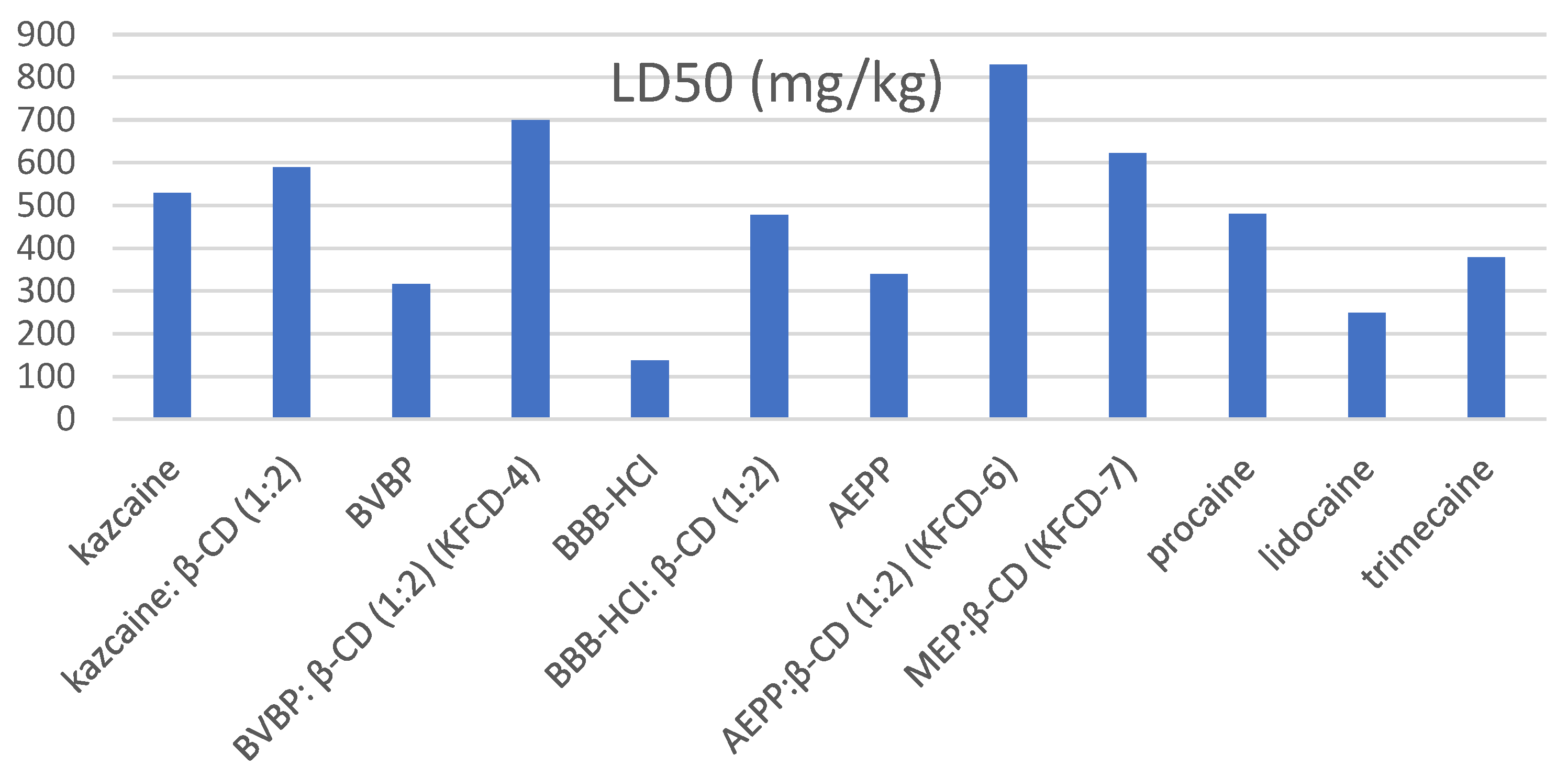2.1. In Silico Pharmacology
In drug development, efficient target binding is not only important, but it also ensures oral bioavailability and drug-like properties. In this regard, the study of the physicochemical properties of compounds is crucial for drug development.
The predictive analysis and in silico studies of possible targets, ADME parameters (absorption, distribution, metabolism, and excretion), and compliance with the bioavailability criteria [
11,
22] were carried out for the studied compounds.
An analysis of the structures for compliance with Lipinski’s rule of five (molecular weights (MW) ≤ 500, cLogP ≤ 5.0, TPSA ≤ 140 Å
2, number of H-acceptors ≤ 10, H-donors ≤ 5) [
23,
24] was performed, using the SwissADME software package [
25]. Compliance with Lipinski’s rule makes the compounds active drug candidates. The substance is unlikely to become an active drug candidate if Lipinski’s rule is violated even by one parameter.
The analysis of lipophilicity (LogP) is provided in
Table 2. Optimal values for LogP (P is the partition coefficient of all forms of the molecule between n-octanol and water) are between 0 and 3. LogP < 0 corresponds to the bad permeability of the lipid bilayer; LogP > 3 indicates poor water solubility [
26]. Compounds with high cLogP values may have difficulty in achieving the therapeutic targets due to their lipophilicity, which potentially limits their effectiveness.
The LogP value shows moderately good (0.33) absorption and permeability for the MEP. For EEHP and MEBP, the cLogP values are 1.07 and 2.42, respectively. For the other compounds, the distribution coefficient is significantly higher and ranges from 3.30 to 4.93. More positive cLogP values usually indicate a higher concentration of the compound in the lipid phase.
