Ginsenoside Rb1, Compound K and 20(S)-Protopanaxadiol Attenuate High-Fat Diet-Induced Hyperlipidemia in Rats via Modulation of Gut Microbiota and Bile Acid Metabolism
Abstract
1. Introduction
2. Results
2.1. Effects of Ginsenoside Rb1, CK, and PPD on Body Weight, Serum Biochemistry, Hepatic Adipose Accumulation in HFD-Fed Rats
2.2. Effects of Ginsenoside Rb1, CK, and PPD on the Composition of Gut Microbiota in HFD-Fed Rats
2.3. Effects of Ginsenoside Rb1, CK, and PPD on the Function of Gut Microbiota in HFD-Fed Rats
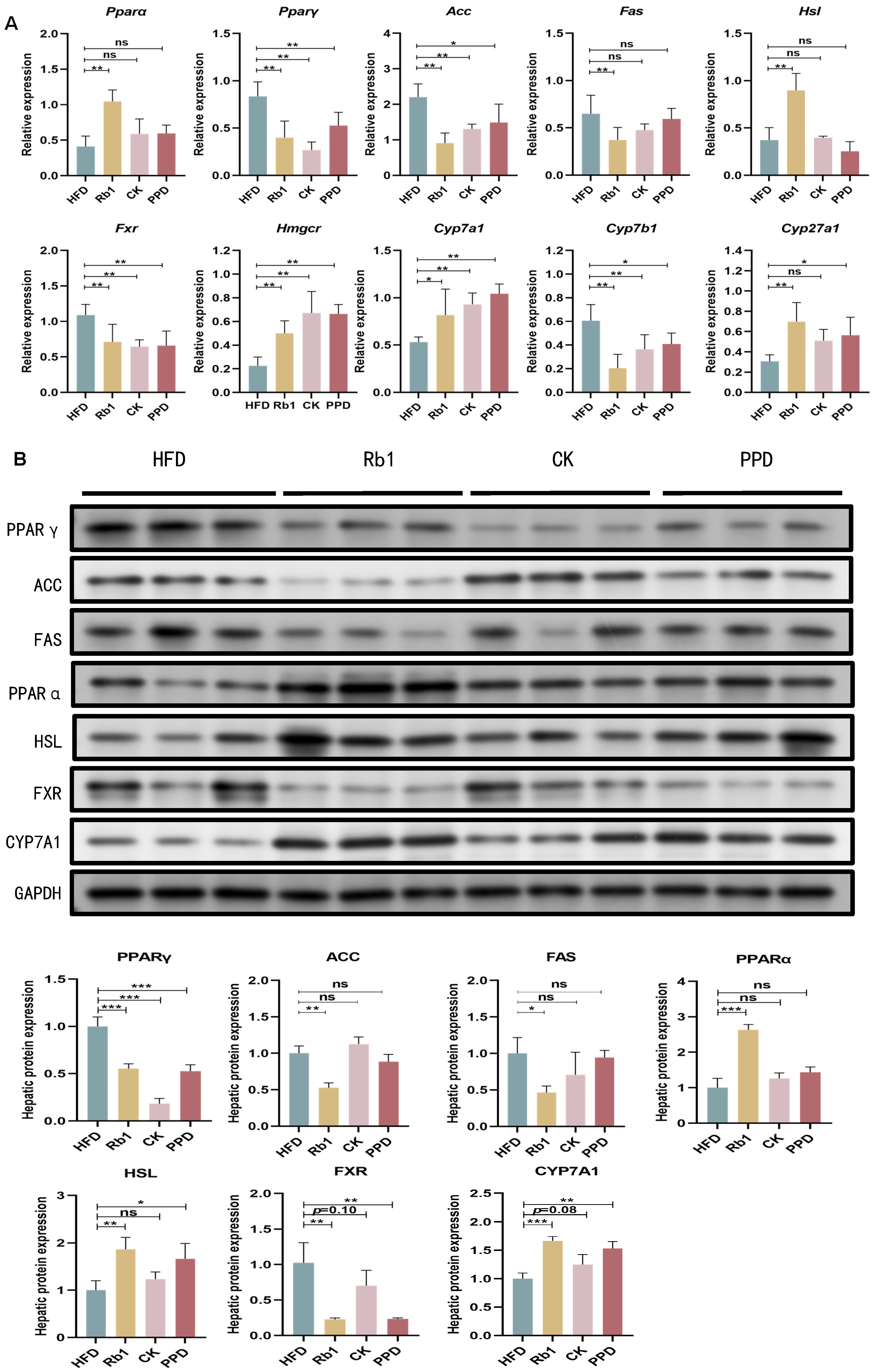
2.4. Effects of Ginsenoside Rb1, CK, and PPD on FA and Cholesterol Metabolism in HFD-Fed Rats
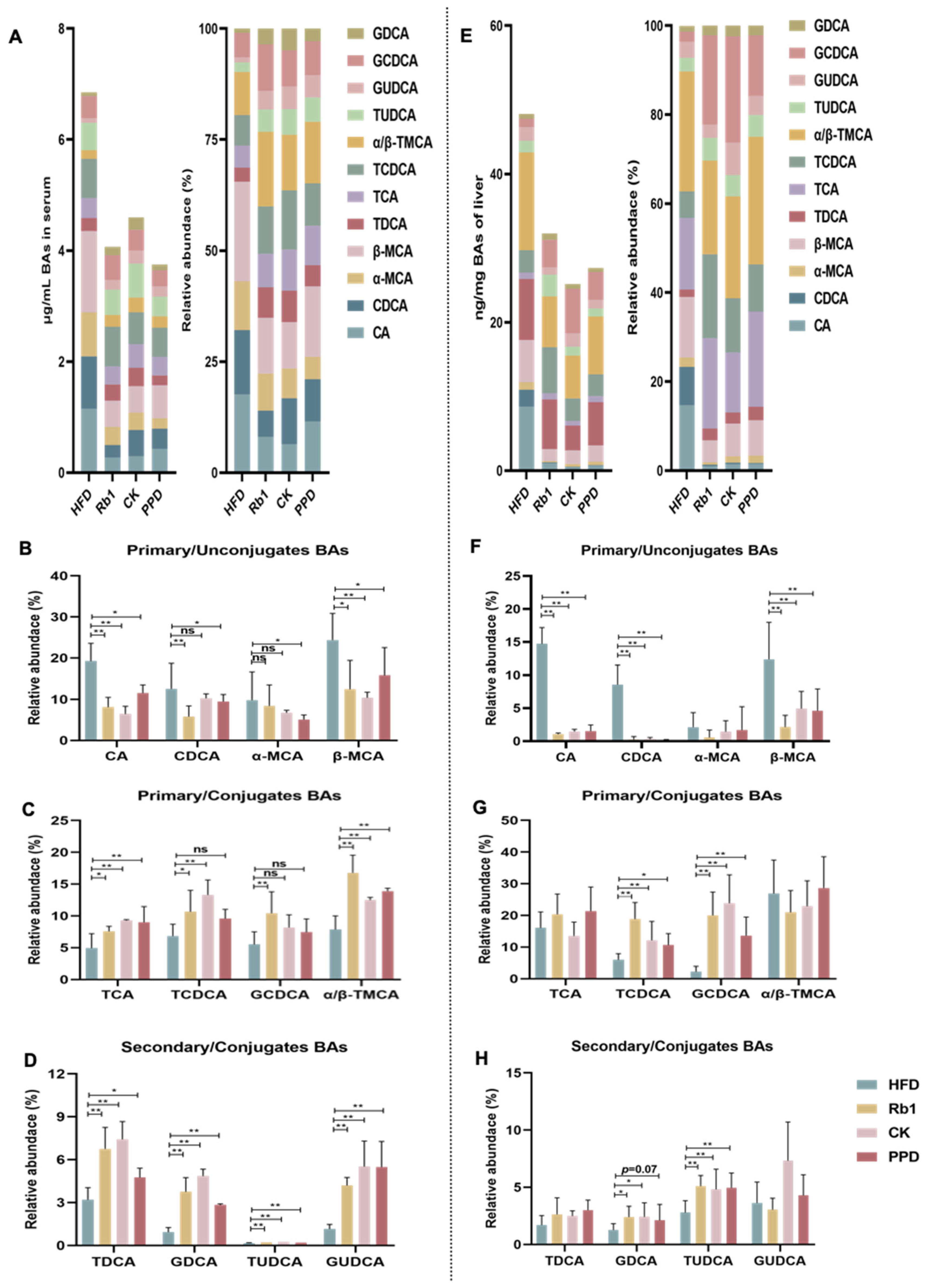
2.5. Effects of Ginsenoside Rb1, CK, and PPD on BA Metabolism in HFD-Fed Rats
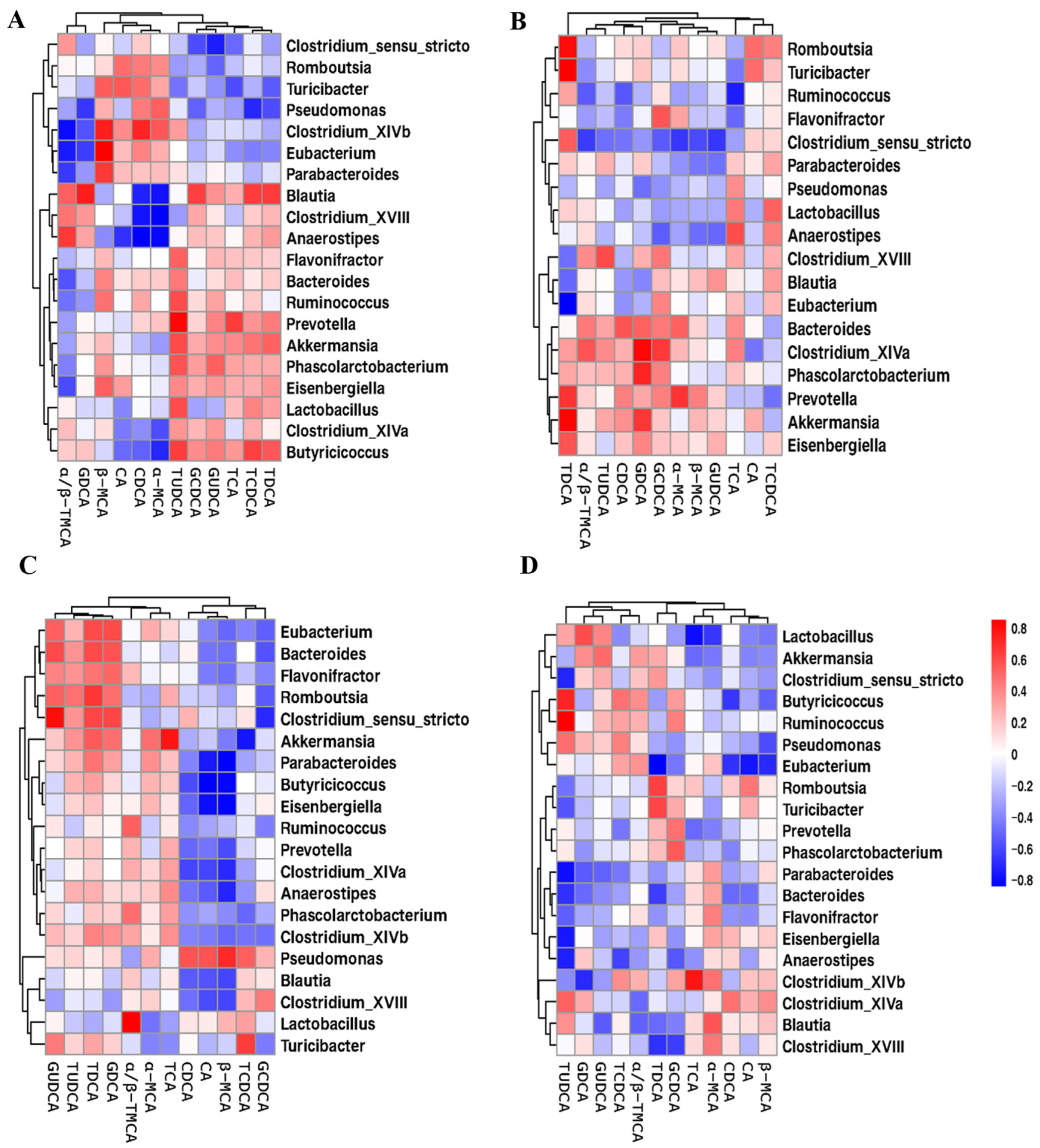
2.6. Correlation Analysis of Gut Microbiota and BA Metabolism
3. Discussion
4. Materials and Methods
4.1. Animal Study Design
4.2. Biochemical Analysis
4.3. Histological Analysis
4.4. Real-Time Quantitative PCR (RT-qPCR) Analysis
4.5. Western Blot Analysis
4.6. Extraction and Sequencing of Fecal DNA Using 16S rRNA Gene
4.7. UHPLC-MS/MS Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leuti, A.; Fazio, D.; Fava, M.; Piccoli, A.; Oddi, S.; Maccarrone, M. Bioactive lipids, inflammation and chronic diseases. Adv. Drug Deliv. Rev. 2020, 159, 133–169. [Google Scholar] [CrossRef]
- Deprince, A.; Haas, J.T.; Staels, B. Dysregulated lipid metabolism links NAFLD to cardiovascular disease. Mol. Metab. 2020, 42, 101092. [Google Scholar] [CrossRef] [PubMed]
- Badmus, O.O.; Hillhouse, S.A.; Anderson, C.D.; Hinds, T.D.; Stec, D.E. Molecular mechanisms of metabolic associated fatty liver disease (MAFLD): Functional analysis of lipid metabolism pathways. Clin. Sci. 2022, 136, 1347–1366. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.Y.; Shi, X.J.; Hu, A.; Wang, J.Q.; Ding, Y.; Jiang, W.; Sun, M.; ZAhao, X.; Luo, J.; Qi, W.; et al. Feeding induces cholesterol biosynthesis via the mTORC1–USP20–HMGCR axis. Nature 2020, 588, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y.L. Bile Acid Metabolism and Signaling. Compr. Physiol. 2013, 3, 1191–1212. [Google Scholar] [PubMed]
