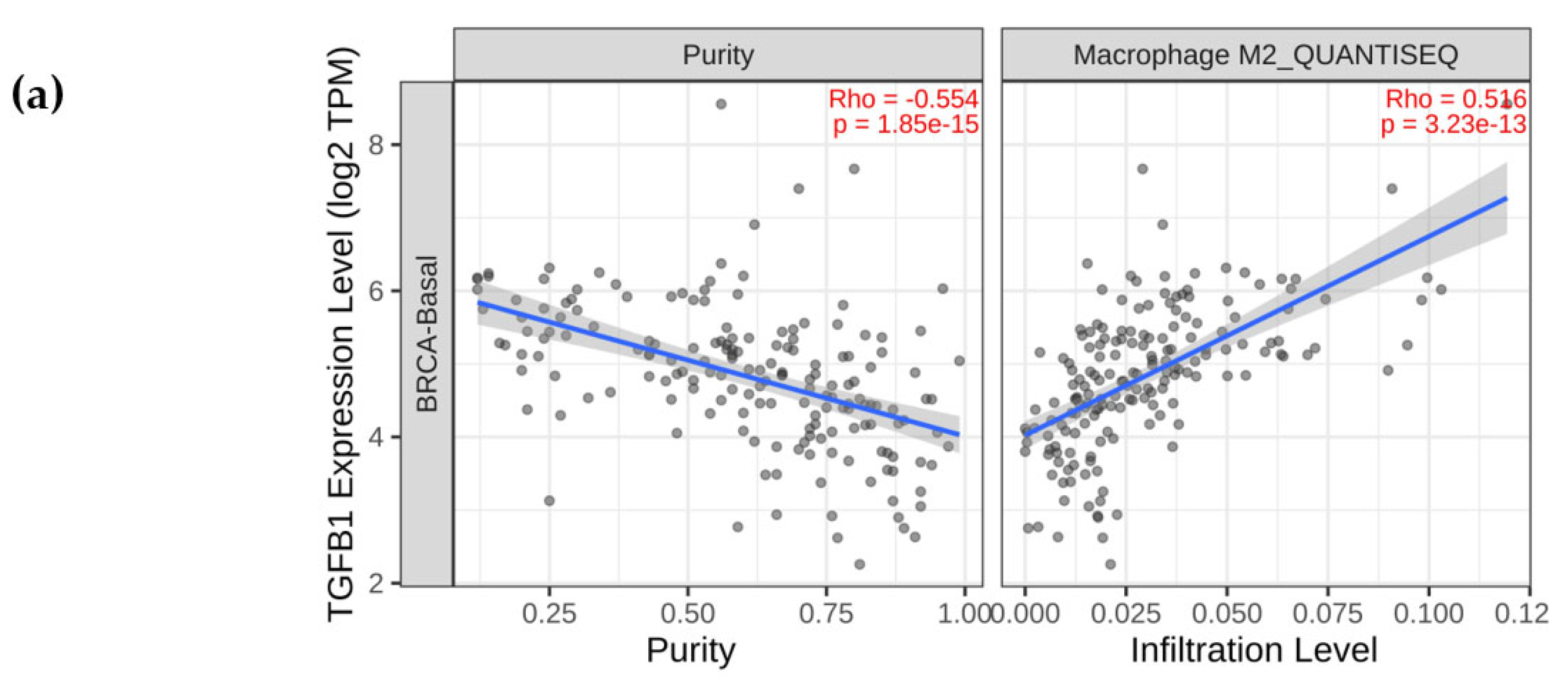
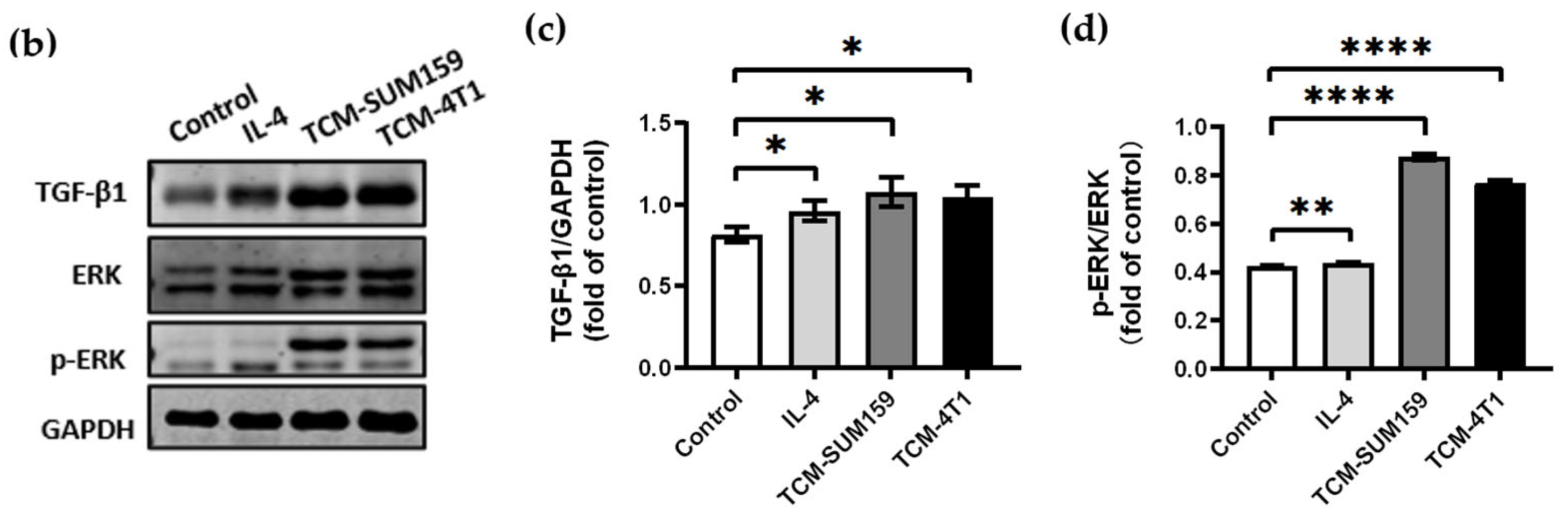
2.2. TGF-β1 Expression Was Upregulated and the ERK Signalling Pathway Was Activated in Macrophages Co-Cultured with TCM-TNBC
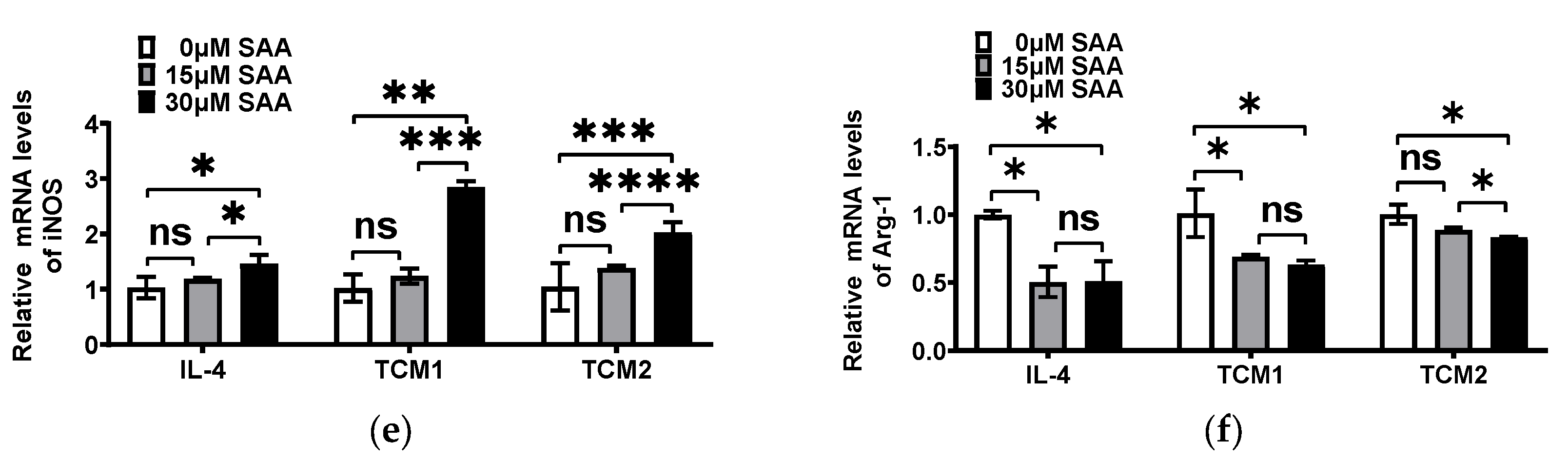
2.3. SAA Mediated the mRNA Expression of Cytokines in M2-like TAMs Induced by the TNBC-Cell-Conditioned Medium
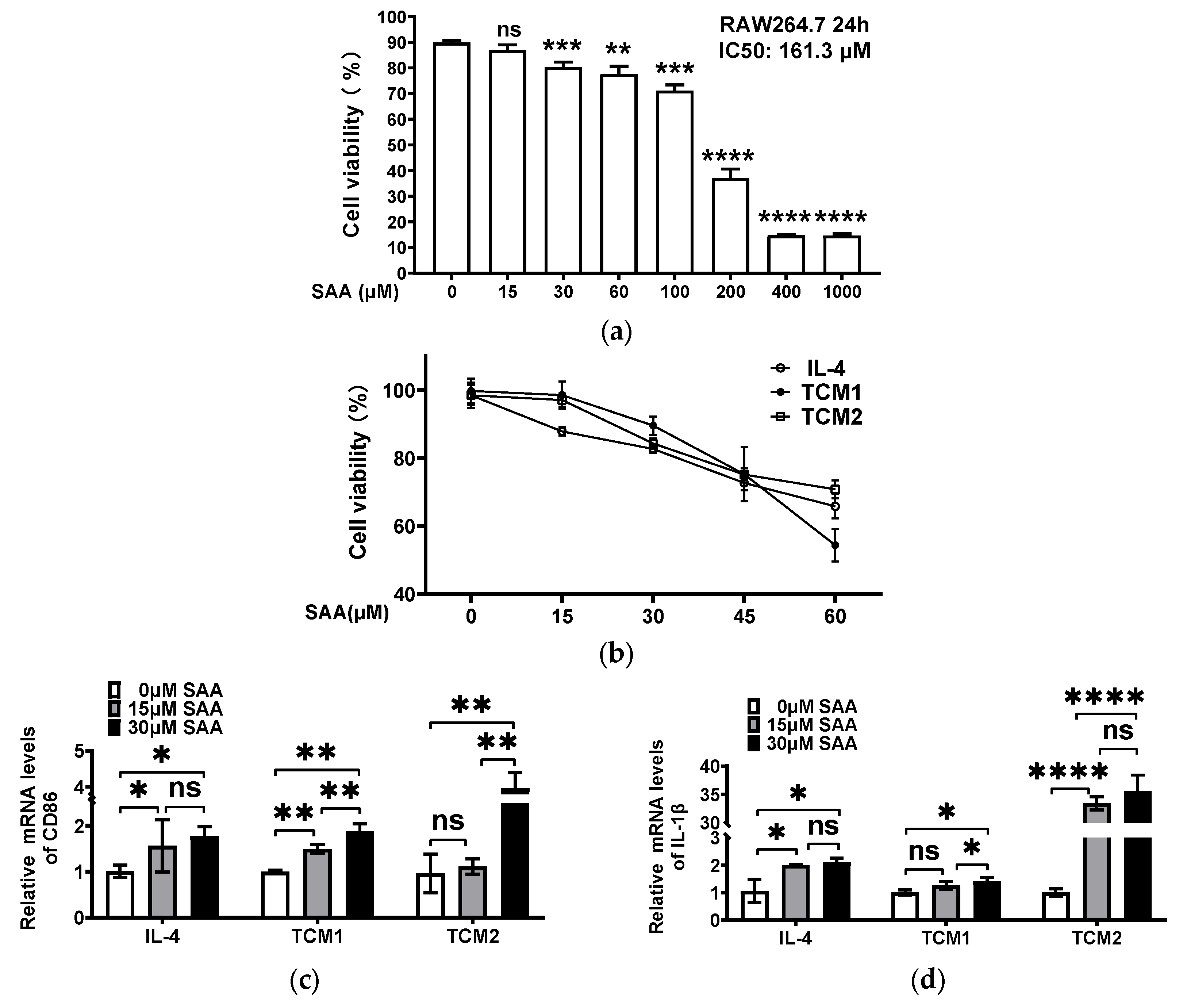
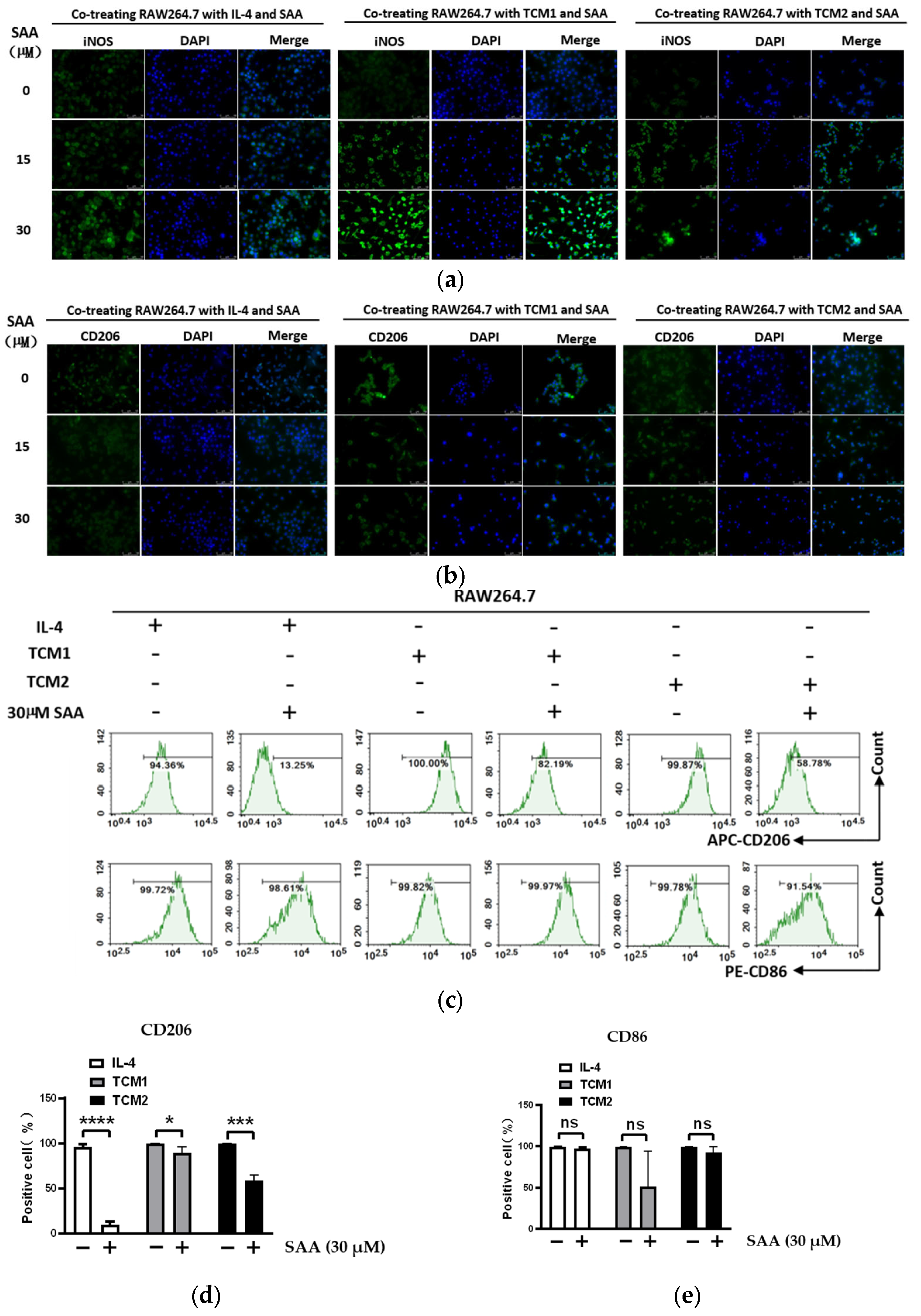
2.4. SAA Inhibited the Progression of RAW264.7 Cells Polarization towards M2-like TAMs
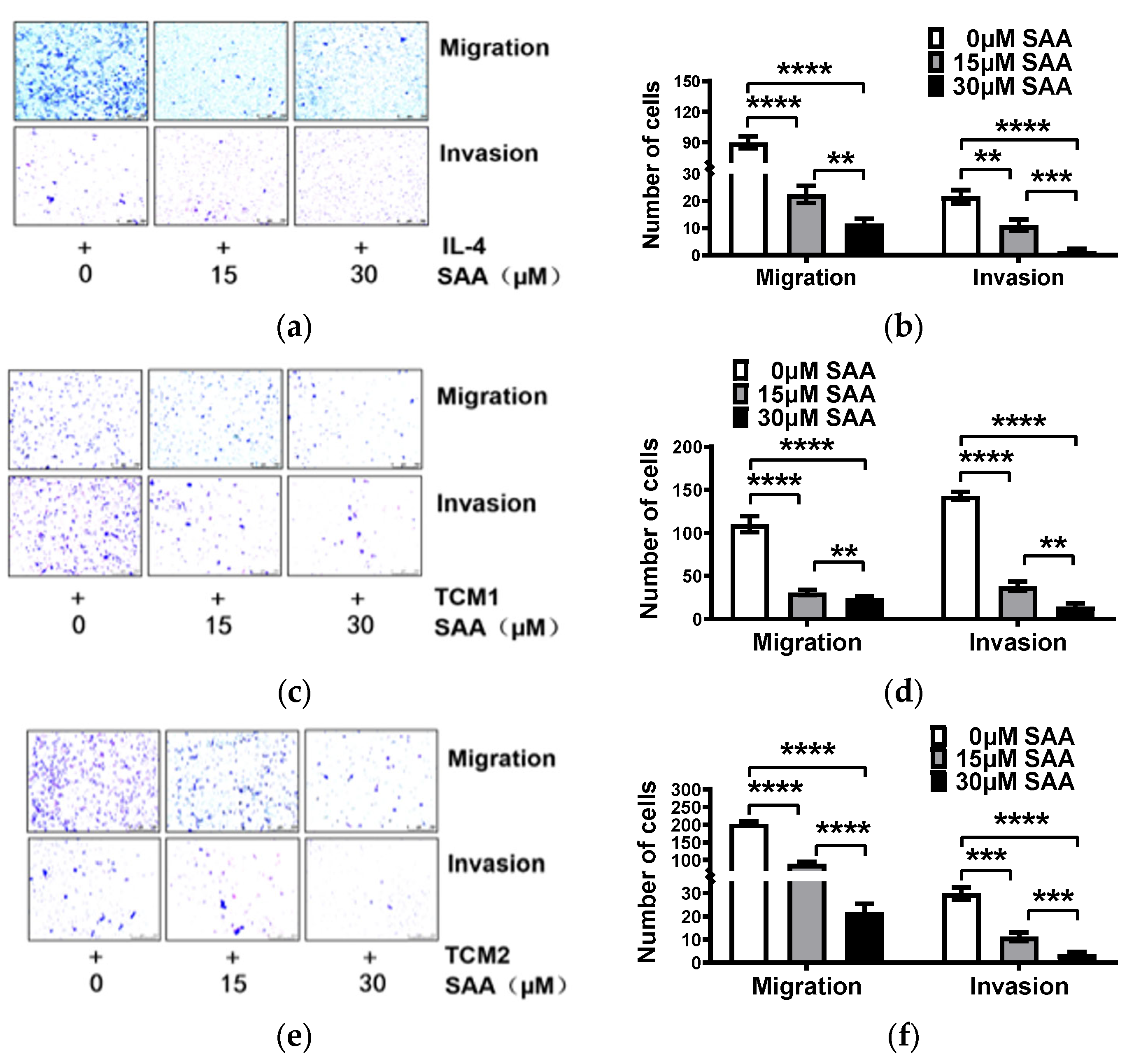
2.5. SAA Inhibited the Migration and Invasion of M2-like TAMs Induced by the TCM-TNBC
2.6. SAA Inhibited TGF-β1 Expression and ERK Signalling in M2-like TAMs
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Lines and Culture
4.3. Preparation of Tumor-Conditioned Medium (TCM)
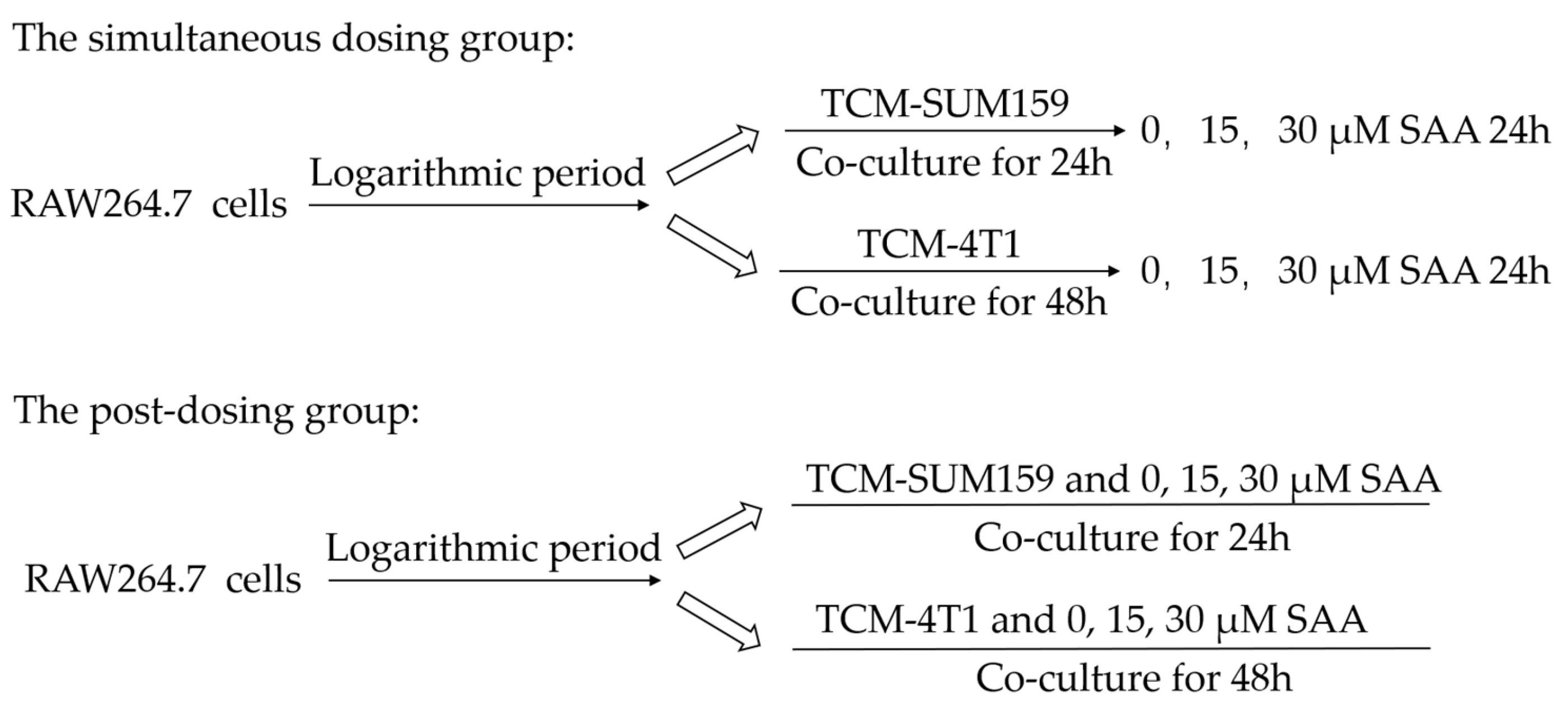
4.4. Polarization of TAMs
4.5. Cell Viability Assay
4.6. RNA Extraction and qRT-PCR
4.7. Enzyme-Linked Immunosorbent Assay (ELISA)
4.8. Immunofluorescence Staining
4.9. Flow Cytometry
4.10. Transwell Assay
4.11. Western Blot
4.12. Bioinformatics Analysis
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TME | tumor microenvironment |
| TAM | tumor-associated macrophage |
| TNBC-TCM | triple-negative-breast-cancer-cell-conditioned medium |
| SAA | salvianolic acid A |
| TCM1 | SUM159-conditioned medium |
| TCM2 | 4T1-conditioned medium |
References
- Pedersen, R.N.; Esen, B.; Mellemkjær, L.; Christiansen, P.; Ejlertsen, B.; Lash, T.L.; Nørgaard, M.; Cronin-Fenton, D. The Incidence of Breast Cancer Recurrence 10-32 Years After Primary Diagnosis. JNCI J. Natl. Cancer Inst. 2022, 114, 391–399. [Google Scholar] [CrossRef]
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Thakur, K.K.; Bordoloi, D.; Kunnumakkara, A.B. Alarming Burden of Triple-Negative Breast Cancer in India. Clin. Breast Cancer 2018, 18, e393–e399. [Google Scholar] [CrossRef]