LogS values (logarithm of water solubility value, expressed in log mol/L) above −4 logmol/L and below 10 µg/mL indicate low solubility. In the range of 10–60 μg/mL, the compounds have moderate solubility. All LogS values higher than 60 µg/mL indicate high solubility [
27].
The TPSA parameter for EEHP, MEBP, and MEP has a low value of 23.47 Å
2 and meets the criteria for oral bioavailability. The MEP compound meets the Lipinski, Egan, and Weber criteria. The Egan filter (Pharmacia filter) is based on the LogP and TPSA parameters. It anticipates drug absorption, depending on the processes involved in the membrane permeability of a small molecule, and considers the molecule drug-like if it has WLOGP ≤ 5.88 and TPSA ≤ 131.6, respectively [
28]. The Muegge filter (the Bayer filter) is the independent pharmacophore point filter that separates drug-like and non-drug-like molecules. The Ghose filter (Amgen) describes small molecules based on their physicochemical properties and the existence of functional groups and substructures [
28]. EEHP only fails to meet the Muegge criteria due to its low molecular weight. BBB·HCl has failed to meet the Ghose criteria because the calculation was carried out for a hydrochloride form. The remaining compounds correspond to all the criteria provided in
Table 3.
All compounds have shown favorable bioavailability values (0.55). This indicates good suitability for oral drug administration and implies achieving a therapeutic result at lower concentrations.
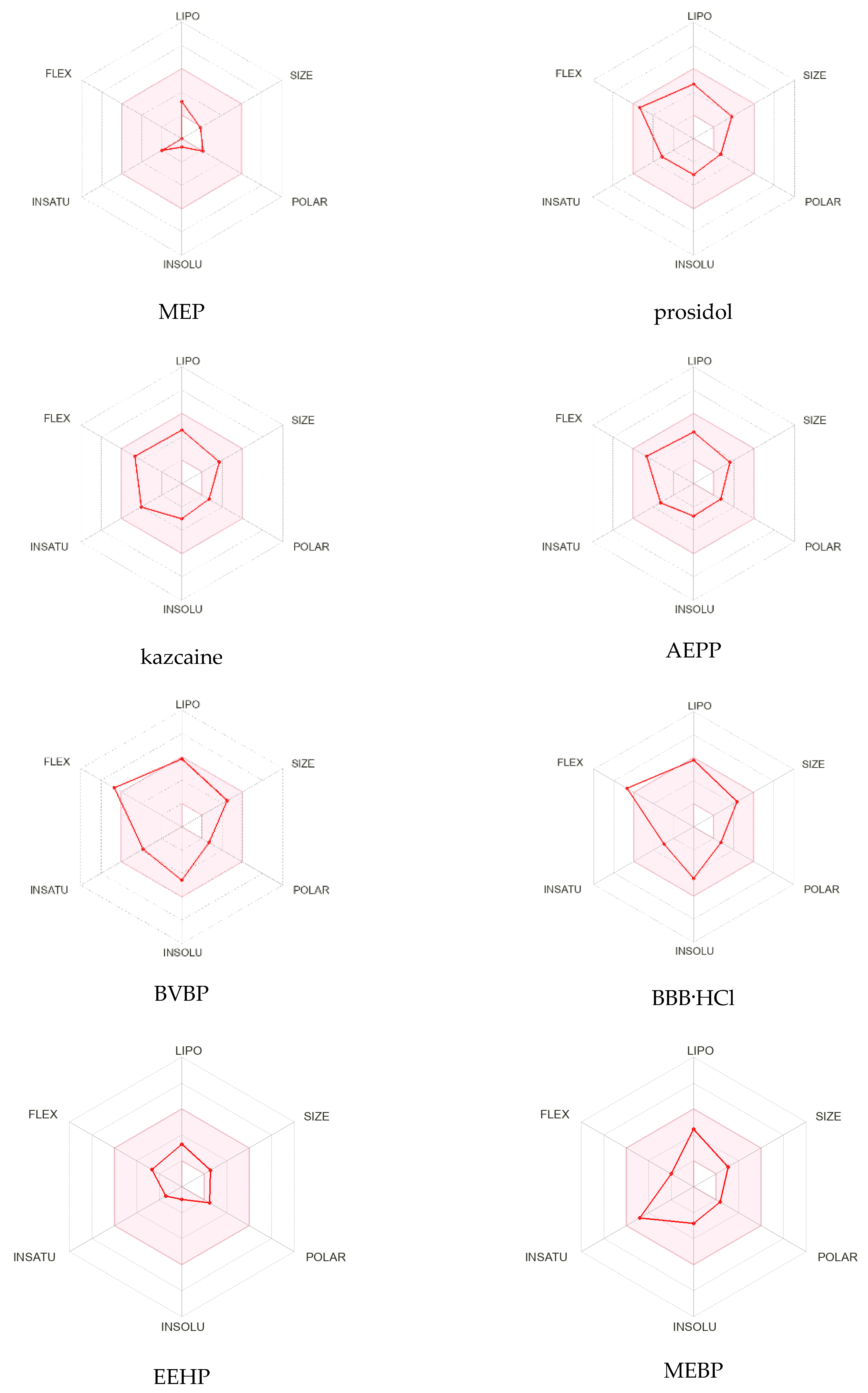
The radar diagrams (
Figure 2) show the distribution of the physicochemical properties of the compounds: lipophilicity (LIPO), size (SIZE), polarity (POLAR), solubility (INSOLU), saturation (INSATU), elasticity (FLEX), presence of donors (nHD), and proton acceptors (nHA). The pink area represents the optimal range for each property (lipophilicity: XLOGP3 −0.7 to +5.0, size: molecular weight 150 to 500 g/mol, polarity: TPSA 20 to 130 Å
2, solubility: log S not above 6, saturation: the fraction of carbons in sp
3 hybridization is at least 0.25 and flexibility no more than nine rotating bonds) [
25]. The analyses of the diagrams show that prosidol, kazcaine, and AEPP have the best distribution of parameters, though all the compounds, in principle, meet the requirements for a medicinal substance. BVBP and BBB have a slight excess in the FLEX parameter, and for MEP, the size, polarity, and flexibility indicators are at the lower limit.
