Mechanistic Insights into the Inhibition of a Common CTLA-4 Gene Mutation in the Cytoplasmic Domain
Abstract
1. Introduction
2. Results
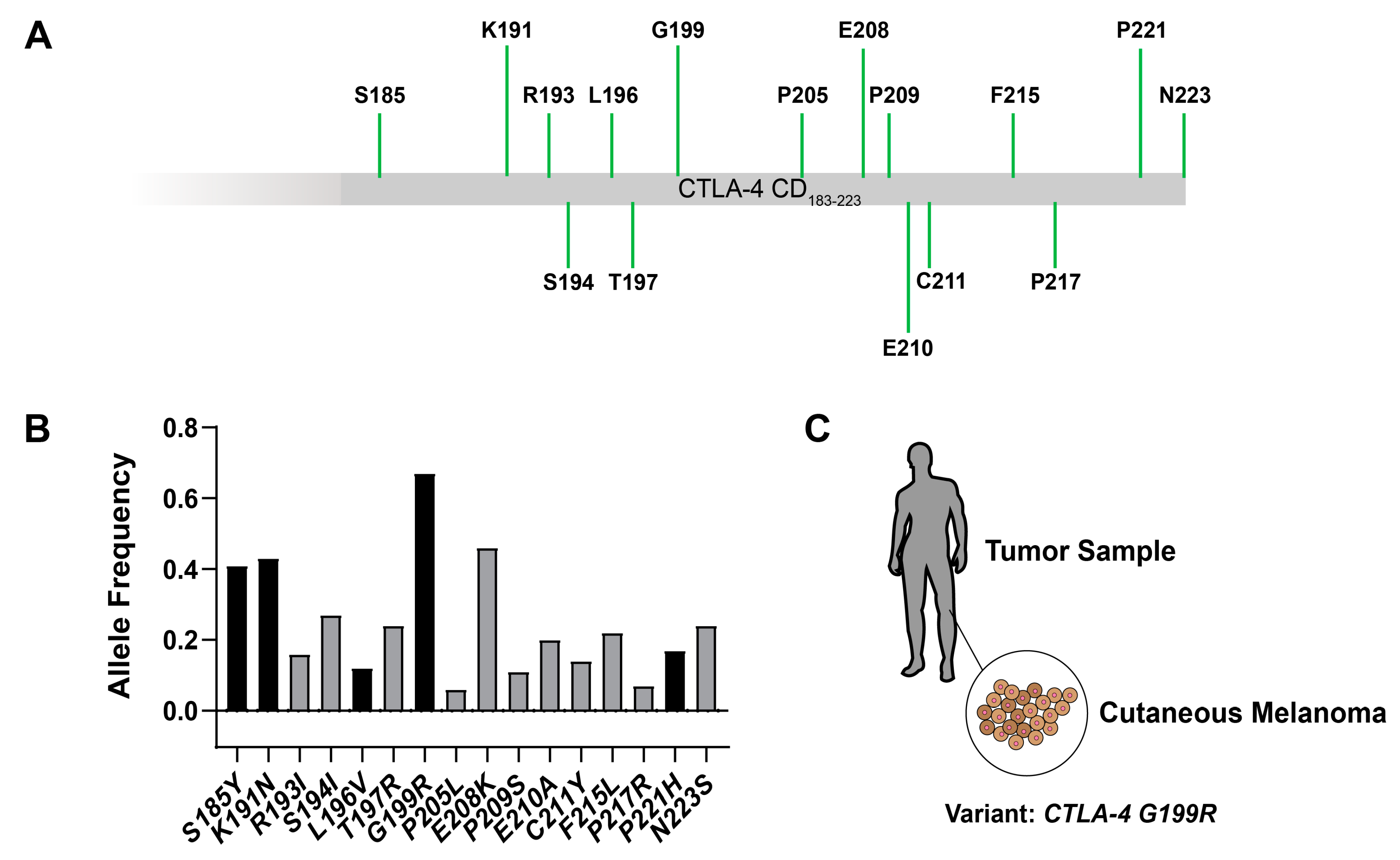
2.1. Bioinformatic Analysis of Genetic Mutations in the Cytoplasmic Domain of CTLA-4
2.2. Membrane Interaction of the Cytoplasmic Domain of CTLA-4 in the Presence of Acidic Lipids
2.3. Membrane Partition of WT in DMPG Bicelles
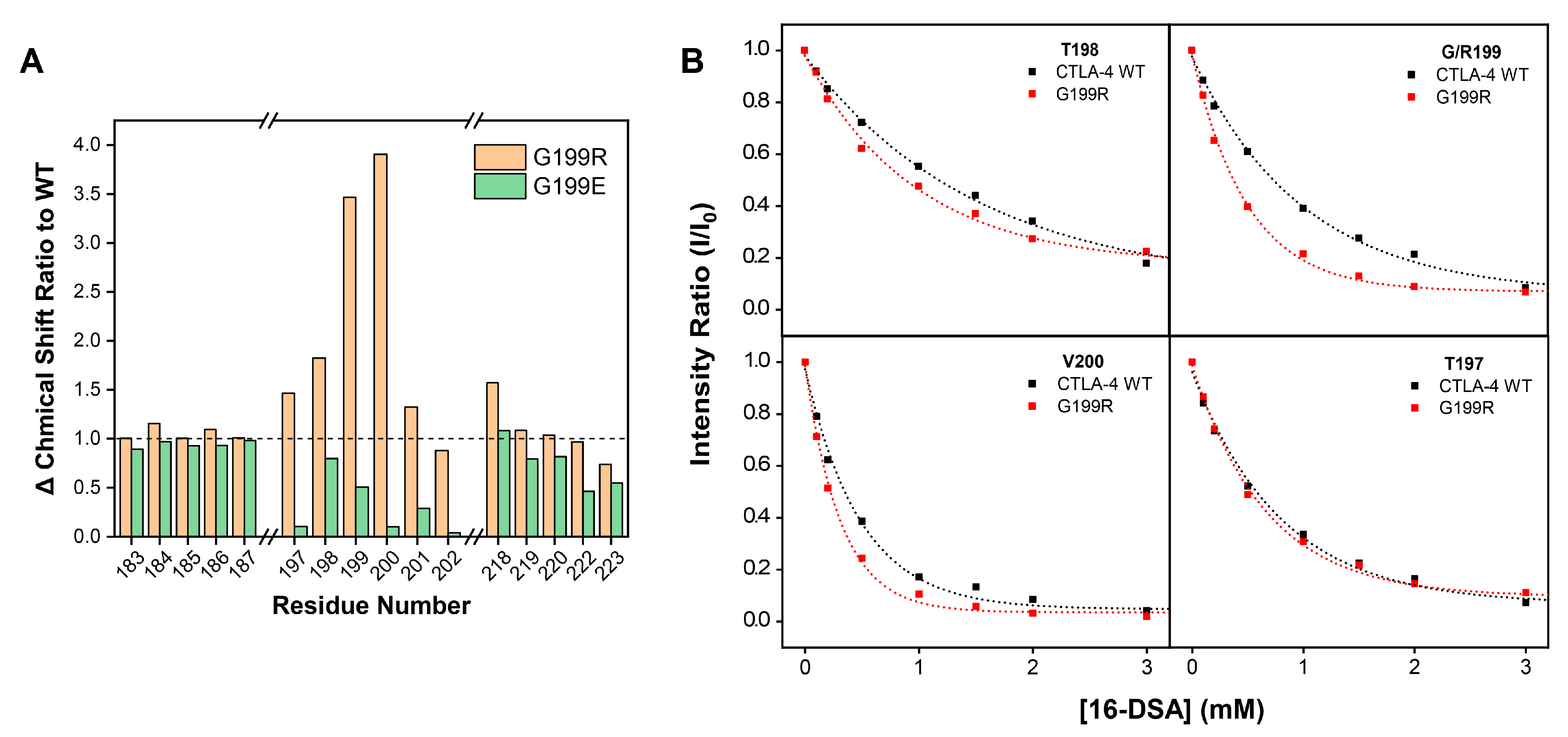
2.4. Characterization of G199R by NMR Spectroscopy
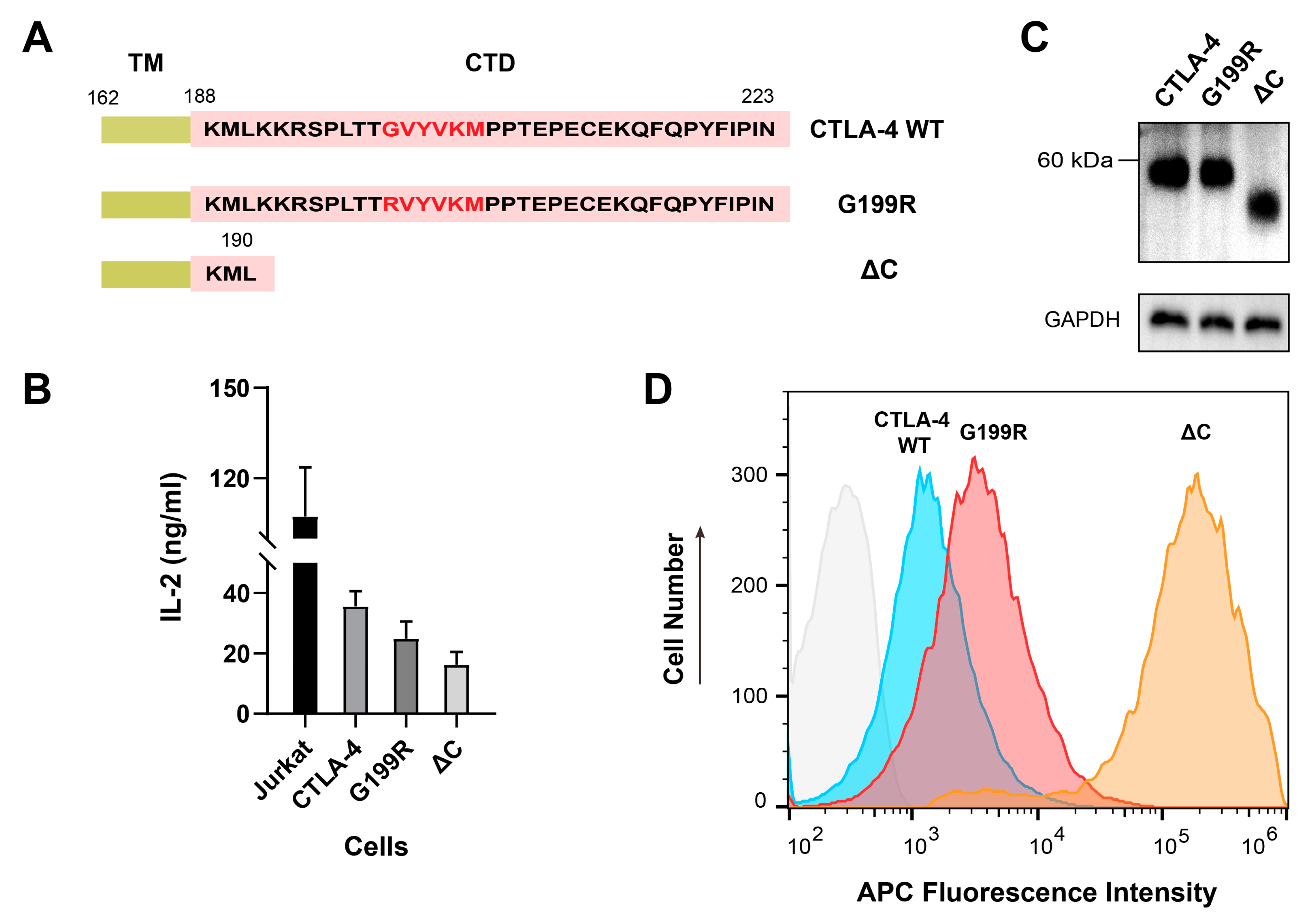
2.5. Functional Investigation of the G199R Mutant
3. Discussion
4. Materials and Methods
4.1. Reagents and Cells
4.2. Expression and Purification of CD183–223 and Its Mutants
4.3. Liposome-Binding Assays
4.4. Reconstitution of CD183–223 into Bicelles
4.5. Assignment of NMR Resonances and Secondary Structure Calculation
4.6. PRE Titration
4.7. Lentivirus Production and Transduction
4.8. Jurkat Stimulation Using Raji B Cells and IL-2 Assays
4.9. Western Blot and Immunoprecipitation
4.10. Flow Cytometry for Detection of CTLA-4
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brunet, J.F.; Denizot, F.; Luciani, M.F.; Roux-Dosseto, M.; Suzan, M.; Mattei, M.G.; Golstein, P. A new member of the immunoglobulin superfamily—CTLA-4. Nature 1987, 328, 267–270. [Google Scholar] [CrossRef]
- Linsley, P.S.; Greene, J.L.; Brady, W.; Bajorath, J.; Ledbetter, J.A.; Peach, R. Human B7-1 (CD80) and B7-2 (CD86) bind with similar avidities but distinct kinetics to CD28 and CTLA-4 receptors. Immunity 1994, 1, 793–801. [Google Scholar] [CrossRef]
- Sharpe, A.H.; Freeman, G.J. The B7-CD28 superfamily. Nat. Rev. Immunol. 2002, 2, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Gross, J.A.; St John, T.; Allison, J.P. The murine homologue of the T lymphocyte antigen CD28. Molecular cloning and cell surface expression. J. Immunol. 1990, 144, 3201–3210. [Google Scholar] [CrossRef] [PubMed]