- de Aguiar Vallim, T.Q.; Tarling, E.J.; Edwards, P.A. Pleiotropic Roles of Bile Acids in Metabolism. Cell Metab. 2013, 17, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Rimal, B.; Jiang, C.; Chiang, J.Y.; Patterson, A.D. Bile acid metabolism and signaling, the microbiota, and metabolic disease. Pharmacol. Ther. 2022, 237, 108238. [Google Scholar] [CrossRef] [PubMed]
- Guzior, D.V.; Quinn, R.A. Review: Microbial transformations of human bile acids. Microbiome 2021, 9, 140. [Google Scholar] [CrossRef]
- Li, X.; Liu, J.; Zuo, T.-T.; Hu, Y.; Li, Z.; Wang, H.-D.; Xu, X.-Y.; Yang, W.-Z.; Guo, D.-A. Advances and challenges in ginseng research from 2011 to 2020: The phytochemistry, quality control, metabolism, and biosynthesis. Nat. Prod. Rep. 2022, 39, 875–909. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Zeng, J.; Wong, A.S.T. Chemical Structures and Pharmacological Profiles of Ginseng Saponins. Molecules 2019, 24, 2443. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Sun, Y.; Chu, Y.; Wang, J.; Zheng, H.; Liu, L.; Cai, W.; Zhang, H. Ginsenoside Rb1 Alleviated High-Fat-Diet-Induced Hepatocytic Apoptosis via Peroxisome Proliferator-Activated Receptor γ. BioMed Res. Int. 2020, 2020, 2315230. [Google Scholar] [CrossRef]
- Yang, X.; Dong, B.; An, L.; Zhang, Q.; Chen, Y.; Wang, H.; Song, Z. Ginsenoside Rb1 ameliorates Glycemic Disorder in Mice with High Fat Diet-Induced Obesity via Regulating Gut Microbiota and Amino Acid Metabolism. Front. Pharmacol. 2021, 12, 756491. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Lee, H.-J. Ginsenoside Compound K: Insights into Recent Studies on Pharmacokinetics and Health-Promoting Activities. Biomolecules 2020, 10, 1028. [Google Scholar] [CrossRef]
- Lee, S.J.; Ko, W.G.; Kim, J.H.; Sung, J.H.; Lee, S.J.; Moon, C.K.; Lee, B.H. Induction of apoptosis by a novel intestinal metabolite of ginseng saponin via cytochrome c-mediated activation of caspase-3 protease. Biochem. Pharmacol. 2000, 60, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-Y.; Cho, J.Y. 20S-dihydroprotopanaxadiol, a ginsenoside derivative, boosts innate immune responses of monocytes and macrophages. J. Ginseng Res. 2013, 37, 293–299. [Google Scholar] [CrossRef]