To predict possible biological effects, the open software products PASS Online, AntiBac-Pred, and AntiFun Pred [
29,
30,
31] were used. Here, and below, the score function F = Pa − Pi is used, which is the difference in the probabilities that a substance will be active (Pa) or inactive (Pi) for the corresponding biological activity.
In
Table 4, the results for MEP are provided (for F > 0.1). Based on these data, the most probable biological activity of MEP is the suppression of ovulation; there is also a very high probability of its influence on the hormones responsible for reproductive functions. The substance can be used as an anticonvulsant. The other activities (anesthetic, anabolic, nootropic, antidepressant, analgesic, and muscle relaxant) have a rather low probability. Comparative data on the major types of activity for all the substances under consideration are provided in
Table 5. The results are provided for the substances in the form of bases since the calculation programs, in most cases, cannot work with the substances in the form of salts and complex compounds (including inclusion complexes).
Possible protein targets (for
Homo sapiens) were evaluated using the Swiss Target Prediction service. The results are shown in
Table 6. The score for each target is called “confidence”, which is the difference between probabilities of chemical compounds interacting and not interacting with a particular target. Higher confidence means a higher chance of a positive prediction being true. The first 5–6 results are listed and the rest are provided in the
Supplementary Materials. The probabilities for MEP are very low, but we can conclude that the substance may affect mechanisms that occur in the central nervous system.
The PASS Targets program provides a slightly different prediction of possible molecular targets. It is advisable to consider results with a confidence value greater than 0.5.
Table 7 shows the values greater than 0.5 for MEP, EEHP, and MEBP and greater than 0.25 for the remaining compounds. The full list is presented in
Table S1.
According to
Table 7, MEP has the largest number of possible targets with a confidence value greater than 0.5. It looks most similar to kazcaine according to the list of possible targets, though the character of the data obtained (a large number of targets and high probability values) should rather be considered an anomaly. The substances MEP, EEHP, and MEBP actively bind to protein kinases.
In silico prediction of acute toxicity values (LD50) for rats for four types of administration (oral, intravenous, intraperitoneal, subcutaneous, and inhalation) was carried out using the GUSAR program [
32]. This program compares the structure of a substance with structures from the SYMYX MDL toxicity database. In order to assess which of these drugs best corresponds to the optimal characteristics required for an ideal drug, the acute toxicity parameter LD50 (known as the “lethal dose, 50%” or oral acute dose for rats) was calculated. High toxicity was indicated by values of 1–50 mg/kg; average toxicity was in the range of 51–500 mg/kg. Low toxicity values were 501–5000 mg/kg [
33]. The GUSAR program could not calculate data for BBB HCl in the form of either hydrochloride (which is expected) or a base.
The acute toxic class is provided according to the OECD. Low concentrations of the substance reduce the risk of side effects and toxicity. Analyzing the data in
Table 8, it can be argued that the acute toxicity values of the compounds exceed the values of the average toxicity range for the compounds prosidol, AEPP, and BVBP. MEP showed a fairly low predicted toxicity risk for intraperitoneal, intravenous, and subcutaneous administration but higher toxicity for all routes of administration compared to the other study drugs.
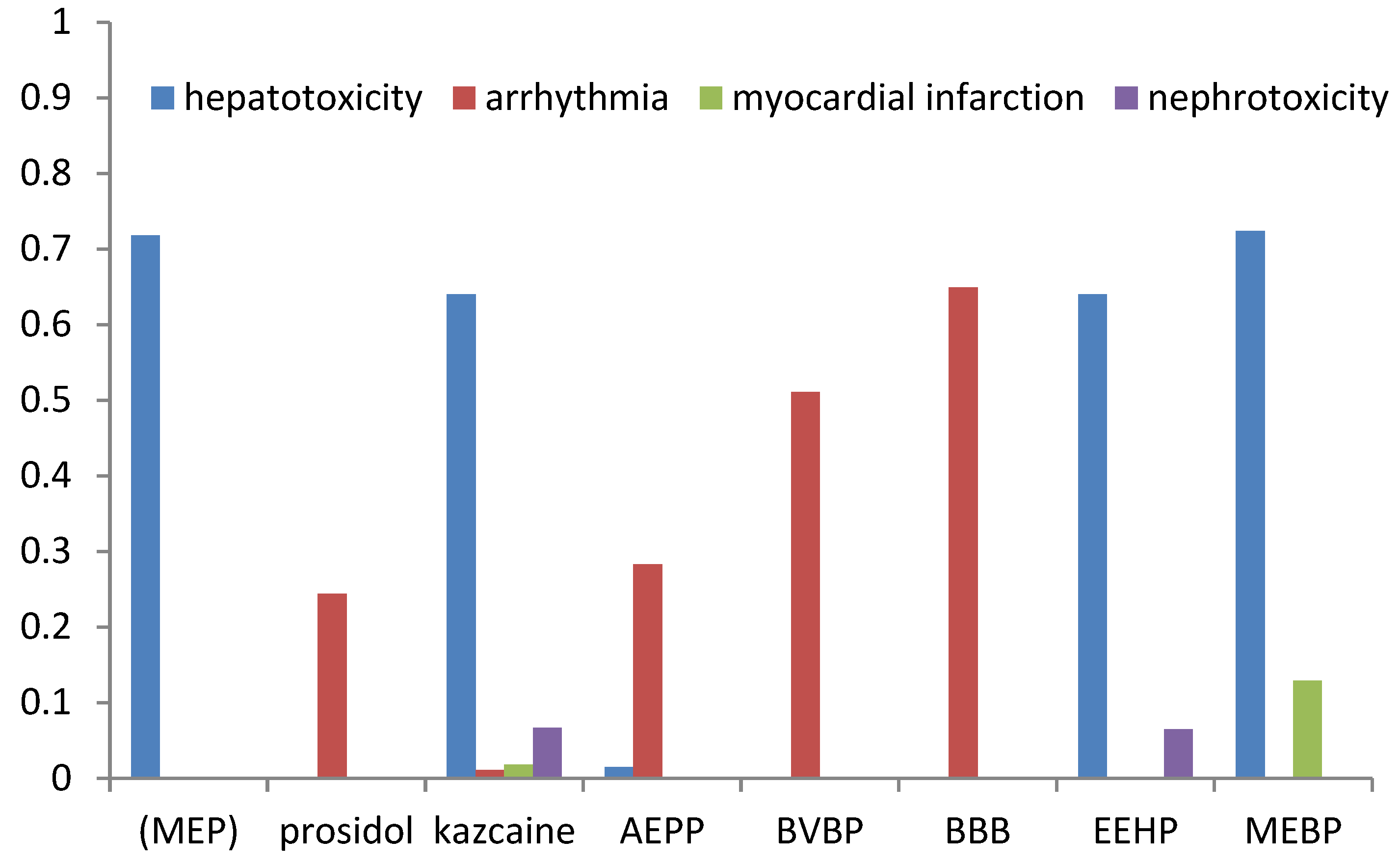