- Balzano, C.; Buonavista, N.; Rouvier, E.; Golstein, P. CTLA-4 and CD28: Similar proteins, neighbouring genes. Int. J. Cancer Suppl. 1992, 7, 28–32. [Google Scholar] [PubMed]
- Krummel, M.F.; Allison, J.P. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J. Exp. Med. 1995, 182, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Perkins, D.; Wang, Z.; Donovan, C.; He, H.; Mark, D.; Guan, G.; Wang, Y.; Walunas, T.; Bluestone, J.; Listman, J.; et al. Regulation of CTLA-4 expression during T cell activation. J. Immunol. 1996, 156, 4154–4159. [Google Scholar] [CrossRef] [PubMed]
- Alegre, M.L.; Noel, P.J.; Eisfelder, B.J.; Chuang, E.; Clark, M.R.; Reiner, S.L.; Thompson, C.B. Regulation of surface and intracellular expression of CTLA4 on mouse T cells. J. Immunol. 1996, 157, 4762–4770. [Google Scholar] [CrossRef] [PubMed]
- Leung, H.T.; Bradshaw, J.; Cleaveland, J.S.; Linsley, P.S. Cytotoxic T lymphocyte-associated molecule-4, a high-avidity receptor for CD80 and CD86, contains an intracellular localization motif in its cytoplasmic tail. J. Biol. Chem. 1995, 270, 25107–25114. [Google Scholar] [CrossRef]
- Teft, W.A.; Kirchhof, M.G.; Madrenas, J. A molecular perspective of CTLA-4 function. Annu. Rev. Immunol. 2006, 24, 65–97. [Google Scholar] [CrossRef]
- Bauer, B.; Steinle, A. HemITAM: A single tyrosine motif that packs a punch. Sci. Signal. 2017, 10, eaan3676. [Google Scholar] [CrossRef]
- Dariavach, P.; Mattei, M.G.; Golstein, P.; Lefranc, M.P. Human Ig superfamily CTLA-4 gene: Chromosomal localization and identity of protein sequence between murine and human CTLA-4 cytoplasmic domains. Eur. J. Immunol. 1988, 18, 1901–1905. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Rudd, C.E.; Schneider, H. Src kinases Fyn and Lck facilitate the accumulation of phosphorylated CTLA-4 and its association with PI-3 kinase in intracellular compartments of T-cells. Biochem. Biophys. Res. Commun. 2001, 288, 573–578. [Google Scholar] [CrossRef] [PubMed]
