Volatile Organic Compounds (VOCs) Produced by Levilactobacillus brevis WLP672 Fermentation in Defined Media Supplemented with Different Amino Acids
Abstract
1. Introduction
2. Results and Discussion
2.1. Development of Defined Medium
2.2. Physiochemical Properties
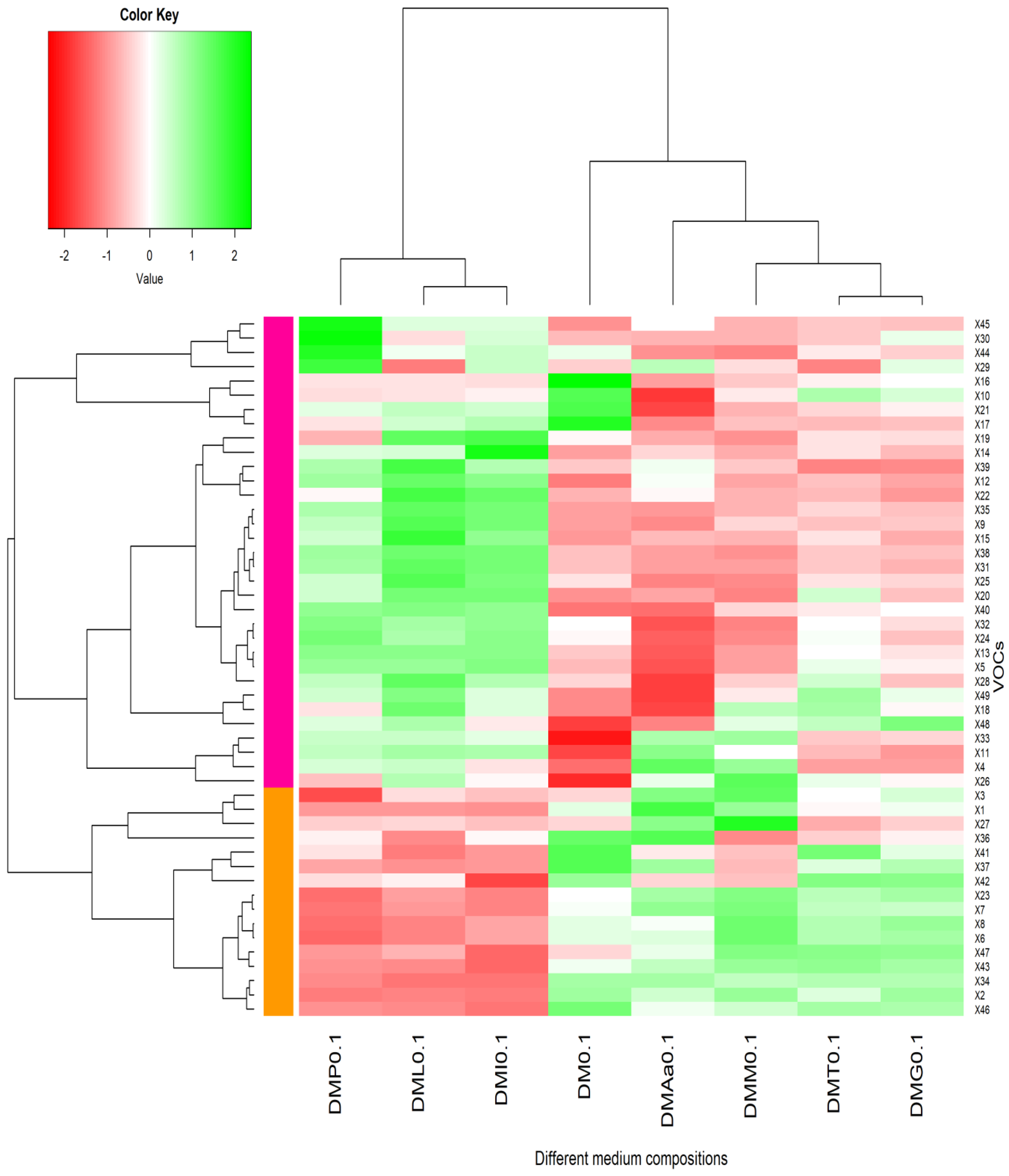
2.3. Volatile Organic Compounds (VOCs) after Fermentation
2.4. Phe-Derived VOCs
2.5. Met-Derived VOCs
2.6. Leu/Ile-Derived VOCs
3. Materials and Methods
3.1. LAB Strain
3.2. Medium Compositions
3.3. Fermentation
3.4. Determination of Volatile Organic Compounds
3.5. Data Analysis
3.5.1. GC-MS Data Extraction
3.5.2. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lea, E.J.; Crawford, D.; Worsley, A. Consumers’ readiness to eat a plant-based diet. Eur. J. Clin. Nutr. 2006, 60, 342–351. [Google Scholar] [CrossRef] [PubMed]
- Szejda, K.; Urbanovich, T.; Wilks, M. Accelerating Consumer Adoption of Plant-Based Meat: An Evidence-Based Guide for Effective Practice; The Good Food Institute: Washington, DC, USA, 2020. [Google Scholar]
- Austgulen, M.; Skuland, S.; Schjøll, A.; Alfnes, F. Consumer readiness to reduce meat consumption for the purpose of environmental sustainability: Insights from Norway. Sustainability 2018, 10, 3058. [Google Scholar] [CrossRef]
- Clem, J.; Barthel, B. A look at plant-based diets. Mo. Med. 2021, 118, 233–238. [Google Scholar] [PubMed]
- Alcorta, A.; Porta, A.; Tarrega, A.; Alvarez, M.D.; Vaquero, M.P. Foods for plant-based diets: Challenges and innovations. Foods 2021, 10, 293. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.; Costa, R.; Rodrigues, I.; Lameiras, J.; Botelho, G. A narrative review of alternative protein sources: Highlights on meat, fish, egg and dairy analogues. Foods 2022, 11, 2053. [Google Scholar] [CrossRef] [PubMed]
- Bryant, C.J. Plant-based animal product alternatives are healthier and more environmentally sustainable than animal products. Future Foods 2022, 6, 100174–100186. [Google Scholar] [CrossRef]
- Ishaq, A.; Irfan, S.; Sameen, A.; Khalid, N. Plant-based meat analogs: A review with reference to formulation and gastrointestinal fate. Curr. Res. Food Sci. 2022, 5, 973–983. [Google Scholar] [CrossRef]
- Michel, F.; Hartmann, C.; Siegrist, M. Consumers’ associations, perceptions and acceptance of meat and plant-based meat alternatives. Food Qual. Prefer. 2021, 87, 104063–104073. [Google Scholar] [CrossRef]
- Aschemann-Witzel, J.; Gantriis, R.F.; Fraga, P.; Perez-Cueto, F.J.A. Plant-based food and protein trend from a business perspective: Markets, consumers, and the challenges and opportunities in the future. Crit. Rev. Food Sci. Nutr. 2021, 61, 3119–3128. [Google Scholar] [CrossRef]
- Pointke, M.; Pawelzik, E. Plant-based alternative products: Are they healthy alternatives? Micro- and macronutrients and nutritional scoring. Nutrients 2022, 14, 601. [Google Scholar] [CrossRef]
- Szenderak, J.; Frona, D.; Rakos, M. Consumer acceptance of plant-based meat substitutes: A narrative review. Foods 2022, 11, 1274. [Google Scholar] [CrossRef]
- Reineccius, G. Flavor Chemistry and Technology, 2nd ed.; Taylor & Francis Group: Boca Raton, FL, USA, 2006. [Google Scholar]
- Lawless, H. The sense of smell in food quality and sensory evaluation. J. Food Qual. 1991, 14, 33–60. [Google Scholar] [CrossRef]
- Astray, G.; García-Río, L.; Mejuto, J.C.; Pastrana, L. Chemistry in food: Flavours. Electron. J. Environ. Agric. Food Chem. 2007, 6, 1742–1763. [Google Scholar]
- Paravisini, L.; Guichard, E. Interactions between aroma compounds and food matrix. In Flavour: From Food to Perception; Wiley: New York, NY, USA, 2016; pp. 208–234. [Google Scholar]
- Small, D.M.; Gerber, J.C.; Mak, Y.E.; Hummel, T. Differential neural responses evoked by orthonasal versus retronasal odorant perception in humans. Neuron 2005, 47, 593–605. [Google Scholar] [CrossRef]