The prognosis of adverse effects (arrhythmia, heart failure, hepatotoxicity, myocardial infarction, and nephrotoxicity) was made using ADVER Pred [
34]. The results are shown in
Table 9 and
Figure 3.
The compounds may exhibit side effects such as arrhythmia (prosidol, kazcaine, AEPP, BVBP, and BBB), hepatotoxicity (MEP, kazcaine, AEPP, EEHP, and MEBP), myocardial infarction (kazkain and MEBP), and nephrotoxicity (MEBP and EEHP). Kazcaine was predicted to cause the highest number of adverse effects compared to the other compounds. However, their probability, excluding hepatotoxicity, was low. The calculated results also indicate a high probability of hepatotoxicity for MEP. For most compounds, a high probability of arrhythmia was predicted as an adverse effect. In order to improve the bioavailability parameters and reduce the toxic side effects, it is advisable to use active compounds in the form of inclusion complexes with cyclodextrin.
2.2. Host–Guest Complexes with β-Cyclodextrin
The severity of adverse effects, such as hepatotoxicity and nephrotoxicity, can be reduced using drug inclusion complexes with β-cyclodextrin. Cyclodextrins usually improve the solubility of guest molecules in water, significantly reduce their toxicity, and increase the period of action due to the slow dissociation of the inclusion complex in the body.
Usually, the drugs are used not in a pure form but in a so-called “dosage form”. For example, water-soluble drugs are used in the form of isotonic solutions containing a local anesthetic, while fat-soluble drugs are administered subcutaneously in the form of an oil solution, from which the drug slowly passes into the interstitial fluid.
Earlier, piperidines have been often used as water-soluble salt forms, such as hydrochlorides, to prepare useful dosage forms.
However, along with a high anesthetic effect, such dosage forms also have significant toxicity. Therefore, the preparation of new dosage forms with minimal adverse effects is an actual problem.
The preparation of new dosage forms based on inclusion (host–guest) complexes of cyclodextrins seems to be a promising solution to the problem.
Inclusion complexes are effective as delivery tools. With the conventional type of administration, only nearly one-tenth of the drug molecules can reach the site of application (nerves, tumors, etc.). When the drug is delivered in the form of an inclusion complex and released directly near the site of application, the effective local concentration is increased. Therefore, less amount of drug is required, which can also reduce overall toxicity.
In our previous works [
7,
13,
14,
15,
16,
17,
18,
19,
20,
21], we reported the preparation of host–guest complexes of the above piperidines with β-CD and studied their structure (
Table 10). All the compounds except MEP formed inclusion complexes with a guest–host ratio of 1:2. For MEP, the 1:1 complex was isolated, which is most likely due to the smaller size of the guest molecule.
The structures of inclusion complexes were studied by NMR during their complex formation in the solutions as well as by X-ray diffraction in their crystalline form. Due to the flexibility of the piperidine ring, piperidines can exist in two main conformations. In inclusion complexes, they can either remain in their starting conformation (for example, BBB) or have a different conformation compared to their free form (kazcaine and prosidol). In addition, in a solution (CDCl3 and D2O), BBB-HCl exists as two isomers in a 2:1 ratio with different orientations of benzoyloxy groups: 1e-(3-n-butoxypropyl)-4a-benzoyloxypiperidine hydrochloride and 1e-(3-n-butoxypropyl)-4e-benzoyloxypiperidine hydrochloride. BBB-HCl forms inclusion complexes with β-CD with a stoichiometry of 2 β-CD:1 BBB-HCl. The same conformation also exists in the inclusion complex isolated in the solid form.