- de Vos, W.M.; Tilg, H.; Van Hul, M.; Cani, P.D. Gut microbiome and health: Mechanistic insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H. Gut microbiota-mediated pharmacokinetics of ginseng saponins. J. Ginseng Res. 2018, 42, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, S.; Duan, F.; Liu, A.; Li, S.; Zhong, W.; Sheng, W.; Chen, J.; Xu, J.; Xiao, S. Prebiotics enhance the biotransformation and bioavailability of ginsenosides in rats by modulating gut microbiota. J. Ginseng Res. 2021, 45, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.W.; Kitai, T.; Hazen, S.L. Gut microbiota in cardiovascular health and disease. Circ. Res. 2017, 120, 1183–1196. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, Y. Gut microbiota derived metabolites in cardiovascular health and disease. Protein Cell 2018, 9, 416–431. [Google Scholar] [CrossRef] [PubMed]
- Ratan, Z.A.; Haidere, M.F.; Hong, Y.H.; Park, S.H.; Lee, J.-O.; Lee, J.; Cho, J.Y. Pharmacological potential of ginseng and its major component ginsenosides. J. Ginseng Res. 2021, 45, 199–210. [Google Scholar] [CrossRef]
- Zou, H.; Zhang, M.; Zhu, X.; Zhu, L.; Chen, S.; Luo, M.; Xie, Q.; Chen, Y.; Zhang, K.; Bu, Q.; et al. Ginsenoside Rb1 Improves Metabolic Disorder in High-Fat Diet-Induced Obese Mice Associated with Modulation of Gut Microbiota. Front. Microbiol. 2022, 13, 826487. [Google Scholar] [CrossRef]
- Yang, S.; Liu, T.; Hu, C.; Li, W.; Meng, Y.; Li, H.; Song, C.; He, C.; Zhou, Y.; Fan, Y. Ginsenoside Compound K Protects against Obesity through Pharmacological Targeting of Glucocorticoid Receptor to Activate Lipophagy and Lipid Metabolism. Pharmaceutics 2022, 14, 1192. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, H.; Zhou, Z.; Li, J.; Chen, H.; Liu, Y.; Huang, C.; Fan, S. Protopanaxadiol alleviates obesity in high-fat diet-fed mice via activation of energy-sensing neuron in the paraventricular nucleus of hypothalamus. Biochem. Biophys. Res. Commun. 2019, 513, 1092–1099. [Google Scholar] [CrossRef] [PubMed]
- Willey, J.Z.; Elkind, M.S. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase inhibitors in the treatment of central nervous system diseases. Arch. Neurol. 2010, 67, 1062–1067. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.; Peng, H.; Wu, X.; Chen, X.; Wang, B.; Su, X. IL-38 in modulating hyperlipidemia and its related cardiovascular diseases. Int. Immunopharmacol. 2022, 108, 108876. [Google Scholar] [CrossRef]