- Dastager, S.G. Aroma Compounds; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Janssens, L.; De Pooter, H.L.; Schamp, N.M.; Vandamme, E.J. Production of flavours by microorganisms. Process Biochem. 1992, 27, 195–215. [Google Scholar] [CrossRef]
- Longo, M.A.; Sanromán, M.A. Production of food aroma compounds: Microbial and enzymatic methodologies. Food Technol. Biotechnol. 2006, 44, 335–353. [Google Scholar] [CrossRef]
- Bamforth, C.W.; Cook, D.J. Food, Fermentation, and Micro-Organisms, 2nd ed.; Wiley: New York, NY, USA, 2019. [Google Scholar] [CrossRef]
- Petrovici, A.R.; Ciolacu, D.E. Natural flavours obtained by microbiological pathway. In Generation of Aromas and Flavours; InTech: London, UK, 2018; pp. 33–52. [Google Scholar]
- Rajendran, S.; Silcock, P.; Bremer, P. Flavour volatiles of fermented vegetable and fruit substrates: A review. Molecules 2023, 28, 3236. [Google Scholar] [CrossRef]
- Szutowska, J. Functional properties of lactic acid bacteria in fermented fruit and vegetable juices: A systematic literature review. Eur. Food Res. Technol. 2020, 246, 357–372. [Google Scholar] [CrossRef]
- Teusink, B.; Molenaar, D. Systems biology of lactic acid bacteria: For food and thought. Curr. Opin. Syst. Biol. 2017, 6, 7–13. [Google Scholar] [CrossRef]
- Hayek, S.A.; Gyawali, R.; Aljaloud, S.O.; Krastanov, A.; Ibrahim, S.A. Cultivation media for lactic acid bacteria used in dairy products. J. Dairy Res. 2019, 86, 490–502. [Google Scholar] [CrossRef]
- Wegkamp, A.; Teusink, B.; de Vos, W.M.; Smid, E.J. Development of a minimal growth medium for Lactobacillus plantarum. Lett. Appl. Microbiol. 2010, 50, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Kwoji, I.D.; Okpeku, M.; Adeleke, M.A.; Aiyegoro, O.A. Formulation of chemically defined media and growth evaluation of Ligilactobacillus salivarius ZJ614 and Limosilactobacillus reuteri ZJ625. Front. Microbiol. 2022, 13, 1450. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.R.; Hammer, K. Minimal requirements for exponential growth of Lactococcus lactis. Appl. Environ. Microbiol. 1993, 59, 4363–4366. [Google Scholar] [CrossRef] [PubMed]
- Niven, C.F. Nutrition of Streptococcus lactis. J. Bacteriol. 1944, 47, 343–350. [Google Scholar] [CrossRef]
- Cocaign-Bousquet, M.; Garrigues, C.; Novak, L.; Lindley, N.D.; Loublere, P. Rational development of a simple synthetic medium for the sustained growth of Lactococcus lactis. J. Appl. Bacteriol. 1995, 79, 108–116. [Google Scholar] [CrossRef]
- van Niel, E.W.J.; Hahn-Hägerdal, B. Nutrient requirements of lactococci in defined growth media. Appl. Microbiol. Biotechnol. 1999, 52, 617–627. [Google Scholar] [CrossRef]
- Pastink, M.I.; Teusink, B.; Hols, P.; Visser, S.; de Vos, W.M.; Hugenholtz, J. Genome-scale model of Streptococcus thermophilus LMG18311 for metabolic comparison of lactic acid bacteria. Appl. Environ. Microbiol. 2009, 75, 3627–3633. [Google Scholar] [CrossRef]
- Kranenburg, R.V.; Kleerebezem, M.; van Hylckama Vlieg, J.; Ursing, B.M.; Boekhorst, J.; Smit, B.A.; Ayad, E.H.E.; Smit, G.; Siezen, R.J. Flavour formation from amino acids by lactic acid bacteria: Predictions from genome sequence analysis. Int. Dairy J. 2002, 12, 111–121. [Google Scholar] [CrossRef]
- Christensen, J.E.; Dudley, E.G.; Pederson, J.A.; Steele, J.L. Peptidases and amino acid catabolism in lactic acid bacteria. Antonie Van Leeuwenhoek 1999, 76, 217–246. [Google Scholar] [CrossRef]
- Fernandez, M.; Zuniga, M. Amino acid catabolic pathways of lactic acid bacteria. Crit. Rev. Microbiol. 2006, 32, 155–183. [Google Scholar] [CrossRef]
- Teixeira, P. Lactobacillus, Lactobacillus brevis. In Encyclopedia of Food Microbiology; Elsevier: Amsterdam, The Netherlands, 2014; Volume 2, pp. 418–424. [Google Scholar] [CrossRef]
- Chen, D.; Chia, J.Y.; Liu, S.Q. Impact of addition of aromatic amino acids on non-volatile and volatile compounds in lychee wine fermented with Saccharomyces cerevisiae MERIT.ferm. Int. J. Food Microbiol. 2014, 170, 12–20. [Google Scholar] [CrossRef]