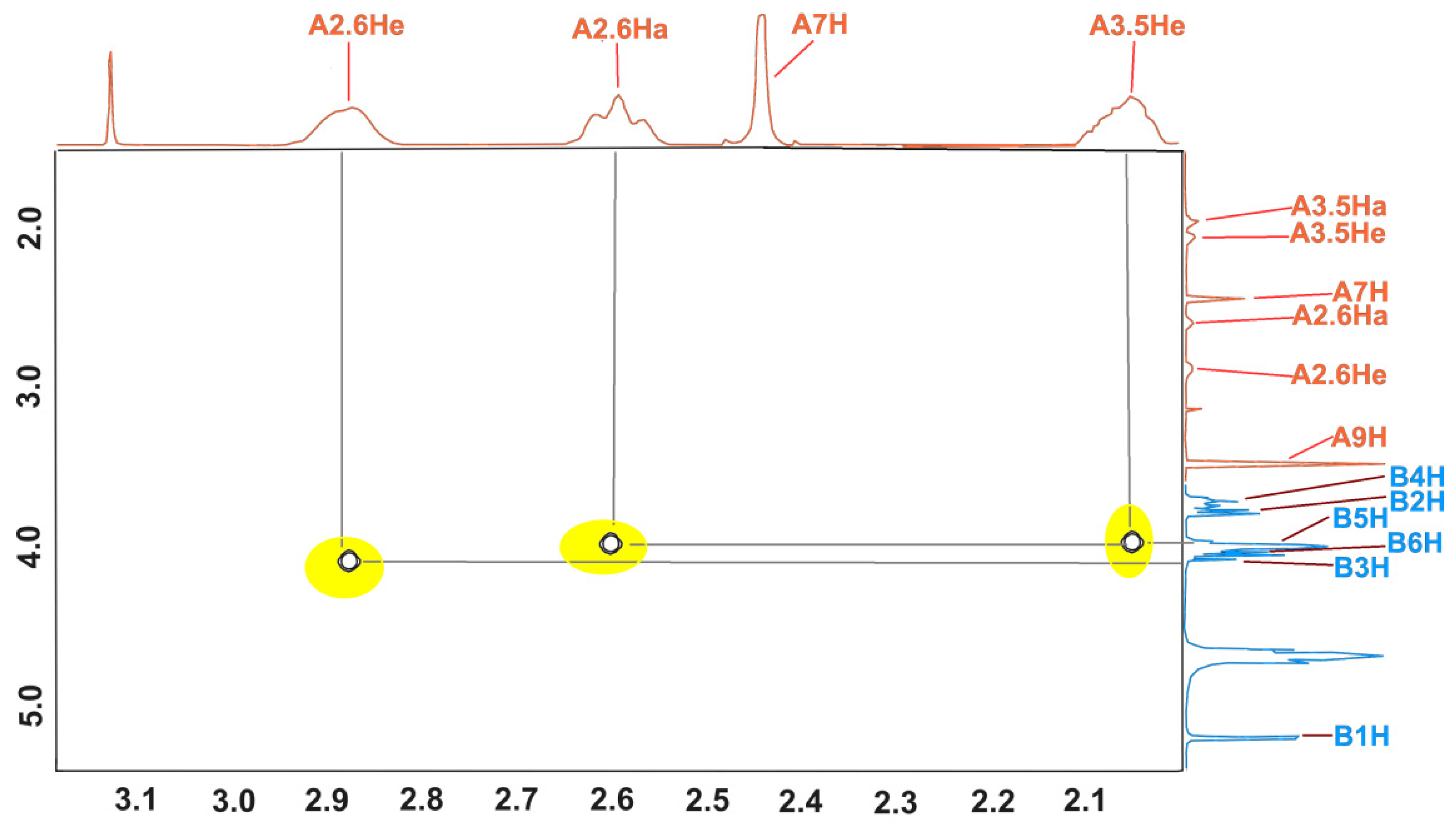
The structure of the inclusion complex of β-CD with MEP (KFCD-7) was studied using NMR and X-ray diffraction [
21]. Below (
Figure 4), the expansion from the ROESY NMR spectrum in addition to the data published earlier are shown. The cross peaks between inner (3 and 5) protons of β-CD and 2 and 6 protons of the piperidine ring clearly show that the structure of the MEP:β-CD complex in the solution corresponds to the one obtained from the X-ray data in the solid state.
An analysis of the predicted biological activity shows that MEP, as well as its β-CD inclusion complex, should be significantly different in biological activity from the other piperidine derivatives and their inclusion complexes. Because of that, we conducted a pharmacological study of KFCD-7 and compared its acute toxicity, infiltration, and conduction anesthesia with the data for previously obtained piperidine derivatives and reference drugs.
2.3. Pharmacological Study
2.3.1. Infiltration Anesthesia
The test was performed using the Bulbring–Wade method. All the compounds were tested as 0.5% aqua solutions. The results are summarized in
Table 11.
As we can see from
Table 11, all the drugs have an anesthetic effect that exceeds both novocaine and lidocaine. KFCD-7 shows a slightly longer duration of complete anesthesia than lidocaine, higher than trimecaine in terms of the anesthesia index (35.4 ± 1.3) and duration of complete anesthesia, but less in total duration of anesthesia. The other piperidine derivatives revealed the best values for all parameters of the infiltration anesthesia.
The only exception is kazcaine which has a duration of complete anesthesia comparable to lidocaine but a higher total duration of anesthesia. The formation of an inclusion complex significantly (two times) increases the duration of complete anesthesia up to the KFCD-6 value.