- Ellulu, M.S.; Patimah, I.; KhazáAi, H.; Rahmat, A.; Abed, Y. Obesity and inflammation: The linking mechanism and the complications. Arch. Med. Sci. 2017, 13, 851–863. [Google Scholar] [CrossRef] [PubMed]
- Moghbeli, M.; Khedmatgozar, H.; Yadegari, M.; Avan, A.; Ferns, G.A.; Mobarhan, M.G. Cytokines and the immune response in obesity-related disorders. Adv. Clin. Chem. 2021, 101, 135–168. [Google Scholar]
- Joh, E.-H.; Lee, I.-A.; Jung, I.-H.; Kim, D.-H. Ginsenoside Rb1 and its metabolite compound K inhibit IRAK-1 activation—The key step of inflammation. Biochem. Pharmacol. 2011, 82, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Cuong, T.T.; Yang, C.-S.; Yuk, J.-M.; Lee, H.-M.; Ko, S.-R.; Cho, B.-G.; Jo, E.-K. Glucocorticoid receptor agonist compound K regulates dectin-1-dependent inflammatory signaling through inhibition of reactive oxygen species. Life Sci. 2009, 85, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-S.; Shin, J.A.; Jung, J.-S.; Hyun, J.-W.; Van Le, T.K.; Kim, D.-H.; Park, E.-M.; Kim, H.-S. Anti-Inflammatory Mechanism of Compound K in Activated Microglia and Its Neuroprotective Effect on Experimental Stroke in Mice. J. Pharmacol. Exp. Ther. 2012, 341, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Rhule, A.; Navarro, S.; Smith, J.R.; Shepherd, D.M. Panax notoginseng attenuates LPS-induced pro-inflammatory mediators in RAW264.7 cells. J. Ethnopharmacol. 2006, 106, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Dabke, K.; Hendrick, G.; Devkota, S. The gut microbiome and metabolic syndrome. J. Clin. Investig. 2019, 129, 4050–4057. [Google Scholar] [CrossRef] [PubMed]
- Schoeler, M.; Caesar, R. Dietary lipids, gut microbiota and lipid metabolism. Rev. Endocr. Metab. Disord. 2019, 20, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Xu, W.; Zhang, L.; Li, X.; Wang, R.; Wu, S. Impact of Gut Microbiota and Microbiota-Related Metabolites on Hyperlipidemia. Front. Cell. Infect. Microbiol. 2021, 11, 634780. [Google Scholar] [CrossRef]
- Liu, X.; Mao, B.; Gu, J.; Wu, J.; Cui, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Blautia—A new functional genus with potential probiotic properties? Gut Microbes 2021, 13, 1875796. [Google Scholar] [CrossRef]