- Fairbairn, S.; McKinnon, A.; Musarurwa, H.T.; Ferreira, A.C.; Bauer, F.F. The impact of single amino acids on growth and volatile aroma production by Saccharomyces cerevisiae strains. Front. Microbiol. 2017, 8, 2554. [Google Scholar] [CrossRef]
- Chua, J.Y.; Tan, S.J.; Liu, S.Q. The impact of mixed amino acids supplementation on Torulaspora delbrueckii growth and volatile compound modulation in soy whey alcohol fermentation. Food Res. Int. 2021, 140, 109901–109913. [Google Scholar] [CrossRef]
- Lee, P.-R.; Yu, B.; Curran, P.; Liu, S.-Q. Impact of amino acid addition on aroma compounds in papaya wine fermented with Williopsis mrakii. S. Afr. J. Enol. Vitic. 2011, 32, 220–228. [Google Scholar] [CrossRef]
- Wang, Y.-Q.; Ye, D.-Q.; Liu, P.-T.; Duan, L.-L.; Duan, C.-Q.; Yan, G.-L. Synergistic effects of branched-chain amino acids and phenylalanine addition on major volatile compounds in wine during alcoholic fermentation. S. Afr. J. Enol. Vitic. 2016, 37, 169–175. [Google Scholar] [CrossRef]
- Meza, J.C.; Christen, P.; Revah, S. Effect of added amino acids on the production of a fruity aroma by Ceratocystis fimbriata. Sci. Aliment. 1998, 18, 627–636. [Google Scholar]
- Gutsche, K.A.; Tran, T.B.; Vogel, R.F. Production of volatile compounds by Lactobacillus sakei from branched chain alpha-keto acids. Food Microbiol. 2012, 29, 224–228. [Google Scholar] [CrossRef]
- Tavaria, F.K.; Dahl, S.; Carballo, F.J.; Malcata, F.X. Amino acid catabolism and generation of volatiles by lactic acid bacteria. J. Dairy Sci. 2002, 85, 2462–2470. [Google Scholar] [CrossRef]
- Canon, F.; Maillard, M.B.; Henry, G.; Thierry, A.; Gagnaire, V. Positive interactions between lactic acid bacteria promoted by nitrogen-based nutritional dependencies. Appl. Environ. Microbiol. 2021, 87, e01055-21. [Google Scholar] [CrossRef]
- Henderson, L.M.; Snell, E.E. A uniform medium for determination of amino acids with various microorganisms. J. Biol. Chem. 1948, 172, 15–29. [Google Scholar] [CrossRef]
- De man, J.C.; Rogosa, M.; Sharpe, M.E. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 1960, 23, 130–135. [Google Scholar] [CrossRef]
- Russell, C.; Bhandari, R.R.; Walker, T.K. Vitamin requirements of thirty-four lactic acid bacteria associated with brewery products. J. Gen. Microbiol. 1954, 10, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Hébert, E.M.; Raya, R.R.; Savoy de Giori, G. Evaluation of Minimal Nutritional Requirements of Lactic Acid Bacteria Used in Functional Foods; Humana Press Inc.: Totowa, NJ, USA, 2004. [Google Scholar]
- MacLeod, R.A.; Snell, E.E. Some mineral requirements of the lactic acid bacteria. J. Biol. Chem. 1947, 170, 351–365. [Google Scholar] [CrossRef]
- Zacharof, M.-P.; Lovitt, R.W. Partially chemically defined liquid medium development for intensive propagation of industrial fermentation lactobacilli strains. Ann. Microbiol. 2012, 63, 1235–1245. [Google Scholar] [CrossRef]
- Liu, M.; Nauta, A.; Francke, C.; Siezen, R.J. Comparative genomics of enzymes in flavor-forming pathways from amino acids in lactic acid bacteria. Appl. Environ. Microbiol. 2008, 74, 4590–4600. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.; Slesarev, A.; Wolf, Y.; Sorokin, A.; Mirkin, B.; Koonin, E.; Pavlov, A.; Pavlov, N.; Karamychev, V.; Polouchine, N.; et al. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 2006, 103, 15611–15616. [Google Scholar] [CrossRef]
- Klompong, V.; Benjakul, S.; Kantachote, D.; Shahidi, F. Characteristics and use of yellow stripe trevally hydrolysate as culture media. J. Food Sci. 2009, 74, 219–225. [Google Scholar] [CrossRef]
- Foudin, A.S.; Wynn, W.K. Growth of Puccinia graminis f. sp. tritici. Phytopathology 1972, 62, 1032–1040. [Google Scholar] [CrossRef]
- Li, T.; Jiang, T.; Liu, N.; Wu, C.; Xu, H.; Lei, H. Biotransformation of phenolic profiles and improvement of antioxidant capacities in jujube juice by select lactic acid bacteria. Food Chem. 2021, 339, 127859–127869. [Google Scholar] [CrossRef]
