Lignin Degradation by Klebsiella aerogenes TL3 under Anaerobic Conditions
Abstract
1. Introduction
2. Results
2.1. Growth of TL3 on Various Carbon Sources
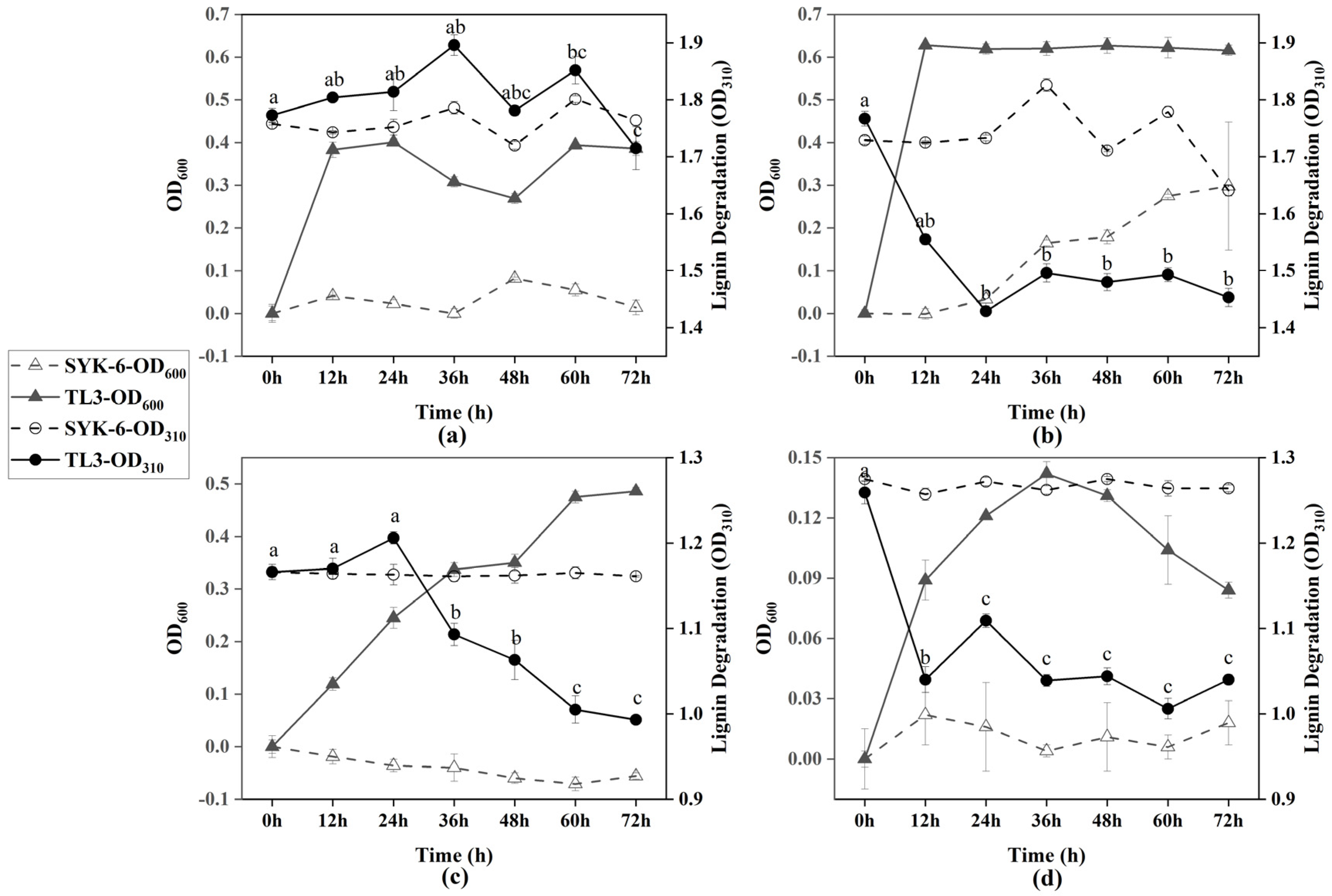
2.2. The Lignin Degradation Curve
2.3. FTIR Plots of Lignin Degradation by TL3 under Various Conditions
2.4. GC-MS Analysis of the Metabolites
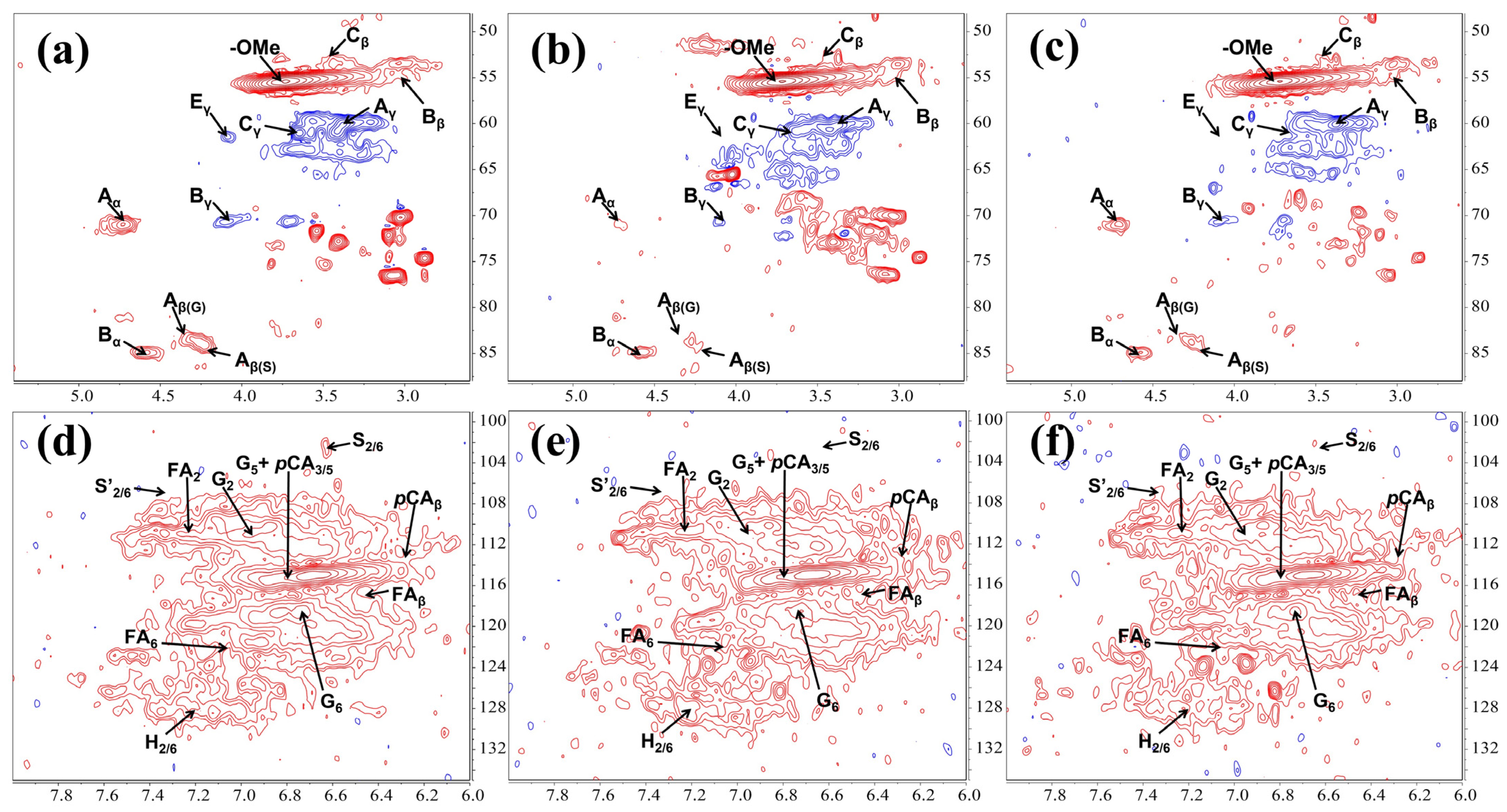
2.5. 2D-HSQC NMR Analysis of the Residual Lignin
3. Discussion
4. Materials and Methods
4.1. Strain and Media
4.2. Metabolic Characterization of Different Carbon Sources
4.3. Time Courses of Bacterial Growth and Lignin Degradation under Anaerobic Conditions
4.4. Fourier-Transform Infrared Spectroscopy (FTIR) Analysis
4.5. Gas Chromatography–Mass Spectrometry (GC-MS) Analysis
4.6. Nuclear Magnetic Resonance (NMR) Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nanda, S.; Azargohar, R.; Dalai, A.K.; Kozinski, J.A. An assessment on the sustainability of lignocellulosic biomass for biorefining. Renew. Sustain. Energy Rev. 2015, 50, 925–941. [Google Scholar] [CrossRef]
- Upton, B.M.; Kasko, A.M. Strategies for the conversion of lignin to high-value polymeric materials: Review and perspective. Chem. Rev. 2016, 116, 2275–2306. [Google Scholar] [CrossRef] [PubMed]
- Andlar, M.; Rezic, T.; Mardetko, N.; Kracher, D.; Ludwig, R.; Santek, B. Lignocellulose degradation: An overview of fungi and fungal enzymes involved in lignocellulose degradation. Eng. Life Sci. 2018, 18, 768–778. [Google Scholar] [CrossRef] [PubMed]
- Weng, C.; Peng, X.; Han, Y. Depolymerization and conversion of lignin to value-added bioproducts by microbial and enzymatic catalysis. Biotechnol. Biofuels 2021, 14, 84. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Li, X.; Zou, L.; Zheng, Z.; Ouyang, J. Biological Valorization of Lignin-Derived Aromatics in Hydrolysate to Protocatechuic Acid by Engineered Pseudomonas putida KT2440. Molecules 2024, 29, 1555. [Google Scholar] [CrossRef] [PubMed]