- Cani, P.D.; Depommier, C.; Derrien, M.; Everard, A.; de Vos, W.M. Akkermansia muciniphila: Paradigm for next-generation beneficial microorganisms. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 625–637. [Google Scholar] [CrossRef]
- Zhang, T.; Li, Q.; Cheng, L.; Buch, H.; Zhang, F. Akkermansia muciniphila is a promising probiotic. Microb. Biotechnol. 2019, 12, 1109–1125. [Google Scholar] [CrossRef]
- Kang, Y.; Kang, X.; Yang, H.; Liu, H.; Yang, X.; Liu, Q.; Tian, H.; Xue, Y.; Ren, P.; Kuang, X.; et al. Lactobacillus acidophilus ameliorates obesity in mice through modulation of gut microbiota dysbiosis and intestinal permeability. Pharmacol. Res. 2022, 175, 106020. [Google Scholar] [CrossRef]
- Gao, X.; Du, L.; Randell, E.; Zhang, H.; Li, K.; Li, D. Effect of different phosphatidylcholines on high fat diet-induced insulin resistance in mice. Food Funct. 2021, 12, 1516–1528. [Google Scholar] [CrossRef]
- Jamar, G.; Santamarina, A.B.; Dias, G.C.; Masquio, D.C.L.; de Rosso, V.V.; Pisani, L.P. Relationship between fatty acids intake and Clostridium coccoides in obese individuals with metabolic syndrome. Food Res. Int. 2018, 113, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zou, Y.; Tang, H.; Zhuang, J.; Ye, Z.; Wei, T.; Lin, J.; Zheng, Q. Cordyceps militaris polysaccharides modulate gut microbiota and improve metabolic disorders in mice withdiet-induced obesity. J. Sci. Food Agric. 2023, 103, 1885–1894. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Z.; Liu, J.; Qi, H.; Li, J.; Chen, J.; Huang, Q.; Liu, Q.; Mi, J.; Li, X. Gut Microbiota: Therapeutic Targets of Ginseng Against Multiple Disorders and Ginsenoside Transformation. Front. Cell. Infect. Microbiol. 2022, 12, 853981. [Google Scholar] [CrossRef]
- Lv, X.-C.; Chen, M.; Huang, Z.-R.; Guo, W.-L.; Ai, L.-Z.; Bai, W.-D.; Yu, X.-D.; Liu, Y.-L.; Rao, P.-F.; Ni, L. Potential mechanisms underlying the ameliorative effect of Lactobacillus paracasei FZU103 on the lipid metabolism in hyperlipidemic mice fed a high-fat diet. Food Res. Int. 2021, 139, 109956. [Google Scholar] [CrossRef] [PubMed]