- Marengere, L.E.; Waterhouse, P.; Duncan, G.S.; Mittrucker, H.W.; Feng, G.S.; Mak, T.W. Regulation of T cell receptor signaling by tyrosine phosphatase SYP association with CTLA-4. Science 1996, 272, 1170–1173. [Google Scholar] [CrossRef] [PubMed]
- Schneider, H.; Smith, X.; Liu, H.; Bismuth, G.; Rudd, C.E. CTLA-4 disrupts ZAP70 microcluster formation with reduced T cell/APC dwell times and calcium mobilization. Eur. J. Immunol. 2008, 38, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Baroja, M.L.; Vijayakrishnan, L.; Bettelli, E.; Darlington, P.J.; Chau, T.A.; Ling, V.; Collins, M.; Carreno, B.M.; Madrenas, J.; Kuchroo, V.K. Inhibition of CTLA-4 function by the regulatory subunit of serine/threonine phosphatase 2A. J. Immunol. 2002, 168, 5070–5078. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.F.; Fu, G.; Zhang, Y.; Yokosuka, T.; Casas, J.; Canonigo-Balancio, A.J.; Becart, S.; Kim, G.; Yates, J.R., 3rd; Kronenberg, M.; et al. Protein kinase C-eta controls CTLA-4-mediated regulatory T cell function. Nat. Immunol. 2014, 15, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Masteller, E.L.; Chuang, E.; Mullen, A.C.; Reiner, S.L.; Thompson, C.B. Structural analysis of CTLA-4 function in vivo. J. Immunol. 2000, 164, 5319–5327. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, H.; Takahashi, S.; Takase, K.; Yamasaki, S.; Yokosuka, T.; Koike, T.; Saito, T. CD25(+)CD4(+) regulatory T cells exert in vitro suppressive activity independent of CTLA-4. Int. Immunol. 2005, 17, 421–427. [Google Scholar] [CrossRef]
- Tai, X.; Van Laethem, F.; Pobezinsky, L.; Guinter, T.; Sharrow, S.O.; Adams, A.; Granger, L.; Kruhlak, M.; Lindsten, T.; Thompson, C.B.; et al. Basis of CTLA-4 function in regulatory and conventional CD4(+) T cells. Blood 2012, 119, 5155–5163. [Google Scholar] [CrossRef]
- Nakaseko, C.; Miyatake, S.; Iida, T.; Hara, S.; Abe, R.; Ohno, H.; Saito, Y.; Saito, T. Cytotoxic T lymphocyte antigen 4 (CTLA-4) engagement delivers an inhibitory signal through the membrane-proximal region in the absence of the tyrosine motif in the cytoplasmic tail. J. Exp. Med. 1999, 190, 765–774. [Google Scholar] [CrossRef]