- McSweeney, P.L.H.; Sousa, M.J. Biochemical pathways for the production of flavour compounds in cheeses during ripening: A review. Le Lait 2000, 80, 293–324. [Google Scholar] [CrossRef]
- Ricci, A.; Cirlini, M.; Levante, A.; Dall’Asta, C.; Galaverna, G.; Lazzi, C. Volatile profile of elderberry juice: Effect of lactic acid fermentation using L. plantarum, L. rhamnosus and L. casei strains. Food Res. Int. 2018, 105, 412–422. [Google Scholar] [CrossRef]
- Valera, M.J.; Boido, E.; Ramos, J.C.; Manta, E.; Radi, R.; Dellacassa, E.; Carrau, F. The mandelate pathway, an alternative to the phenylalanine ammonia lyase pathway for the synthesis of benzenoids in Ascomycete Yeasts. Appl. Environ. Microbiol. 2020, 86, e00701-20. [Google Scholar] [CrossRef]
- Yvon, M.; Rijnen, L. Cheese flavour formation by amino acid catabolism. Int. Dairy J. 2001, 11, 185–201. [Google Scholar] [CrossRef]
- Hu, S.; Zhang, X.; Lu, Y.; Lin, Y.-C.; Xie, D.-F.; Fang, H.; Huang, J.; Mei, L.-H. Cloning, expression and characterization of an aspartate aminotransferase gene from Lactobacillus brevis CGMCC 1306. Biotechnol. Biotechnol. Equip. 2017, 31, 544–553. [Google Scholar] [CrossRef]
- Marilley, L.; Casey, M.G. Flavours of cheese products: Metabolic pathways, analytical tools and identification of producing strains. Int. J. Food Microbiol. 2004, 90, 139–159. [Google Scholar] [CrossRef] [PubMed]
- Ardö, Y. Flavour formation by amino acid catabolism. Biotechnol. Adv. 2006, 24, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Hols, P.; Ramos, A.; Hugenholtz, J.; Delcour, J.; de Vos, W.M.; Santos, H.; Kleerebezem, M. Acetate utilization in Lactococcus lactis deficient in lactate dehydrogenase: A rescue pathway for maintaining redox balance. J. Bacteriol. 1999, 181, 5521–5526. [Google Scholar] [CrossRef]
- Feyereisen, M.; Mahony, J.; Kelleher, P.; Roberts, R.J.; O’Sullivan, T.; Geertman, J.A.; van Sinderen, D. Comparative genome analysis of the Lactobacillus brevis species. BMC Genom. 2019, 20, 416–431. [Google Scholar] [CrossRef]
- Barker, H.A.; Kamen, M.D.; Bornstein, B.T. The synthesis of butyric and caproic acids from ethanol and acetic acid by Clostridium kluyveri. Proc. Natl. Acad. Sci. USA 1945, 31, 373–381. [Google Scholar] [CrossRef]
- Reed, L.J.; DeBusk, B.G.; Johnston, P.M.; Getzendaner, M.E. Acetate-replacing factors for lactic acid bacteria. J. Biol. Chem. 1951, 192, 851–858. [Google Scholar] [CrossRef]
- Lindinger, W.; Hansel, A.; Jordan, A. Proton-transfer-reaction mass spectrometry (PTR-MS): On-line monitoring of volatile organic compounds at pptv levels. Chem. Soc. Rev. 1998, 27, 347–354. [Google Scholar] [CrossRef]
- Ba, V.H.; Hwang, I.; Jeong, D.; Touseef, A. Principle of meat aroma flavors and future prospect. In Latest Research into Quality Control; InTech: London, UK, 2012; pp. 145–176. [Google Scholar]
- Marsili, R. Flavors and off-flavors in dairy foods. In Encyclopedia of Dairy Sciences; Elsevier: Amsterdam, The Netherlands, 2022; pp. 560–578. [Google Scholar] [CrossRef]
- Smit, G.; Smit, B.A.; Engels, W.J. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 2005, 29, 591–610. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, L.G.; Skou, P.B.; Khakimov, B.; Bro, R. Gas chromatography—Mass spectrometry data processing made easy. J. Chromatogr. A 2017, 1503, 57–64. [Google Scholar] [CrossRef]
- Baccolo, G.; Quintanilla-Casas, B.; Vichi, S.; Augustijn, D.; Bro, R. From untargeted chemical profiling to peak tables—A fully automated AI driven approach to untargeted GC-MS. Trends Anal. Chem. 2021, 145, 116451–116459. [Google Scholar] [CrossRef]
- Halang, W.A.; Langlais, R.; Kugler, E. Cubic spline interpolation for the calculation of retention indices in temperature programmed gas-liquid chromatography. Anal. Chem. 1978, 50, 1829–1832. [Google Scholar] [CrossRef]

 and 1.2%
and 1.2%  acetate. Values are presented as mean ± standard error (n = 6). Different letters represent significant difference between the different medium compositions according to Tukey’s test at p < 0.05.