- Lew, L.-C.; Hor, Y.-Y.; Jaafar, M.-H.; Lau, A.-S.; Lee, B.-K.; Chuah, L.-O.; Yap, K.-P.; Azlan, A.; Azzam, G.; Choi, S.-B.; et al. Lactobacillus Strains Alleviated Hyperlipidemia and Liver Steatosis in Aging Rats via Activation of AMPK. Int. J. Mol. Sci. 2020, 21, 5872. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Guan, W.-Y.; Wang, C.; Yu, H.-S.; Li, X.; Wang, Y.-H. Effect of Lactobacillus plantarum LP104 on hyperlipidemia in high-fat diet induced C57BL/6N mice via alteration of intestinal microbiota. J. Funct. Foods 2022, 95, 105176. [Google Scholar] [CrossRef]
- Zhuang, T.; Li, W.; Yang, L.; Wang, Z.; Ding, L.; Zhou, M. Gut Microbiota: Novel Therapeutic Target of Ginsenosides for the Treatment of Obesity and Its Complications. Front. Pharmacol. 2021, 12, 731288. [Google Scholar] [CrossRef] [PubMed]
- Gross, B.; Pawlak, M.; Lefebvre, P.; Staels, B. PPARs in obesity-induced T2DM, dyslipidaemia and NAFLD. Nat. Rev. Endocrinol. 2017, 13, 36–49. [Google Scholar] [CrossRef]
- Peters, J.M.; Hennuyer, N.; Staels, B.; Fruchart, J.-C.; Fievet, C.; Gonzalez, F.J.; Auwerx, J. Alterations in Lipoprotein Metabolism in Peroxisome Proliferator-activated Receptor α-deficient Mice. J. Biol. Chem. 1997, 272, 27307–27312. [Google Scholar] [CrossRef]
- Montaigne, D.; Butruille, L.; Staels, B. PPAR control of metabolism and cardiovascular functions. Nat. Rev. Cardiol. 2021, 18, 809–823. [Google Scholar] [CrossRef] [PubMed]
- Lorente-Cebrián, S.; Kulyté, A.; Hedén, P.; Näslund, E.; Arner, P.; Rydén, M. Relationship between Site-Specific HSL Phosphorylation and Adipocyte Lipolysis in Obese Women. Obes. Facts 2011, 4, 365–371. [Google Scholar] [CrossRef]
- Pushpass, R.-A.G.; Alzoufairi, S.; Jackson, K.G.; Lovegrove, J.A. Circulating bile acids as a link between the gut microbiota and cardiovascular health: Impact of prebiotics, probiotics and polyphenol-rich foods. Nutr. Res. Rev. 2022, 35, 161–180. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhao, M.; Bo, T.; Ma, S.; Yuan, Z.; Chen, W.; He, Z.; Hou, X.; Liu, J.; Zhang, Z.; et al. Blocking FSH inhibits hepatic cholesterol biosynthesis and reduces serum cholesterol. Cell Res. 2019, 29, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, P.; Cariou, B.; Lien, F.; Kuipers, F.; Staels, B. Role of Bile Acids and Bile Acid Receptors in Metabolic Regulation. Physiol. Rev. 2009, 89, 147–191. [Google Scholar] [CrossRef] [PubMed]