- Cinek, T.; Sadra, A.; Imboden, J.B. Cutting edge: Tyrosine-independent transmission of inhibitory signals by CTLA-4. J. Immunol. 2000, 164, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, J.D.; Lu, P.; Leytze, G.; Rodgers, J.; Schieven, G.L.; Bennett, K.L.; Linsley, P.S.; Kurtz, S.E. Interaction of the cytoplasmic tail of CTLA-4 (CD152) with a clathrin-associated protein is negatively regulated by tyrosine phosphorylation. Biochemistry 1997, 36, 15975–15982. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Allison, J.P. Interaction of CTLA-4 with AP50, a clathrin-coated pit adaptor protein. Proc. Natl. Acad. Sci. USA 1997, 94, 9273–9278. [Google Scholar] [CrossRef] [PubMed]
- Shiratori, T.; Miyatake, S.; Ohno, H.; Nakaseko, C.; Isono, K.; Bonifacino, J.S.; Saito, T. Tyrosine phosphorylation controls internalization of CTLA-4 by regulating its interaction with clathrin-associated adaptor complex AP-2. Immunity 1997, 6, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Stumpf, M.; Zhou, X.; Chikuma, S.; Bluestone, J.A. Tyrosine 201 of the cytoplasmic tail of CTLA-4 critically affects T regulatory cell suppressive function. Eur. J. Immunol. 2014, 44, 1737–1746. [Google Scholar] [CrossRef] [PubMed]
- Kozik, P.; Francis, R.W.; Seaman, M.N.; Robinson, M.S. A screen for endocytic motifs. Traffic 2010, 11, 843–855. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.S.; Sansom, D.M. Confusing signals: Recent progress in CTLA-4 biology. Trends Immunol. 2015, 36, 63–70. [Google Scholar] [CrossRef]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Krawczak, M.; Ball, E.V.; Fenton, I.; Stenson, P.D.; Abeysinghe, S.; Thomas, N.; Cooper, D.N. Human gene mutation database-a biomedical information and research resource. Hum. Mutat. 2000, 15, 45–51. [Google Scholar] [CrossRef]