acetate. Values are presented as mean ± standard error (n = 6). Different letters represent significant difference between the different medium compositions according to Tukey’s test at p < 0.05.
 and 1.2%
and 1.2%  acetate. Values are presented as mean ± standard error (n = 6). Different letters represent significant difference between the different medium compositions according to Tukey’s test at p < 0.05.
acetate. Values are presented as mean ± standard error (n = 6). Different letters represent significant difference between the different medium compositions according to Tukey’s test at p < 0.05.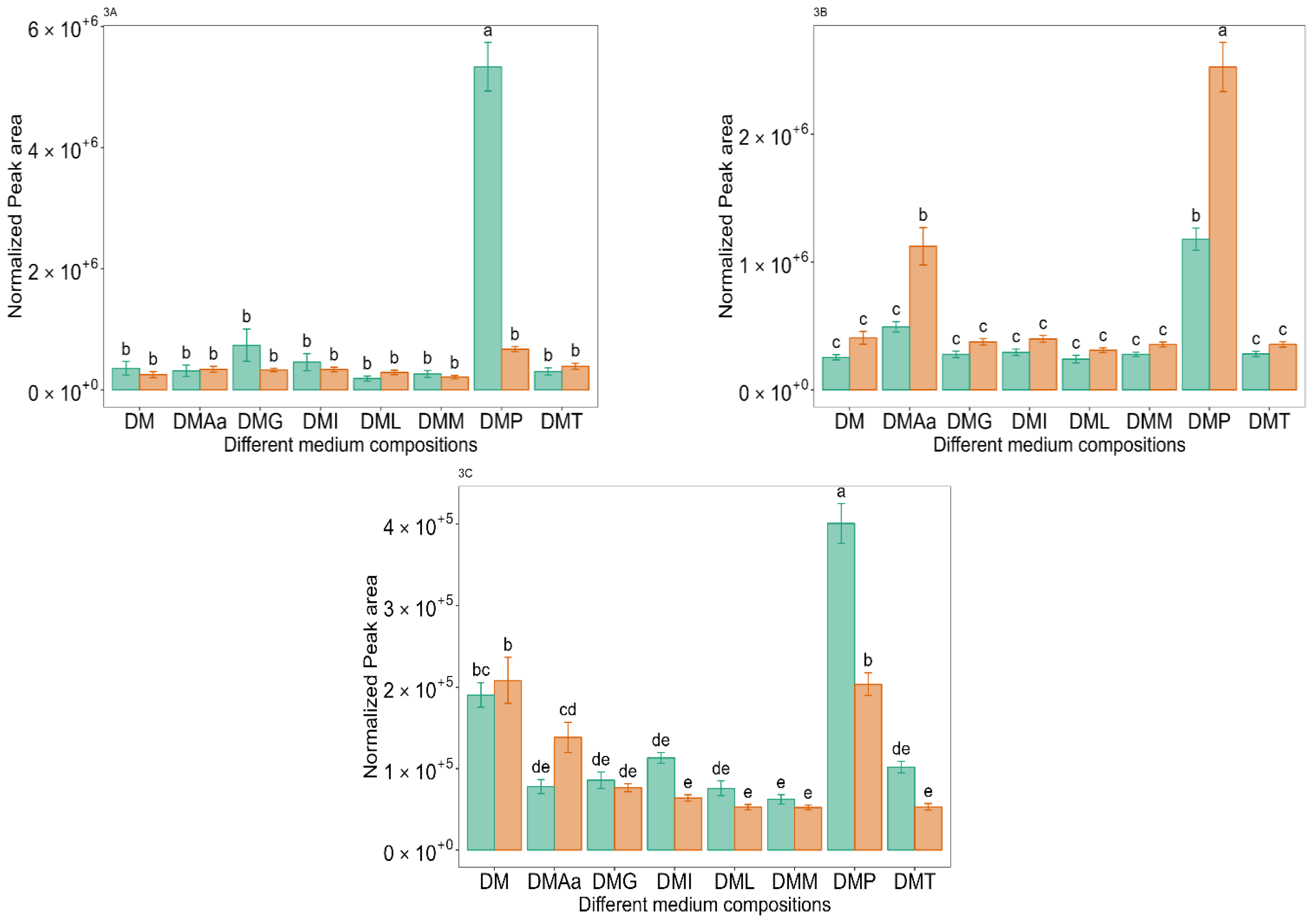
 and 1.2%
and 1.2%  acetate. Values are presented as mean ± standard error (n = 6). Different letters represent significant difference between the different medium compositions according to Tukey’s test at p < 0.05.
acetate. Values are presented as mean ± standard error (n = 6). Different letters represent significant difference between the different medium compositions according to Tukey’s test at p < 0.05.
 and 1.2%
and 1.2%  acetate. Values are presented as mean ± standard error (n = 6). Different letters represent significant difference between the different medium compositions according to Tukey’s test at p < 0.05.