- Kim, J.; Bae, J.-S. Tumor-Associated Macrophages and Neutrophils in Tumor Microenvironment. Mediat. Inflamm. 2016, 2016, 6058147. [Google Scholar] [CrossRef]
- Pittet, M.J.; Michielin, O.; Migliorini, D. Clinical relevance of tumour-associated macrophages. Nat. Rev. Clin. Oncol. 2022, 19, 402–421. [Google Scholar] [CrossRef]
- Ojalvo, L.S.; Whittaker, C.A.; Condeelis, J.S.; Pollard, J.W. Gene expression analysis of macrophages that facilitate tumor invasion supports a role for Wnt-signaling in mediating their activity in primary mammary tumors. J. Immunol. 2010, 184, 702–712. [Google Scholar] [CrossRef]
- Kowal, J.; Kornete, M.; A Joyce, J. Re-education of macrophages as a therapeutic strategy in cancer. Immunotherapy 2019, 11, 677–689. [Google Scholar] [CrossRef]
- Lian-Niang, L.; Rui, T.; Wei-Ming, C. Salvianolic Acid A, a New Depside from Roots of Salvia miltiorrhiza. Planta Med. 1984, 50, 227–228. [Google Scholar] [CrossRef]
- Qin, T.; Rasul, A.; Sarfraz, A.; Sarfraz, I.; Hussain, G.; Anwar, H.; Riaz, A.; Liu, S.; Wei, W.; Li, J.; et al. Salvianolic acid A & B: Potential cytotoxic polyphenols in battle against cancer via targeting multiple signaling pathways. Int. J. Biol. Sci. 2019, 15, 2256–2264. [Google Scholar] [CrossRef]
- Martinez, F.O.; Helming, L.; Gordon, S. Alternative activation of macrophages: An immunologic functional perspective. Annu. Rev. Immunol. 2009, 27, 451–483. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, X.; Wan, C.; Liu, Y.; Wang, Y.; Meng, C.; Zhang, Y.; Jiang, C. NLRP3 inflammasome mediates M1 macrophage polarization and IL-1β production in inflammatory root resorption. J. Clin. Periodontol. 2020, 47, 451–460. [Google Scholar] [CrossRef]
- Wisitpongpun, P.; Potup, P.; Usuwanthim, K. Oleamide-Mediated Polarization of M1 Macrophages and IL-1β Production by Regulating NLRP3-Inflammasome Activation in Primary Human Monocyte-Derived Macrophages. Front. Immunol. 2022, 13, 856296. [Google Scholar] [CrossRef]
- Sica, A.; Bronte, V. Altered macrophage differentiation and immune dysfunction in tumor development. J. Clin. Investig. 2007, 117, 1155–1166. [Google Scholar] [CrossRef]