- Desai, M.S.; Mathur, B.; Eblimit, Z.; Vasquez, H.; Taegtmeyer, H.; Karpen, S.J.; Penny, D.J.; Moore, D.D.; Anakk, S. Bile acid excess induces cardiomyopathy and metabolic dysfunctions in the heart. Hepatology 2016, 65, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Sun, L.; Ma, P.; Xu, L.; Xiao, P. Dihydromyricetin prevents obesity via regulating bile acid metabolism associated with the farnesoid X receptor inob/ob mice. Food Funct. 2022, 13, 2491–2503. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Xie, G.; Jia, W. Bile acid–microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Xie, C.; Li, F.; Zhang, L.; Nichols, R.G.; Krausz, K.W.; Cai, J.; Qi, Y.; Fang, Z.-Z.; Takahashi, S.; et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J. Clin. Investig. 2015, 125, 386–402. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y.L.; Ferrell, J.M. Bile Acid Metabolism in Liver Pathobiology. Gene Expr. 2018, 18, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Münzker, J.; Haase, N.; Till, A.; Sucher, R.; Haange, S.-B.; Nemetschke, L.; Gnad, T.; Jäger, E.; Chen, J.; Riede, S.J.; et al. Functional changes of the gastric bypass microbiota reactivate thermogenic adipose tissue and systemic glucose control via intestinal FXR-TGR5 crosstalk in diet-induced obesity. Microbiome 2022, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; An, Y.; Tang, H.; Wang, Y. Alterations of Bile Acids and Gut Microbiota in Obesity Induced by High Fat Diet in Rat Model. J. Agric. Food Chem. 2019, 67, 3624–3632. [Google Scholar] [CrossRef] [PubMed]

 representing more abundance in the Rb1, CK and PPD groups compared with the HFD group,
representing more abundance in the Rb1, CK and PPD groups compared with the HFD group,  representing less abundance in the Rb1, CK and PPD groups compared with the HFD group, (F) change in the relative abundance of representative genera. Values are the means ± SEMs. * p < 0.05, ** p < 0.01 and *** p < 0.001 versus the HFD group, ns means no significantly difference.
representing less abundance in the Rb1, CK and PPD groups compared with the HFD group, (F) change in the relative abundance of representative genera. Values are the means ± SEMs. * p < 0.05, ** p < 0.01 and *** p < 0.001 versus the HFD group, ns means no significantly difference.