acetate. Values are presented as mean ± standard error (n = 6). Different letters represent significant difference between the different medium compositions according to Tukey’s test at p < 0.05.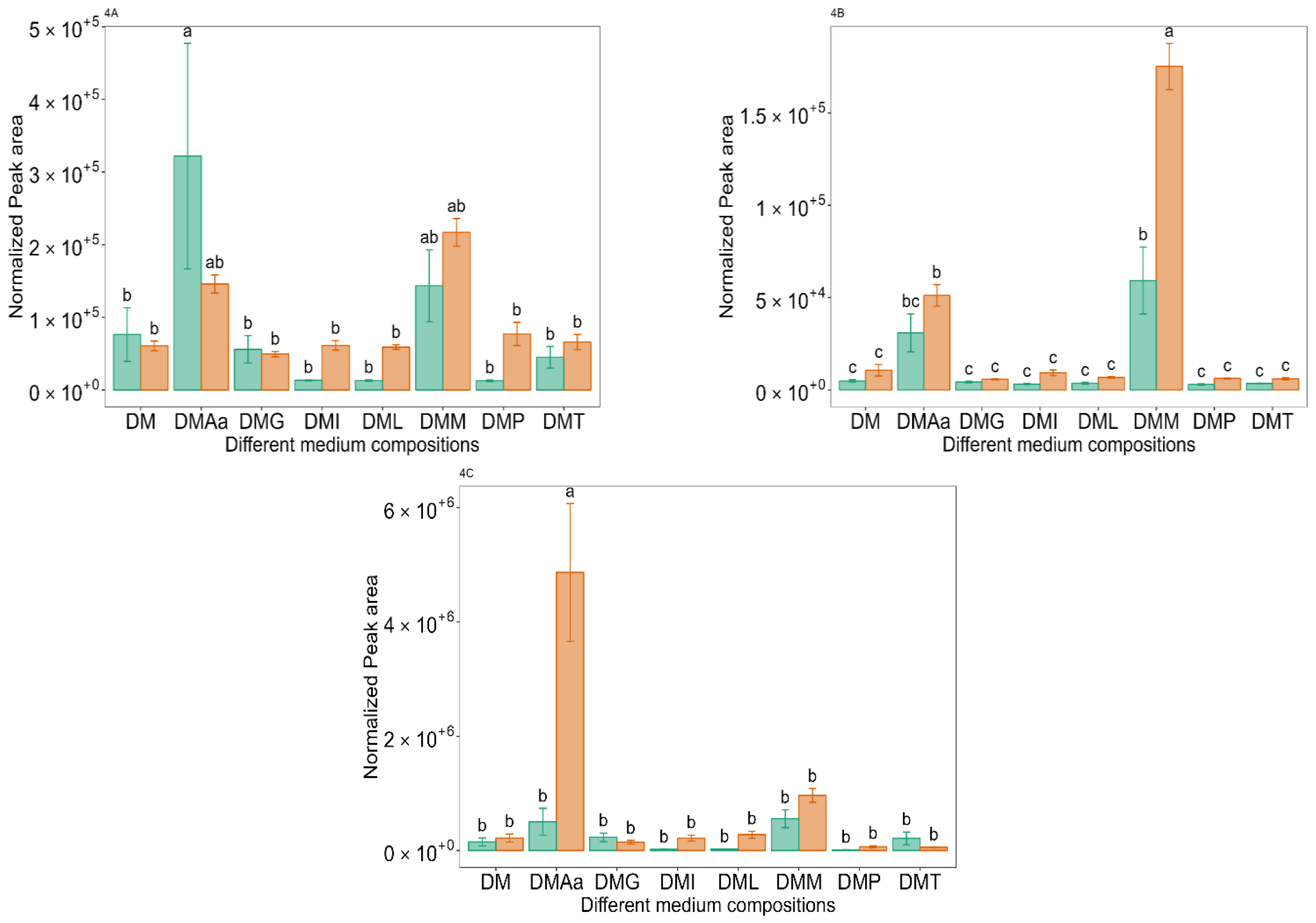
 and 1.2%
and 1.2%  acetate. Values are presented as mean ± standard error (n = 6). Different letters represent significant difference between different medium compositions according to Tukey’s test at p < 0.05.
acetate. Values are presented as mean ± standard error (n = 6). Different letters represent significant difference between different medium compositions according to Tukey’s test at p < 0.05.
 and 1.2%
and 1.2%  acetate. Values are presented as mean ± standard error (n = 6). Different letters represent significant difference between different medium compositions according to Tukey’s test at p < 0.05.
acetate. Values are presented as mean ± standard error (n = 6). Different letters represent significant difference between different medium compositions according to Tukey’s test at p < 0.05.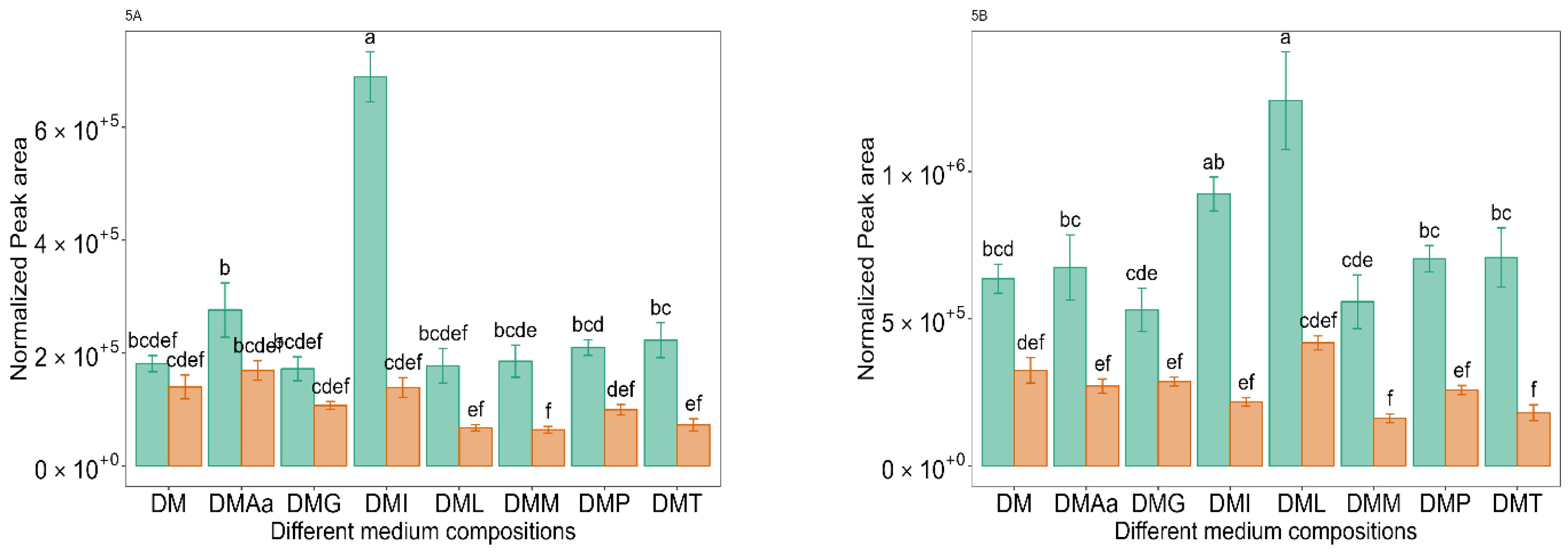
 and 1.2%
and 1.2%  acetate. Values are presented as mean ± standard error (n = 6). Different letters represent significant difference between the different medium compositions according to Tukey’s test at p < 0.05.