- Fuxe, J.; Karlsson, M.C. TGF-β-induced epithelial-mesenchymal transition: A link between cancer and inflammation. Semin. Cancer Biol. 2012, 22, 455–461. [Google Scholar] [CrossRef]
- Huang, X.; Li, Y.; Fu, M.; Xin, H.B. Polarizing Macrophages In Vitro. Methods Mol. Biol. 2018, 1784, 119–126. [Google Scholar]
- Dong, P.; Ma, L.; Liu, L.; Zhao, G.; Zhang, S.; Dong, L.; Xue, R.; Chen, S. CD86+/CD206+, Diametrically Polarized Tumor-Associated Macrophages, Predict Hepatocellular Carcinoma Patient Prognosis. Int. J. Mol. Sci. 2016, 17, 320. [Google Scholar] [CrossRef]
- Sun, D.; Luo, T.; Dong, P.; Zhang, N.; Chen, J.; Zhang, S.; Liu, L.; Dong, L.; Zhang, S. CD86(+)/CD206(+) tumor-associated macrophages predict prognosis of patients with intrahepatic cholangiocarcinoma. PeerJ 2020, 8, e8458. [Google Scholar] [CrossRef]
- Kinouchi, M.; Miura, K.; Mizoi, T.; Ishida, K.; Fujibuchi, W.; Ando, T.; Yazaki, N.; Saito, K.; Shiiba, K.-I.; Sasaki, I. Infiltration of CD14-positive macrophages at the invasive front indicates a favorable prognosis in colorectal cancer patients with lymph node metastasis. Hepatogastroenterology 2011, 58, 352–358. [Google Scholar]
- Beider, K.; Bitner, H.; Leiba, M.; Gutwein, O.; Koren-Michowitz, M.; Ostrovsky, O.; Abraham, M.; Wald, H.; Galun, E.; Peled, A.; et al. Multiple myeloma cells recruit tumor-supportive macrophages through the CXCR4/CXCL12 axis and promote their polarization toward the M2 phenotype. Oncotarget 2014, 5, 11283–11296. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, X.; Shen, Z.; Xu, J.; Qin, J.; Sun, Y. Infiltration of diametrically polarized macrophages predicts overall survival of patients with gastric cancer after surgical resection. Gastric Cancer 2015, 18, 740–750. [Google Scholar] [CrossRef]
- Hu, W.; Qian, Y.; Yu, F.; Liu, W.; Wu, Y.; Fang, X.; Hao, W. Alternatively activated macrophages are associated with metastasis and poor prognosis in prostate adenocarcinoma. Oncol. Lett. 2015, 10, 1390–1396. [Google Scholar] [CrossRef]