- Egg, D.; Rump, I.C.; Mitsuiki, N.; Rojas-Restrepo, J.; Maccari, M.E.; Schwab, C.; Gabrysch, A.; Warnatz, K.; Goldacker, S.; Patino, V.; et al. Therapeutic options for CTLA-4 insufficiency. J. Allergy Clin. Immunol. 2022, 149, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- de Bruijn, I.; Kundra, R.; Mastrogiacomo, B.; Tran, T.N.; Sikina, L.; Mazor, T.; Li, X.; Ochoa, A.; Zhao, G.; Lai, B.; et al. Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 2023, 83, 3861–3867. [Google Scholar] [CrossRef] [PubMed]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018, 173, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Consortium, I.T.P.-C.A.o.W.G. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef]
- Wen, M.; Cao, Y.; Wu, B.; Xiao, T.; Cao, R.; Wang, Q.; Liu, X.; Xue, H.; Yu, Y.; Lin, J.; et al. PD-L1 degradation is regulated by electrostatic membrane association of its cytoplasmic domain. Nat. Commun. 2021, 12, 5106. [Google Scholar] [CrossRef]
- Yang, W.; Pan, W.; Chen, S.; Trendel, N.; Jiang, S.; Xiao, F.; Xue, M.; Wu, W.; Peng, Z.; Li, X.; et al. Dynamic regulation of CD28 conformation and signaling by charged lipids and ions. Nat. Struct. Mol. Biol. 2017, 24, 1081–1092. [Google Scholar] [CrossRef]
- Shen, Y.; Delaglio, F.; Cornilescu, G.; Bax, A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 2009, 44, 213–223. [Google Scholar] [CrossRef]
- Piai, A.; Fu, Q.; Dev, J.; Chou, J.J. Optimal Bicelle Size q for Solution NMR Studies of the Protein Transmembrane Partition. Chemistry 2017, 23, 1361–1367. [Google Scholar] [CrossRef]
- Qureshi, O.S.; Kaur, S.; Hou, T.Z.; Jeffery, L.E.; Poulter, N.S.; Briggs, Z.; Kenefeck, R.; Willox, A.K.; Royle, S.J.; Rappoport, J.Z.; et al. Constitutive clathrin-mediated endocytosis of CTLA-4 persists during T cell activation. J. Biol. Chem. 2012, 287, 9429–9440. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Gagnon, E.; Call, M.E.; Schnell, J.R.; Schwieters, C.D.; Carman, C.V.; Chou, J.J.; Wucherpfennig, K.W. Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell 2008, 135, 702–713. [Google Scholar] [CrossRef] [PubMed]