- Mate, D.M.; Alcalde, M. Laccase engineering: From rational design to directed evolution. Biotechnol. Adv. 2015, 33, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Blaschek, L.; Murozuka, E.; Serk, H.; Ménard, D.; Pesquet, E. Different combinations of laccase paralogs nonredundantly control the amount and composition of lignin in specific cell types and cell wall layers in Arabidopsis. Plant Cell 2023, 35, 889–909. [Google Scholar] [CrossRef] [PubMed]
- Ragauskas, A.J.; Beckham, G.T.; Biddy, M.J.; Chandra, R.; Chen, F.; Davis, M.F.; Davison, B.H.; Dixon, R.A.; Gilna, P.; Keller, M. Lignin valorization: Improving lignin processing in the biorefinery. Science 2014, 344, 1246843. [Google Scholar] [CrossRef] [PubMed]
- Janusz, G.; Pawlik, A.; Sulej, J.; Świderska-Burek, U.; Jarosz-Wilkołazka, A.; Paszczyński, A. Lignin degradation: Microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 2017, 41, 941–962. [Google Scholar] [CrossRef]
- De Gonzalo, G.; Colpa, D.I.; Habib, M.H.M.; Fraaije, M.W. Bacterial enzymes involved in lignin degradation. J. Biotechnol. 2016, 236, 110–119. [Google Scholar] [CrossRef]
- Neumann, G.T.; Pimentel, B.R.; Rensel, D.J.; Hicks, J.C. Correlating lignin structure to aromatic products in the catalytic fast pyrolysis of lignin model compounds containing β–O–4 linkages. Catal. Sci. Technol. 2014, 4, 3953–3963. [Google Scholar] [CrossRef]
- Duan, J.; Huo, X.; Du, W.J.; Liang, J.D.; Wang, D.Q.; Yang, S.C. Biodegradation of kraft lignin by a newly isolated anaerobic bacterial strain, Acetoanaerobium sp. WJDL-Y2. Lett. Appl. Microbiol. 2016, 62, 55–62. [Google Scholar] [CrossRef]
- Rashid, G.M.M.; Duran-Pena, M.J.; Rahmanpour, R.; Sapsford, D.; Bugg, T.D.H. Delignification and enhanced gas release from soil containing lignocellulose by treatment with bacterial lignin degraders. J. Appl. Microbiol. 2017, 123, 159–171. [Google Scholar] [CrossRef]
- Deangelis, K.M.; Sharma, D.; Varney, R.; Simmons, B.; Isern, N.G.; Markilllie, L.M.; Nicora, C.; Norbeck, A.D.; Taylor, R.C.; Aldrich, J.T.; et al. Evidence supporting dissimilatory and assimilatory lignin degradation in Enterobacter lignolyticus SCF1. Front. Microbiol. 2013, 4, 280. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.L.; Ballor, N.R.; Hazen, T.C.; Fortney, J.L.; Simmons, B.; Davenport, K.W.; Goodwin, L.; Ivanova, N.; Kyrpides, N.C.; Mavromatis, K. Complete genome sequence of the lignin-degrading bacterium Klebsiella sp. strain BRL6-2. Stand. Genom. Sci. 2014, 9, 19. [Google Scholar] [CrossRef]