- van Dalen, F.J.; Van Stevendaal, M.H.; Fennemann, F.L.; Verdoes, M.; Ilina, O. Molecular Repolarisation of Tumour-Associated Macrophages. Molecules 2018, 24, 9. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003, 3, 23–35. [Google Scholar] [CrossRef]
- Martinez, F.O.; Sica, A.; Mantovani, A.; Locati, M. Macrophage activation and polarization. Front. Biosci. 2008, 13, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cao, J.; Ma, S.; Dong, R.; Meng, W.; Ying, M.; Weng, Q.; Chen, Z.; Ma, J.; Fang, Q.; et al. Tumor hypoxia enhances Non-Small Cell Lung Cancer metastasis by selectively promoting macrophage M2 polarization through the activation of ERK signaling. Oncotarget 2014, 5, 9664–9677. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Shi, W.; Xu, Y.; Xu, C.; Zhao, T.; Geng, B.; Yang, J.; Pan, J.; Hu, S.; Zhang, C.; et al. Tumor-derived lactate induces M2 macrophage polarization via the activation of the ERK/STAT3 signaling pathway in breast cancer. Cell Cycle 2018, 17, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Garg, K.; Pullen, N.A.; Oskeritzian, C.A.; Ryan, J.J.; Bowlin, G.L. Macrophage functional polarization (M1/M2) in response to varying fiber and pore dimensions of electrospun scaffolds. Biomaterials 2013, 34, 4439–4451. [Google Scholar] [CrossRef] [PubMed]
- Locati, M.; Curtale, G.; Mantovani, A. Diversity, Mechanisms, and Significance of Macrophage Plasticity. Annu. Rev. Pathol. Mech. Dis. 2020, 15, 123–147. [Google Scholar] [CrossRef]
- Ohno, S.; Inagawa, H.; Dhar, D.K.; Fujii, T.; Ueda, S.; Tachibana, M.; Suzuki, N.; Inoue, M.; Soma, G.-I.; Nagasue, N. The degree of macrophage infiltration into the cancer cell nest is a significant predictor of survival in gastric cancer patients. Anticancer Res. 2003, 23, 5015–5022. [Google Scholar]
- Ruffell, B.; Affara, N.I.; Coussens, L.M. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 2012, 33, 119–126. [Google Scholar] [CrossRef]