 representing more abundance in the Rb1, CK and PPD groups compared with the HFD group,
representing more abundance in the Rb1, CK and PPD groups compared with the HFD group,  representing less abundance in the Rb1, CK and PPD groups compared with the HFD group, (F) change in the relative abundance of representative genera. Values are the means ± SEMs. * p < 0.05, ** p < 0.01 and *** p < 0.001 versus the HFD group, ns means no significantly difference.
representing less abundance in the Rb1, CK and PPD groups compared with the HFD group, (F) change in the relative abundance of representative genera. Values are the means ± SEMs. * p < 0.05, ** p < 0.01 and *** p < 0.001 versus the HFD group, ns means no significantly difference.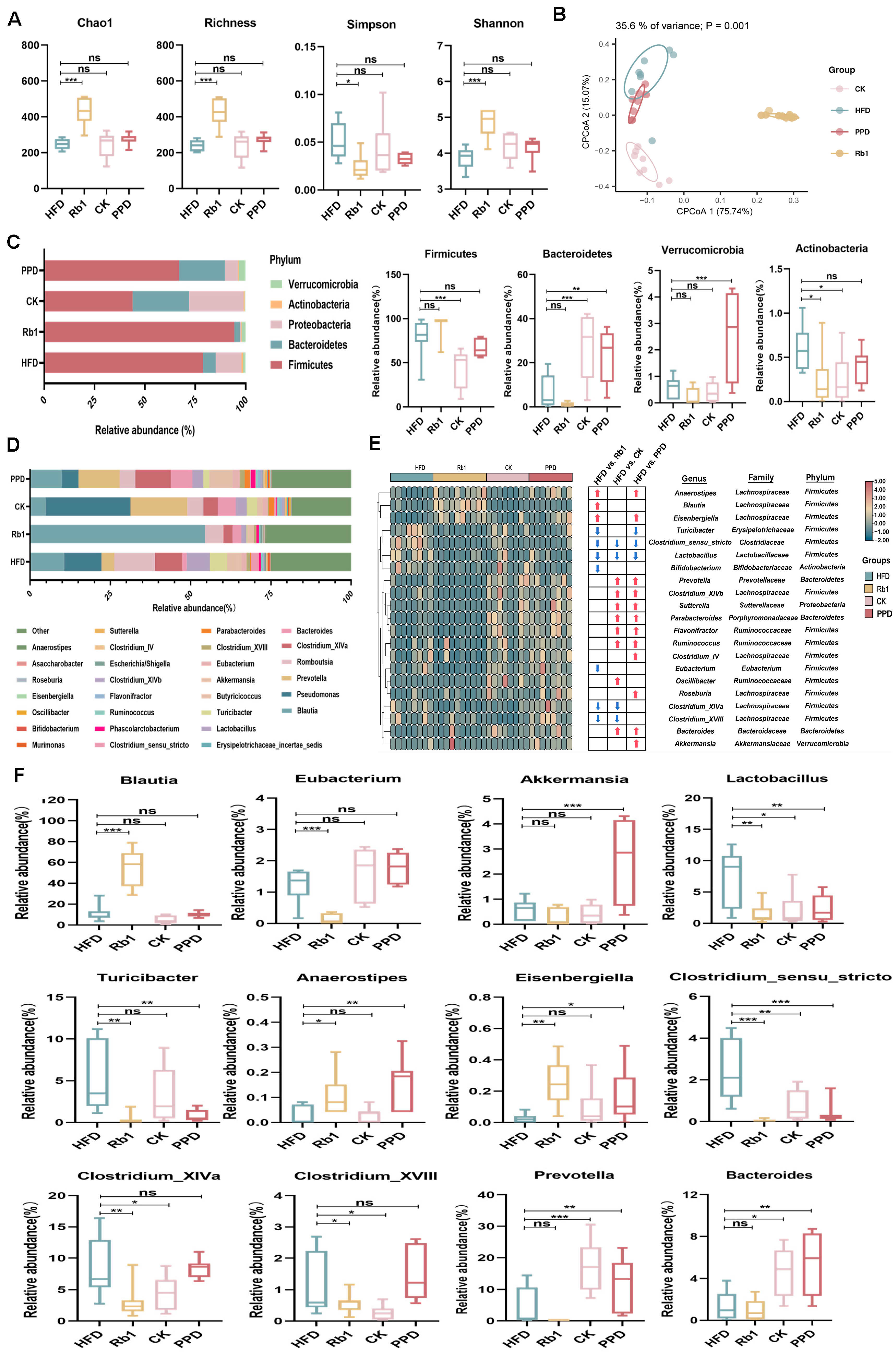

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, K.-X.; Zhu, Y.; Song, S.-X.; Bu, Q.-Y.; You, X.-Y.; Zou, H.; Zhao, G.-P. Ginsenoside Rb1, Compound K and 20(S)-Protopanaxadiol Attenuate High-Fat Diet-Induced Hyperlipidemia in Rats via Modulation of Gut Microbiota and Bile Acid Metabolism. Molecules 2024, 29, 1108. https://doi.org/10.3390/molecules29051108
Zhang K-X, Zhu Y, Song S-X, Bu Q-Y, You X-Y, Zou H, Zhao G-P. Ginsenoside Rb1, Compound K and 20(S)-Protopanaxadiol Attenuate High-Fat Diet-Induced Hyperlipidemia in Rats via Modulation of Gut Microbiota and Bile Acid Metabolism. Molecules. 2024; 29(5):1108. https://doi.org/10.3390/molecules29051108
Chicago/Turabian StyleZhang, Kang-Xi, Yue Zhu, Shu-Xia Song, Qing-Yun Bu, Xiao-Yan You, Hong Zou, and Guo-Ping Zhao. 2024. "Ginsenoside Rb1, Compound K and 20(S)-Protopanaxadiol Attenuate High-Fat Diet-Induced Hyperlipidemia in Rats via Modulation of Gut Microbiota and Bile Acid Metabolism" Molecules 29, no. 5: 1108. https://doi.org/10.3390/molecules29051108
APA StyleZhang, K.-X., Zhu, Y., Song, S.-X., Bu, Q.-Y., You, X.-Y., Zou, H., & Zhao, G.-P. (2024). Ginsenoside Rb1, Compound K and 20(S)-Protopanaxadiol Attenuate High-Fat Diet-Induced Hyperlipidemia in Rats via Modulation of Gut Microbiota and Bile Acid Metabolism. Molecules, 29(5), 1108. https://doi.org/10.3390/molecules29051108