For BVBP and BBB-HCl, the formation of an inclusion complex does not improve their local anesthetic activity. In the case of BBB-HCl, the formation of an inclusion complex significantly (more than two times) reduces the duration of complete anesthesia, while for BVBP, this effect is not so dramatic. The duration of complete anesthesia increases in the following order: procaine < lidocaine < kazcaine < trimecaine < KFCD-7 < BBB-HCl:β-CD < KFCD-4 < KFCD-6 < kazcaine:β-CD < BVBP < BBB-HCl. Total duration of effect: procaine < lidocaine < KFCD-7 < trimecaine < KFCD-4 < kazcaine < KFCD-6 < BVBP < BBB-HCl:β-CD < BBB-HCl < kazcaine:β-CD.
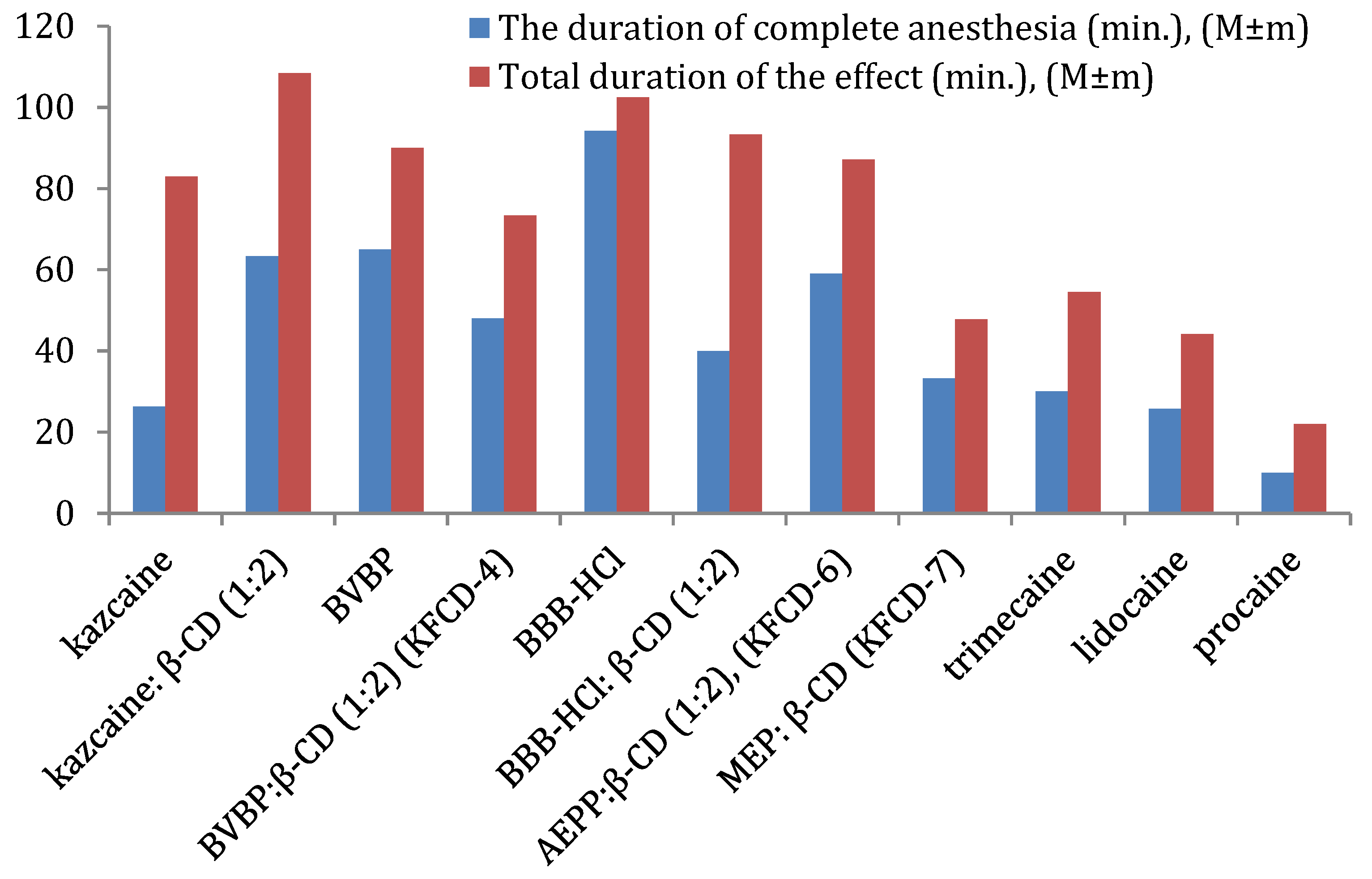
Overall (
Figure 5), the kazcaine:β-CD inclusion complex is comparable to BVBP, while KFCD-6 has a slightly shorter duration of complete anesthesia. KFCD-6 and KFCD-4 are better than procaine by 5.9 and 4.8 times, lidocaine by 2.3 and 1.9 times, and trimecaine by 2.0 and 1.6 times, respectively. They have a longer total duration of effect than trimecaine by approximately 2 and 1.7 times, lidocaine by 2.3 and 1.3 times, and procaine by 4.0 and 3.3 times, respectively (statistically significant at
p < 0.05).