- Billings, A.F.; Fortney, J.L.; Hazen, T.C.; Simmons, B.; Davenport, K.W.; Goodwin, L.; Ivanova, N.; Kyrpides, N.C.; Mavromatis, K.; Woyke, T.; et al. Genome sequence and description of the anaerobic lignin-degrading bacterium Tolumonas lignolytica sp. nov. Stand. Genom. Sci. 2015, 10, 106. [Google Scholar] [CrossRef] [PubMed]
- Chaput, G.; Ford, J.; DeDiego, L.; Narayanan, A.; Tam, W.Y.; Whalen, M.; Huntemann, M.; Clum, A.; Spunde, A.; Pillay, M. Sodalis ligni strain 159R isolated from an anaerobic lignin-degrading consortium. Microbiol. Spectr. 2022, 10, e02346-21. [Google Scholar] [CrossRef]
- Vishnivetskaya, T.A.; Raman, B.; Phelps, T.J.; Podar, M.; Elkins, J.G. Cellulolytic microorganisms from thermal environments. In Extremophiles: Microbiology and Biotechnology, 1st ed.; Anitori, R.P., Ed.; Caister Academic Press: Norfolk, UK, 2012; pp. 131–158. [Google Scholar]
- Silva, J.P.; Ticona, A.R.P.; Hamann, P.R.V.; Quirino, B.F.; Noronha, E.F. Deconstruction of lignin: From enzymes to microorganisms. Molecules 2021, 26, 2299. [Google Scholar] [CrossRef] [PubMed]
- Bugg, T.D.; Ahmad, M.; Hardiman, E.M.; Singh, R. The emerging role for bacteria in lignin degradation and bio-product formation. Curr. Opin. Biotechnol. 2011, 22, 394–400. [Google Scholar] [CrossRef]
- Khan, M.U.; Ahring, B.K. Lignin degradation under anaerobic digestion: Influence of lignin modifications—A review. Biomass Bioenerg. 2019, 128, 105325. [Google Scholar] [CrossRef]
- Nawaz, M.Z.; Shang, H.; Sun, J.; Geng, A.; Ali, S.S.; Zhu, D. Genomic insights into the metabolic potential of a novel lignin-degrading and polyhydroxyalkanoates producing bacterium Pseudomonas sp. Hu109A. Chemosphere 2023, 310, 136754. [Google Scholar] [CrossRef] [PubMed]
- Bugg, T.D.H.; Ahmad, M.; Hardiman, E.M.; Rahmanpour, R. Pathways for degradation of lignin in bacteria and fungi. Nat. Prod. Rep. 2011, 28, 1883–1896. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Liang, J.; Wang, Y.; Du, W.; Wang, D. Kraft Lignin Biodegradation by Dysgonomonas sp. WJDL-Y1, a new anaerobic bacterial strain isolated from sludge of a pulp and paper mill. J. Microbiol. Biotechnol. 2016, 26, 1765–1773. [Google Scholar] [CrossRef] [PubMed]