acetate. Values are presented as mean ± standard error (n = 6). Different letters represent significant difference between the different medium compositions according to Tukey’s test at p < 0.05.
 and 1.2%
and 1.2%  acetate. Values are presented as mean ± standard error (n = 6). Different letters represent significant difference between the different medium compositions according to Tukey’s test at p < 0.05.
acetate. Values are presented as mean ± standard error (n = 6). Different letters represent significant difference between the different medium compositions according to Tukey’s test at p < 0.05.
| Media | Glucose | Peptone | Vitamins | Salt | Sodium Acetate | Glu | Leu | Ile | Phe | Thr | Met |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DML0.1 | 2% | 0.5% | √ | √ | 0.1% | 0.2% | 0.2% | - | - | - | - |
| DMI0.1 | 2% | 0.5% | √ | √ | 0.1% | 0.2% | - | 0.2% | - | - | - |
| DMP0.1 | 2% | 0.5% | √ | √ | 0.1% | 0.2% | - | - | 0.2% | - | - |
| DMT0.1 | 2% | 0.5% | √ | √ | 0.1% | 0.2% | - | - | - | 0.2% | - |
| DMM0.1 | 2% | 0.5% | √ | √ | 0.1% | 0.2% | - | - | - | - | 0.2% |
| DMG0.1 | 2% | 0.5% | √ | √ | 0.1% | 0.2% | - | - | - | - | - |
| DM0.1 | 2% | 0.5% | √ | √ | 0.1% | - | - | - | - | - | - |
| DMAa0.1 | 2% | 0.5% | √ | √ | 0.1% | 0.2% | 0.04% | 0.04% | 0.04% | 0.04% | 0.04% |
| DML1.2 | 2% | 0.5% | √ | √ | 1.2% | 0.2% | 0.2% | - | - | - | - |
| DMI1.2 | 2% | 0.5% | √ | √ | 1.2% | 0.2% | - | 0.2% | - | - | - |
| DMP1.2 | 2% | 0.5% | √ | √ | 1.2% | 0.2% | - | - | 0.2% | - | - |
| DMT1.2 | 2% | 0.5% | √ | √ | 1.2% | 0.2% | - | - | - | 0.2% | - |
| DMM1.2 | 2% | 0.5% | √ | √ | 1.2% | 0.2% | - | - | - | - | 0.2% |
| DMG1.2 | 2% | 0.5% | √ | √ | 1.2% | 0.2% | - | - | - | - | - |
| DM1.2 | 2% | 0.5% | √ | √ | 1.2% | - | - | - | - | - | - |
| DMAa1.2 | 2% | 0.5% | √ | √ | 1.2% | 0.2% | 0.04% | 0.04% | 0.04% | 0.04% | 0.04% |
| Media | Initial pH | After 16 Days of Fermentation | |
|---|---|---|---|
| pH | OD600 | ||
| DML0.1 | 6.66 | 6.24 | 0.269 de |
| DMI0.1 | 6.64 | 6.30 | 0.271 de |
| DMP0.1 | 6.65 | 6.17 | 0.223 e |
| DMT0.1 | 6.68 | 6.10 | 0.297 de |
| DMM0.1 | 6.67 | 6.09 | 0.249 de |
| DMG0.1 | 6.83 | 6.20 | 0.286 de |
| DM0.1 | 7.85 | 7.28 | 0.391 cd |
| DMAa0.1 | 6.63 | 6.45 | 0.233 e |
| DML1.2 | 5.27 | 4.63 | 0.511 bc |
| DMI1.2 | 5.28 | 4.64 | 0.536 bc |
| DMP1.2 | 5.29 | 4.72 | 0.542 bc |
| DMT1.2 | 5.26 | 4.66 | 0.517 bc |
| DMM1.2 | 5.3 | 4.66 | 0.515 bc |
| DMG1.2 | 5.4 | 4.89 | 0.605 ab |
| DM1.2 | 7.01 | 5.0 | 0.711 a |
| DMAa1.2 | 5.31 | 4.87 | 0.492 bc |
| No. | Compound Name | RI (Calc.) | RI (Lit.) | R Match | Identification Method |
|---|---|---|---|---|---|
| Alcohols | |||||
| 1 | Ethanol | 930 | 932 | 942 | MS, RI |
| 2 | 2-Methyl propanol | 1086 | 1092 | 827 | MS, RI |
| 3 | 2-Pentanol | 1116 | 1119 | 881 | MS, RI |
| 4 | 2-Methyl butanol | 1199 | 1208 | 905 | MS, RI |
| 5 | 3-Methyl butanol (isoamyl alcohol) | 1200 | 1209 | 962 | MS, RI |
| 6 | 3-Heptanol | 1287 | 1290 | 959 | MS, RI |
| 7 | 2-Heptanol | 1310 | 1320 | 953 | MS, RI |
| 8 | 3-Methyl-2-buten-1-ol (prenol) | 1312 | 1320 | 815 | MS, RI |
| 9 | 1-Hexanol | 1343 | 1355 | 832 | MS, RI |
| 10 | 4-Methyl-2-heptanol | 1349 | 1372 | 900 | MS, RI |
| 11 | 2-Octanol | 1410 | 1412 | 849 | MS, RI |