- DeNardo, D.G.; Ruffell, B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 2019, 19, 369–382. [Google Scholar] [CrossRef]
- Zhao, C.; Pang, X.; Yang, Z.; Wang, S.; Deng, H.; Chen, X. Nanomaterials targeting tumor associated macrophages for cancer immunotherapy. J. Control. Release 2022, 341, 272–284. [Google Scholar] [CrossRef]
- Pan, Y.; Yu, Y.; Wang, X.; Zhang, T. Tumor-Associated Macrophages in Tumor Immunity. Front. Immunol. 2020, 11, 583084. [Google Scholar] [CrossRef]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef]
- Russell, J.S.; Brown, J.M. The irradiated tumor microenvironment: Role of tumor-associated macrophages in vascular recovery. Front. Physiol. 2013, 4, 157. [Google Scholar] [CrossRef]
- Yuan, R.; Li, S.; Geng, H.; Wang, X.; Guan, Q.; Li, X.; Ren, C.; Yuan, X. Reversing the polarization of tumor-associated macrophages inhibits tumor metastasis. Int. Immunopharmacol. 2017, 49, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Kawanishi, N.; Yano, H.; Yokogawa, Y.; Suzuki, K. Exercise training inhibits inflammation in adipose tissue via both suppression of macrophage infiltration and acceleration of phenotypic switching from M1 to M2 macrophages in high-fat-diet-induced obese mice. Exerc. Immunol. Rev. 2010, 16, 105–118. [Google Scholar]
- Hartley, J.W.; Evans, L.H.; Green, K.Y.; Naghashfar, Z.; Macias, A.R.; Zerfas, P.M.; Ward, J.M. Expression of infectious murine leukemia viruses by RAW264.7 cells, a potential complication for studies with a widely used mouse macrophage cell line. Retrovirology 2008, 5, 1. [Google Scholar] [CrossRef]
- Mosser, D.M.; Zhang, X. Activation of murine macrophages. Curr. Protoc. Immunol. 2008, 14, 14.2.1–14.2.8. [Google Scholar] [CrossRef] [PubMed]
- Junttila, I.S.; Mizukami, K.; Dickensheets, H.; Meier-Schellersheim, M.; Yamane, H.; Donnelly, R.P.; Paul, W.E. Tuning sensitivity to IL-4 and IL-13: Differential expression of IL-4Ralpha, IL-13Ralpha1, and gammac regulates relative cytokine sensitivity. J. Exp. Med. 2008, 205, 2595–2608. [Google Scholar] [CrossRef]