- Lankiewicz, T.S.; Choudhary, H.; Gao, Y.; Amer, B.; Lillington, S.P.; Leggieri, P.A.; Brown, J.L.; Swift, C.L.; Lipzen, A.; Na, H.; et al. Lignin deconstruction by anaerobic fungi. Nat. Microbiol. 2023, 8, 596–610. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Peng, B.; Kitata, R.B.; Nicora, C.D.; Weitz, K.K.; Pu, Y.; Shi, T.; Cort, J.R.; Ragauskas, A.J.; Yang, B. Understanding of bacterial lignin extracellular degradation mechanisms by Pseudomonas putida KT2440 via secretomic analysis. Biotechnol. Biofuels Bioprod. 2022, 15, 117. [Google Scholar] [CrossRef] [PubMed]
- Masai, E.; Katayama, Y.; Fukuda, M. Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci. Biotech. Bioch. 2007, 71, 1–15. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Cheng, Y.; Zang, H.; Li, C. Biodegradation characteristics of lignin in pulping wastewater by the thermophilic Serratia sp. AXJ-M: Performance, genetic background, metabolic pathway and toxicity assessment. Environ. Pollut. 2023, 322, 121230. [Google Scholar] [CrossRef] [PubMed]
- Suman, S.K.; Dhawaria, M.; Tripathi, D.; Raturi, V.; Adhikari, D.K.; Kanaujia, P.K. Investigation of lignin biodegradation by Trabulsiella sp. isolated from termite gut. Int. Biodeter. Biodegr. 2016, 112, 12–17. [Google Scholar] [CrossRef]
- Lee, S.; Kang, M.; Bae, J.-H.; Sohn, J.-H.; Sung, B.H. Bacterial valorization of lignin: Strains, enzymes, conversion pathways, biosensors, and perspectives. Front. Bioeng. Biotechnol. 2019, 7, 209. [Google Scholar] [CrossRef]
- Li, N.; Geng, A.; Tu, Z.; Fan, Y.; Xie, R.; Li, X.; Sun, J. Isolation of Lactococcus sp. X1 from termite gut, and its application in lactic acid production. Fermentation 2023, 9, 85. [Google Scholar] [CrossRef]
- Ji, Q.; Yu, X.; Wu, P.; Yagoub, A.E.-G.A.; Chen, L.; Taiye, M.A.; Zhou, C. Pretreatment of sugarcane bagasse with deep eutectic solvents affect the structure and morphology of lignin. Ind. Crop Prod. 2021, 173, 114108. [Google Scholar] [CrossRef]

| Methods | Fermentation Products (g/L) | Carbon Sources | ||||