| 12 | 2-Nonanol | 1508 | 1521 | 952 | MS, RI |
| 13 | 1-Octanol | 1545 | 1557 | 912 | MS, RI |
| 14 | 1-Nonanol | 1647 | 1660 | 883 | MS, RI |
| 15 | 1-Decanol | 1751 | 1760 | 824 | MS, RI |
| 16 | Citronellol | 1752 | 1765 | 801 | MS, RI |
| 17 | Geraniol | 1833 | 1847 | 879 | MS, RI |
| 18 | Benzyl alcohol | 1868 | 1870 | 924 | MS, RI |
| 19 | Phenylethyl alcohol | 1904 | 1906 | 914 | MS, RI |
| Acids | |||||
| 20 | Acetic acid | 1429 | 1449 | 946 | MS, RI |
| 21 | Butanoic acid | 1620 | 1625 | 905 | MS, RI |
| 22 | 2-Methyl butanoic acid | 1659 | 1662 | 929 | MS, RI |
| 23 | 3-Methyl butanoic acid (isovaleric acid) | 1658 | 1666 | 956 | MS, RI |
| 24 | Hexanoic acid | 1836 | 1846 | 903 | MS, RI |
| 25 | Heptanoic acid | 1944 | 1950 | 864 | MS, RI |
| 26 | Octanoic acid | 2049 | 2060 | 920 | MS, RI |
| 27 | Nonanoic acid | 2154 | 2171 | 912 | MS, RI |
| 28 | n-Decanoic acid | 2261 | 2276 | 801 | MS, RI |
| Esters | |||||
| 29 | Ethyl acetate | 889 | 888 | 957 | MS, RI |
| 30 | Butyl acetate | 1069 | 1074 | 959 | MS, RI |
| 31 | 3-Methylbutyl acetate | 1120 | 1122 | 867 | MS, RI |
| 32 | Ethyl hexanoate | 1230 | 1233 | 860 | MS, RI |
| 33 | Ethyl heptanoate * | 1331 | 1331 | 771 | MS, RI |
| 34 | 2-Phenylethyl acetate | 1813 | 1813 | 800 | MS, RI |
| Sulphur compounds | |||||
| 35 | Methanethiol * | 690 | 692 | 734 | MS, RI |
| 36 | Dimethyl disulphide | 1072 | 1077 | 965 | MS, RI |
| 37 | Dimethyl trisulphide | 1386 | 1377 | 935 | MS, RI |
| 38 | Methional | 1455 | 1454 | 809 | MS, RI |
| 39 | 5-Ethenyl-4-methyl thiazole | 1527 | 1520 | 917 | MS, RI |
| Ketones | |||||
| 40 | 4-Methyl-4-penten-2-one | 1069 | 1110 | 804 | MS, RI |
| 41 | 2-Heptanone | 1183 | 1182 | 921 | MS, RI |
| 42 | 2-Nonanone | 1390 | 1390 | 954 | MS, RI |
| 43 | 2-Undecanone | 1600 | 1598 | 939 | MS, RI |
| Aldehydes | |||||
| 44 | Benzaldehyde | 1530 | 1520 | 961 | MS, RI |
| Unknown compounds | |||||
| 45 | Unknown 1 | 1027 | NA | ||
| 46 | Unknown 2 | 1139 | NA | ||
| 47 | Unknown 3 | 1181 | NA | ||
| 48 | Unknown 4 | 1597 | NA | ||
| 49 | Unknown 5 | 1803 | NA | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajendran, S.; Silcock, P.; Bremer, P. Volatile Organic Compounds (VOCs) Produced by Levilactobacillus brevis WLP672 Fermentation in Defined Media Supplemented with Different Amino Acids. Molecules 2024, 29, 753. https://doi.org/10.3390/molecules29040753
Rajendran S, Silcock P, Bremer P. Volatile Organic Compounds (VOCs) Produced by Levilactobacillus brevis WLP672 Fermentation in Defined Media Supplemented with Different Amino Acids. Molecules. 2024; 29(4):753. https://doi.org/10.3390/molecules29040753
Chicago/Turabian StyleRajendran, Sarathadevi, Patrick Silcock, and Phil Bremer. 2024. "Volatile Organic Compounds (VOCs) Produced by Levilactobacillus brevis WLP672 Fermentation in Defined Media Supplemented with Different Amino Acids" Molecules 29, no. 4: 753. https://doi.org/10.3390/molecules29040753
APA StyleRajendran, S., Silcock, P., & Bremer, P. (2024). Volatile Organic Compounds (VOCs) Produced by Levilactobacillus brevis WLP672 Fermentation in Defined Media Supplemented with Different Amino Acids. Molecules, 29(4), 753. https://doi.org/10.3390/molecules29040753