- Wang, W.; Yuan, H.Y.; Liu, G.M.; Ni, W.H.; Wang, F.; Tai, G.X. Escherichia coli Maltose-Binding Protein Induces M1 Polarity of RAW264.7 Macrophage Cells via a TLR2- and TLR4-Dependent Manner. Int. J. Mol. Sci. 2015, 16, 9896–9909. [Google Scholar] [CrossRef]
- Wang, S.; Cao, M.; Xu, S.; Shi, J.; Mao, X.; Yao, X.; Liu, C. Luteolin Alters Macrophage Polarization to Inhibit Inflammation. Inflammation 2019, 43, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Dumitriu, I.E.; Dunbar, D.R.; Howie, S.E.; Sethi, T.; Gregory, C.D. Human dendritic cells produce TGF-beta 1 under the influence of lung carcinoma cells and prime the differentiation of CD4+CD25+Foxp3+ regulatory T cells. J. Immunol. 2009, 182, 2795–2807. [Google Scholar] [CrossRef] [PubMed]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E.; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef] [PubMed]
- Jing, Z.; Fei, W.; Zhou, J.; Zhang, L.; Chen, L.; Zhang, X.; Liang, X.; Xie, J.; Fang, Y.; Sui, X.; et al. Salvianolic acid B, a novel autophagy inducer, exerts antitumor activity as a single agent in colorectal cancer cells. Oncotarget 2016, 7, 61509–61519. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Guo, Y.; Gu, X. Salvianolic Acid B, a potential chemopreventive agent, for head and neck squamous cell cancer. J. Oncol. 2011, 2011, 534548. [Google Scholar] [CrossRef]
- Kan, S.; Cheung, W.M.; Zhou, Y.; Ho, W.S. Enhancement of Doxorubicin Cytotoxicity by Tanshinone IIA in HepG2 Human Hepatoma Cells. Planta Medica 2014, 80, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Guo, J.; Bao, J.; Lu, J.; Wang, Y. The anticancer properties of Salvia miltiorrhiza Bunge (Danshen): A systematic review. Med. Res. Rev. 2013, 34, 768–794. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, S.; Yang, Q.; Cai, J.; Zhang, W.; You, H.; Xing, J.; Dong, Y. Salvianolic acid A reverses the paclitaxel resistance and inhibits the migration and invasion abilities of human breast cancer cells by inactivating transgelin 2. Cancer Biol. Ther. 2015, 16, 1407–1414. [Google Scholar] [CrossRef]