- Follows, E.R.; McPheat, J.C.; Minshull, C.; Moore, N.C.; Pauptit, R.A.; Rowsell, S.; Stacey, C.L.; Stanway, J.J.; Taylor, I.W.; Abbott, W.M. Study of the interaction of the medium chain mu 2 subunit of the clathrin-associated adapter protein complex 2 with cytotoxic T-lymphocyte antigen 4 and CD28. Biochem. J. 2001, 359, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Restrepo, J.; Sindram, E.; Zenke, S.; Haberstroh, H.; Mitsuiki, N.; Gabrysch, A.; Huebscher, K.; Posadas-Cantera, S.; Krausz, M.; Kobbe, R.; et al. Functional Relevance of CTLA4 Variants: An Upgraded Approach to Assess CTLA4-Dependent Transendocytosis by Flow Cytometry. J. Clin. Immunol. 2023, 43, 2076–2089. [Google Scholar] [CrossRef] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Bartels, C.; Xia, T.H.; Billeter, M.; Guntert, P.; Wuthrich, K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 1995, 6, 1–10. [Google Scholar] [CrossRef]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Zhang, Y.; Shen, L.; Du, L.; Xue, H.; Wu, B.; OuYang, B. Mechanistic Insights into the Inhibition of a Common CTLA-4 Gene Mutation in the Cytoplasmic Domain. Molecules 2024, 29, 1330. https://doi.org/10.3390/molecules29061330
Xu J, Zhang Y, Shen L, Du L, Xue H, Wu B, OuYang B. Mechanistic Insights into the Inhibition of a Common CTLA-4 Gene Mutation in the Cytoplasmic Domain. Molecules. 2024; 29(6):1330. https://doi.org/10.3390/molecules29061330
Chicago/Turabian StyleXu, Jikang, Yu Zhang, Lijuan Shen, Lingyu Du, Hongjuan Xue, Bin Wu, and Bo OuYang. 2024. "Mechanistic Insights into the Inhibition of a Common CTLA-4 Gene Mutation in the Cytoplasmic Domain" Molecules 29, no. 6: 1330. https://doi.org/10.3390/molecules29061330
APA StyleXu, J., Zhang, Y., Shen, L., Du, L., Xue, H., Wu, B., & OuYang, B. (2024). Mechanistic Insights into the Inhibition of a Common CTLA-4 Gene Mutation in the Cytoplasmic Domain. Molecules, 29(6), 1330. https://doi.org/10.3390/molecules29061330