|---|---|---|---|---|---|---|
| Avicel | Glucose | Xylose | Corn Cob | Lignin | ||
| Anaerobic | Lactic Acid | ND a | 0.73 ± 0.01 | 0.85 ± 0.02 | 0.23 ± 0.01 | 0.13 ± 0.02 |
| Acetic Acid | ND | 1.25 ± 0.02 | 0.52 ± 0.01 | ND | ND | |
| Succinic Acid | 0.64 ± 0.03 | 1.07 ± 0.00 | 1.69 ± 0.01 | 0.82 ± 0.02 | 1.43 ± 0.00 | |
| Propanoic acid | 0.26 ± 0.02 | 2.70 ± 0.03 | 0.32 ± 0.03 | ND | 0.26 ± 0.01 | |
| Butyric Acid | ND | 1.38 ± 0.02 | ND | ND | ND | |
| Aerobic | Lactic Acid | ND | ND | ND | ND | ND |
| Acetic Acid | ND | ND | ND | ND | ND | |
| Succinic Acid | ND | ND | ND | ND | ND | |
| Propanoic acid | ND | 3.11 ± 0.01 | ND | ND | ND | |
| Butyric Acid | ND | ND | 1.81 ± 0.02 | ND | ND | |
| NO. | RT (min) | Compounds | Treatment | ||
|---|---|---|---|---|---|
| Control | TL3 | TL3 + Glucose | |||
| 1 | 1.344 | Acetic acid, (propylthio)- | − | + | − |
| 2 | 2.879 | 3-Ethylheptanoic acid | − | − | + |
| 3 | 3.256 | Propanoic acid, ethyl ester | − | + | + |
| 4 | 3.400 | Formic acid, butyl ester | − | + | + |
| 5 | 6.400 | p-Xylene | − | + | − |
| 6 | 9.766 | L-(+)-Lactic acid | − | + | − |
| 7 | 11.921 | 1,3-Butanediol, diacetate | − | + | − |
| 8 | 13.398 | Benzaldehyde, 3,4-dimethyl- | + | + | − |
| 9 | 13.944 | Oxalic acid, 6-ethyloct-3-yl isobutyl ester | − | + | − |
| 10 | 14.132 | Heptadecane, 8-methyl- | + | + | − |
| 11 | 15.287 | Butyric acid, 4-pentadecyl ester | − | + | − |
| 12 | 15.587 | Butanoic acid, octyl ester | − | + | − |
| 13 | 17.032 | Octacosane | + | + | − |
| 14 | 17.288 | Phenol, 2,5-bis(1,1-dimethylethyl)- | − | + | + |
| 15 | 18.943 | Pentadecane, 2,6,10-trimethyl- | − | + | − |
| 16 | 19.532 | Hexane, 2,4-dimethyl- | − | + | − |
| 17 | 19.543 | Heneicosane | + | + | − |
| 18 | 20.786 | 3-Vanilpropanol, bis(trimethylsilyl)- | − | + | − |
| 19 | 21.286 | Phthalic acid, isobutyl undecyl ester | − | + | + |
| 20 | 21.798 | Heptacosane | − | + | − |
| 21 | 21.863 | 7,9-Di-tert-butyl-1-oxaspiro(4,5)deca-6,9-diene-2,8-dione | + | − | − |
| 22 | 21.942 | Methoxyacetic acid, 3-tridecyl ester | + | + | − |
| 23 | 22.020 | Benzenepropanoic acid, 3,5-bis(1,1-dimethylethyl)-4-hydroxy-, methyl ester | − | + | − |