- Chuang, C.-Y.; Ho, Y.-C.; Lin, C.-W.; Yang, W.-E.; Yu, Y.-L.; Tsai, M.-C.; Yang, S.-F.; Su, S.-C. Salvianolic acid A suppresses MMP-2 expression and restrains cancer cell invasion through ERK signaling in human nasopharyngeal carcinoma. J. Ethnopharmacol. 2020, 252, 112601. [Google Scholar] [CrossRef]
- Fang, C.; Wu, C.; Chen, P.; Chang, Y.; Chuang, C.; Lai, C.; Yang, S.; Tsai, L. Antimetastatic potentials of salvianolic acid A on oral squamous cell carcinoma by targeting MMP-2 and the c-Raf/MEK/ERK pathway. Environ. Toxicol. 2018, 33, 545–554. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, Y.Y.; Jiang, Q.; Li, K.R.; Zhao, Y.X.; Cao, C.; Yao, J. Salvianolic acid A protects RPE cells against oxidative stress through activation of Nrf2/HO-1 signaling. Free Radic. Biol. Med. 2014, 69, 219–228. [Google Scholar] [CrossRef]
- Li, L.; Yang, L.; Yang, S.; Wang, R.; Gao, H.; Lin, Z.; Zhao, Y.; Tang, W.; Han, R.; Wang, W.; et al. Andrographolide suppresses breast cancer progression by modulating tumor-associated macrophage polarization through the Wnt/β-catenin pathway. Phytother. Res. 2022, 36, 4587–4603. [Google Scholar] [CrossRef]
- Jiang, M.; Qi, Y.; Huang, W.; Lin, Y.; Li, B. Curcumin Reprograms TAMs from a Protumor Phenotype towards an Antitumor Phenotype via Inhibiting MAO-A/STAT6 Pathway. Cells 2022, 11, 3473. [Google Scholar] [CrossRef]
- Weigert, A.; Brüne, B. Nitric oxide, apoptosis and macrophage polarization during tumor progression. Nitric Oxide 2008, 19, 95–102. [Google Scholar] [CrossRef]
- Deswal, B.; Bagchi, U.; Kapoor, S. Curcumin Suppresses M2 Macrophage-derived Paclitaxel Chemoresistance through Inhibition of PI3K-AKT/STAT3 Signaling. Anti-Cancer Agents Med. Chem. 2024, 24, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Sun, X.; Sang, L.; Zhang, M.; Yan, X.; Ju, Q.; Ma, X.; Xu, M. Curcumin inhibits malignant behavior of colorectal cancer cells by regulating M2 polarization of tumor-associated macrophages and metastasis associated in colon cancer 1 (MACC1) expression. Chem. Biol. Drug Des. 2023, 102, 1202–1212. [Google Scholar] [CrossRef] [PubMed]