2.3.2. Conduction Anesthesia
A modified “tail flick” method was used in the study of conduction anesthesia [
36]. It was developed at the Department of Pharmacology of the St. Petersburg Medical University, named after Academician I.P. Pavlov. The principle of the method is to determine the latent period of tail withdrawal during the thermal exposure of its middle part with a focused beam of light from an optoelectronic analgesimeter TF-003 before and after anesthesia. The intensity of the thermal nociceptive stimulus is adjusted so that initial tail flick responses occur with a latency ranging from 3 to 6 s.
The activity of compounds and reference drugs for the conduction anesthesia wasstudied in 1% solutions. The following parameters were determined: the rate of onset of anesthesia, the duration of the complete anesthesia, and the total duration of effect.
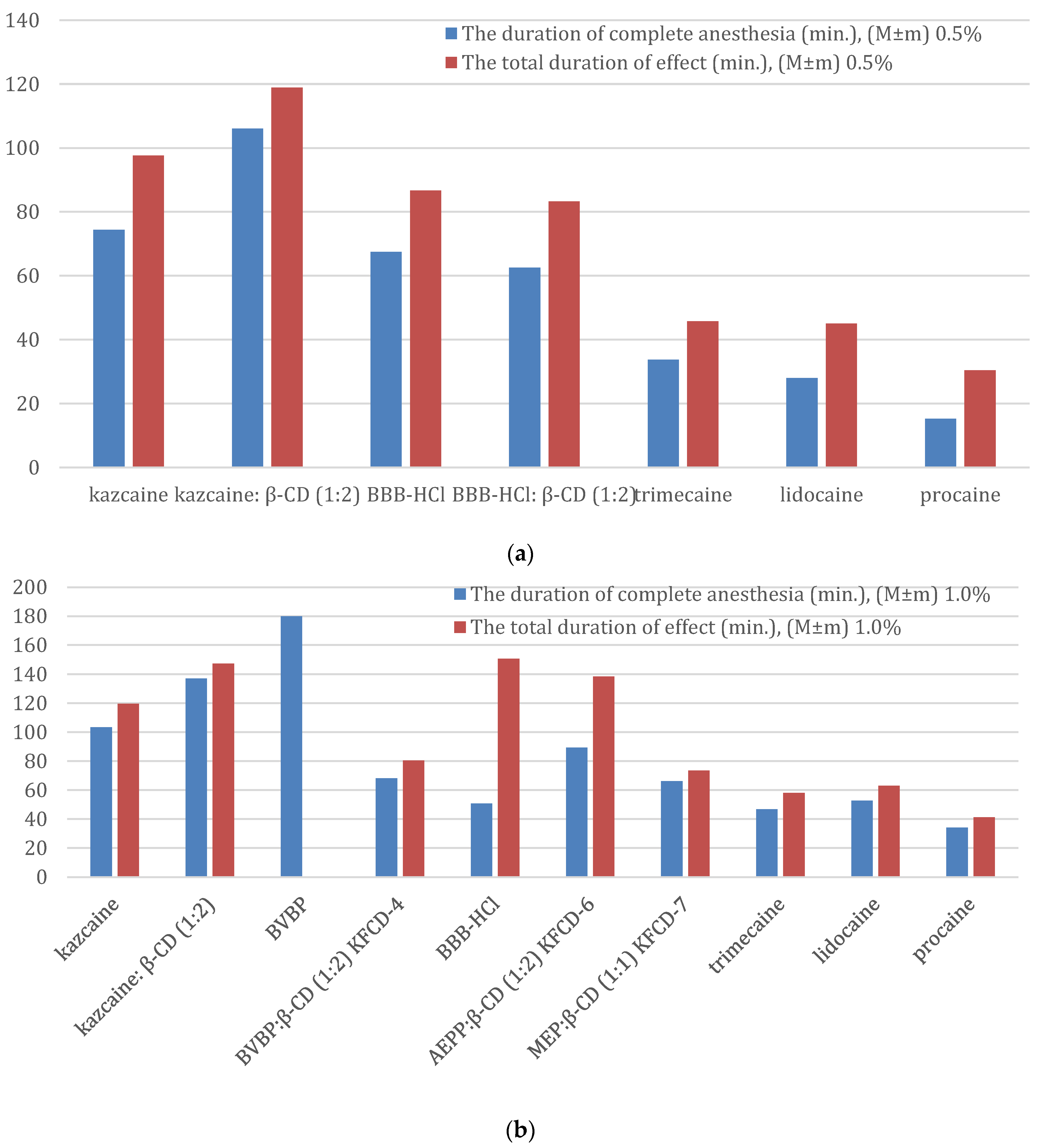
The results are shown in
Table 12. A comparison of the duration of the complete anesthesia and the total duration of effect is shown in
Figure 6a,b.
As can be seen from
Table 12, all the complexes have an apparent local anesthetic effect, and the rate of anesthesia induction is comparable in all cases.
The duration of complete anesthesia (0.5%): procaine < lidocaine < trimecaine < BBB-HCl:β-CD < BBB-HCl < kazcaine < kazcaine:β-CD
The total duration of effect (0.5%): procaine < lidocaine < trimecaine < BBB-HCl:β-CD < BBB-HCl < kazcaine < kazcaine: β-CD.
The duration of complete anesthesia (1%): procaine < trimecaine < BBB-HCl < lidocaine < KFCD-4 < KFCD-6 < kazcaine < kazcaine:β-CD < BVBP
The total duration of effect (1%): procaine < trimecaine < lidocaine < KFCD-7 < KFCD-4 << kazcaine < KFCD-6 < kazcaine: β-CD ≈ BBB-HCl.
KFCD-7 outperformed all three reference anesthetics for the duration of anesthesia and total anesthetic effect and acted like KFCD-4 (
Table 12).