| 24 | 22.287 | n-Hexadecanoic acid | + | + | − |
| 25 | 22.565 | Benzoic acid, 3-[(2,2-dimethyl-1-oxopropyl)amino]- | − | + | − |
| 26 | 24.187 | Hexadecanoic acid, 2-hydroxy-, methyl ester | − | + | − |
| 27 | 24.508 | Octadecanoic acid | + | + | − |
| 28 | 26.109 | Hexanedioic acid, bis(2-ethylhexyl) ester | − | + | − |
| 29 | 26.309 | Phenol, 2,2′-methylenebis[6-(1,1-dimethylethyl)-4-methyl- | − | + | + |
| 30 | 26.686 | 2-Nonenoic acid, methyl ester | − | + | − |
| 31 | 26.799 | Benzenepropanoic acid, 3,5-bis(1,1-dimethylethyl)-4-hydroxy-, octadecyl ester | − | − | + |
| 32 | 27.142 | Hexadecanoic acid, 2-hydroxy-1-(hydroxymethyl)ethyl ester | − | + | − |
| 33 | 27.253 | 1,2-Benzenedicarboxylic acid, dioctyl ester | − | + | − |
| 34 | 27.797 | Benzoic acid, 2,5-bis(trimethylsiloxy)-, trimethylsilyl ester | + | + | − |
| 35 | 28.508 | Butanedioic acid, 2-hydroxy-2-methyl-, dimethyl ester | − | + | − |
| 36 | 28.819 | Octadecanoic acid, 2-hydroxy-1-(hydroxymethyl)ethyl ester | − | + | − |
| 37 | 28.897 | Terephthalic acid, di(2-ethylhexyl) ester | − | + | − |
| 38 | 28.842 | Fumaric acid, dodecyl nonyl ester | − | + | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tu, Z.; Geng, A.; Xiang, Y.; Zayas-Garriga, A.; Guo, H.; Zhu, D.; Xie, R.; Sun, J. Lignin Degradation by Klebsiella aerogenes TL3 under Anaerobic Conditions. Molecules 2024, 29, 2177. https://doi.org/10.3390/molecules29102177
Tu Z, Geng A, Xiang Y, Zayas-Garriga A, Guo H, Zhu D, Xie R, Sun J. Lignin Degradation by Klebsiella aerogenes TL3 under Anaerobic Conditions. Molecules. 2024; 29(10):2177. https://doi.org/10.3390/molecules29102177
Chicago/Turabian StyleTu, Zhuowei, Alei Geng, Yuhua Xiang, Anaiza Zayas-Garriga, Hao Guo, Daochen Zhu, Rongrong Xie, and Jianzhong Sun. 2024. "Lignin Degradation by Klebsiella aerogenes TL3 under Anaerobic Conditions" Molecules 29, no. 10: 2177. https://doi.org/10.3390/molecules29102177
APA StyleTu, Z., Geng, A., Xiang, Y., Zayas-Garriga, A., Guo, H., Zhu, D., Xie, R., & Sun, J. (2024). Lignin Degradation by Klebsiella aerogenes TL3 under Anaerobic Conditions. Molecules, 29(10), 2177. https://doi.org/10.3390/molecules29102177