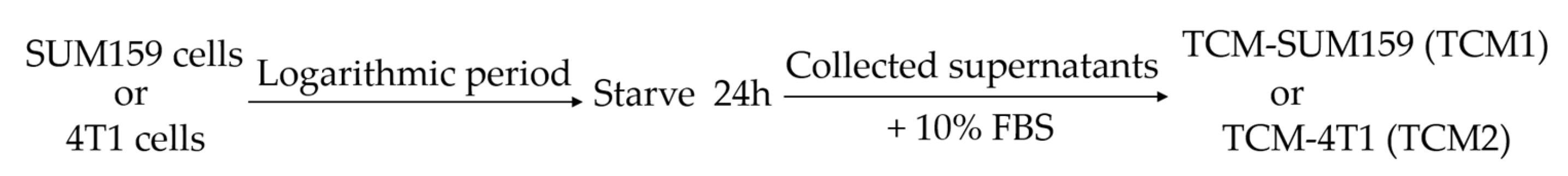
| Gene | Primer (5′-3′) |
|---|---|
| β-actin | F: TGC CGC ATC CTC TTC CTC R: GCC ACA GGA TTC CAT ACC C |
| CD86 | F: ATG GGC TCG TAT GAT TGT TT |
| R: CGG GTG ACC TTG CTT AGA C | |
| IL-1β | F: CAA CCA ACA AGT GAT ATT CTC CAT G |
| R: GAT CCA CAC TCT CCA GCT GCA | |
| iNOS | F: GAA ACG CTT CAC TTC CAA TG |
| R: GTG GTG CGG CTG GAC TTT | |
| IL-6 | F: CTT GGG ACT GAT GCT GGT G |
| R: TTG CCA TTGCAC AAC TCT TT | |
| Arg-1 | F: TGC TCA CAC TGA CAT CAA CAC T |
| R: CTA CGT CTC GCA AGC CAA T | |
| TGF-β1 | F: GCG GAC TAC TAT GCT AAA GAG G |
| R: CAC TGC TTC CCG AAT GTC T |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, C.; Jiang, S.-T.; Li, C.-X.; Jia, X.-F.; Yang, W.-L. The Effect of Salvianolic Acid A on Tumor-Associated Macrophage Polarization and Its Mechanisms in the Tumor Microenvironment of Triple-Negative Breast Cancer. Molecules 2024, 29, 1469. https://doi.org/10.3390/molecules29071469
Tang C, Jiang S-T, Li C-X, Jia X-F, Yang W-L. The Effect of Salvianolic Acid A on Tumor-Associated Macrophage Polarization and Its Mechanisms in the Tumor Microenvironment of Triple-Negative Breast Cancer. Molecules. 2024; 29(7):1469. https://doi.org/10.3390/molecules29071469
Chicago/Turabian StyleTang, Chao, Shi-Ting Jiang, Cheng-Xia Li, Xiao-Fang Jia, and Wen-Li Yang. 2024. "The Effect of Salvianolic Acid A on Tumor-Associated Macrophage Polarization and Its Mechanisms in the Tumor Microenvironment of Triple-Negative Breast Cancer" Molecules 29, no. 7: 1469. https://doi.org/10.3390/molecules29071469
APA StyleTang, C., Jiang, S.-T., Li, C.-X., Jia, X.-F., & Yang, W.-L. (2024). The Effect of Salvianolic Acid A on Tumor-Associated Macrophage Polarization and Its Mechanisms in the Tumor Microenvironment of Triple-Negative Breast Cancer. Molecules, 29(7), 1469. https://doi.org/10.3390/molecules29071469