At the above-mentioned concentrations, KFCD-4 and KFCD-7 exceeded procaine for the duration of complete anesthesia by 2 and 1.9 times, trimecaine by 1.3 and 1.4 times, respectively, and acted slightly longer than lidocaine. These solutions also exceeded novocaine and trimecaine in the total duration of a local anesthetic effect (approximately 1.9 and 1.4 times, respectively) and slightly exceeded the effect of lidocaine.
As for the other drugs under consideration, the best result of conduction anesthesia at a 1% solution was exhibited by BVBP, almost three times longer than its complex with CD (KFCD-4); that is, the same picture was observed as for infiltration anesthesia.
The duration of complete anesthesia for KFCD-6 was 89.4 ± 13.4 min, 46.9 ± 8.1 min for trimecaine, 52.7 ± 6.2 for lidocaine, and 34.2 ± 6.9 min for procaine. Thus, the KFCD-6 complex exceeded procaine by 2.3 times, lidocaine by 1.2 times, and trimecaine by 1.7 times (statistically significant at p < 0.001). When comparing the total duration of effect, the KFCD-6 reliably (p < 0.001) exceeded trimecaine by 2.3 times, lidocaine by 2.2 times, and procaine by 3.4 times, respectively.
Kazcaine initially had a good activity (three times better than procaine), and its complex with CD improved the duration of complete anesthesia and the total duration of a local anesthetic effect (but not so dramatically, approximately 30%).
The results for BBB-HCl look interesting. Similar to the infiltration anesthesia, the formation of the complex did not provide an increase in the activity for a 0.5% concentration. However, what is unexpected is that for the 1% concentration, the duration of complete anesthesia was shorter, but the total duration of anesthesia was longer than for the 0.5% concentration. This may probably be due to the different measurement methods used for the 1% solution.
2.3.3. Acute Toxicity
Behavior changes, reflector breath excitability, rate of development and mitigation of external poisoning symptoms, and mortality (LD50) were registered (
Figure 7).
The toxic reactions were of the same character for KFCD-4, KFCD-6, and KFCD-7. The higher the dose, the faster poisoning was evident. The phenomena of intoxication began to develop after 20–30 min. The initial stage started with general oppression and resulted in a deferred response, absence of reflex to exogenous irritants, and dyspnea, which later developed into a short period of motional excitation, followed by muscular twitching and clonic–tonic spasms. Mice assumed a lateral position and their breathing became slower and irregular. Death was caused by primary respiratory standstill 30–90 min after injection. The surviving mice recovered from stagnation in 2–2.5 h and were as active as the untreated mice by the end of the first day.
An analysis of the data obtained for the entire group of the drugs under consideration (
Table 13) showed that the formation of the inclusion complexes significantly decreases the acute toxicity of substances. The resulting inclusion complexes of piperidine derivatives with β-CD were significantly less toxic than the guests themselves.
The KFCD-6 compound turned out to be the most active and less toxic than procaine by 1.7 times, lidocaine by 3.3 times, and trimecaine by 2.2 times in all experiments (
Table 13). KFCD-7 was less toxic than the reference anesthetics.
The most toxic was BBB-HCl, but in the form of an inclusion complex, its toxicity dropped by more than three times and became comparable to procaine. The formation of an inclusion complex reduced the toxicity of AEPP by 2.4 times and BVBP by 2.2 times. The toxicity of kazcaine (which is slightly less than procaine) remained virtually unchanged upon the formation of the inclusion complex, becoming comparable to KFCD-7.
2.3.4. Terminal Anesthesia
The comparison of the activity of the tested compounds with the reference anesthetic, dicaine, was carried out using the Rainier indices, duration of the complete anesthesia, and total duration of effect.
All the studied compounds were tested in 1% and 3% solutions. The experimental results showed that the compounds KFCD-4, KFCD-6, and KFCD-7 in all tested concentrations were significantly inferior both in strength (the Ragnier index) and in the duration of the local anesthetic effect to dicaine and in all concentrations they did not show irritating effects.
At the same time, the formation of inclusion complexes does not always lead to higher activity and depends both on the characteristics of the “guest” and on the type of anesthesia. The most effective in this sense was the inclusion complex of cyclodextrin with 1-(2-ethoxyethyl)-4-ethynyl-4-benzoyloxypiperidine, which is two times better for infiltration anesthesia and 30% better for conduction anesthesia than its salt form (1-(hydrochloride 2-ethoxyethyl)-4-ethynyl-4-benzoyloxypiperidine).
According to the literature, the extension of the alkyl chain at the N atom of the piperidine derivative to the ethoxyethyl substituent leads to the anesthesia index exceeding trimecaine by 1.5 times, lidocaine by 5.1, procaine by 5.3 times, including piperidine derivatives with butoxypropyl substituent. The EC50 value for conduction anesthesia of 1-(3-n-butoxypropyl)-4-benzoyloxypiperidine hydrochloride exceeds the ethoxyethyl homologue by 140 times, and the reference drugs pyromecaine, trimecaine, and procaine by 270, 446, and 670 times, respectively [
38].
This pattern is also confirmed by good results for BVBP and BBB-HCl. Elongation of the radical at the nitrogen atom of the piperidine ring from ethoxyethyl to butoxypropyl led to a significant increase in activity during infiltration, especially during the conduction of anesthesia. However, these same drugs have the highest toxicity among those considered. The formation of inclusion complexes leads to a significant reduction in toxicity (comparable to trimecaine) but, at the same time, to a significant reduction in the anesthesia time.