Abstract
The enmein-type diterpenoids are a class of anticancer ent-Kaurane diterpnoids that have received much attention in recent years. Herein, a novel 1,14-epoxy enmein-type diterpenoid 4, was reported in this project for the first time. A series of novel enmein-type diterpenoid derivatives were also synthesized and tested for anticancer activities. Among all the derivatives, compound 7h exhibited the most significant inhibitory effect against A549 cells (IC50 = 2.16 µM), being 11.03-folds better than its parental compound 4. Additionally, 7h exhibited relatively weak anti-proliferative activity (IC50 > 100 µM) against human normal L-02 cells, suggesting that it had excellent anti-proliferative selectivity for cancer cells. Mechanism studies suggested that 7h induced G0/G1 arrest and apoptosis in A549 cells by inhibiting the PI3K/AKT/mTOR pathway. This process was associated with elevated intracellular ROS levels and collapsed MMP. In summary, these data identified 7h as a promising lead compound that warrants further investigation of its anticancer properties.
1. Introduction
Cancer, the disease of abnormal cell proliferation, stands as one of the principal threats to human life in the 21st century [1]. According to recent statistics, the global cancer-related death toll reached 10 million in 2020, with lung cancer claiming the highest number of lives, accounting for 18.0% of total cancer deaths, followed by liver cancer (8.3%) and stomach cancer (7.7%) [2]. The global cancer mortality rate is increasing annually, leading to an estimated 16 million future deaths per year by 2040 [3]. With advancements in science and technology, state-of-the-art diagnostic techniques and therapeutic methods have been applied to control various types of cancers, improving the survival of patients to some extent [4,5,6,7]. Chemotherapy is one of the broadest cancer treatment strategies in clinical application, but the conventional chemotherapeutic agents are often prone to developing cancer cell resistance or causing serious poisonous side effects to patients [8]. This urgent health crisis is propelling the need for innovative therapeutic agents with higher targeting efficiency and minimal side effects [9,10].
Natural products are a precious gift from nature to humankind, playing an irreplaceable role in drug discovery [11]. On the one hand, natural products can bind easily with biomolecules in cells and outwardly exhibit diverse biological activities [12,13]. On the other hand, natural products have the characteristic of structural diversity, which can provide rich sources of reference for the design of small-molecule drugs [14,15,16]. At present, more than 50% of clinical anticancer drugs are derived directly or indirectly from natural products [17,18,19].
ent-Kaurane diterpenoids, a widespread natural tetracyclic diterpenoid product, have received much attention because of their wide range of biological activities [20,21]. Oridonin (1, Figure 1), the most well-known natural ent-Kaurane diterpenoid product, has been reported in numerous studies for its inhibition of various cancer cells [22,23,24,25]. The enmein-type (1,7-lactone) diterpenoid is a sub-type of the ent-Kaurane diterpenoids. Partial enmein-type diterpenoids, such as Enmein, Rabdosin A, Sculponin A, Serrin B, and Isodocarpin (Figure 1), have shown micro-molar inhibitory effects against various cancer cells in vitro [26,27,28,29]. Although they demonstrate attractive anticancer potential, their lower natural yields have limited their further research and application. To solve this problem, Xu’s team constructed a semi-synthetic strategy for the synthesis of enmein-type diterpenoids from odidonin, and also obtained a variety of derivatives with potential for clinical anticancer applications [30]. In summary, enmein-type diterpenoids and their derivatives have the potential to be developed as anticancer drugs for clinical use.
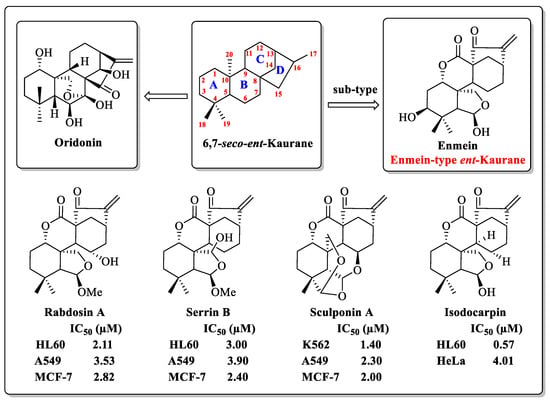
Figure 1.
The structure of oridonin and partial enmein-type dipterpenoids.
To diversify the enmein-type diterpenoids and explore their potential anticancer mechanisms, a novel 1,14-epoxy enmein-type diterpenoid was designed and synthesized. After that, a compound library containing 21 novel enmein-type diterpenoid derivatives was also constructed in this project. The anticancer activities of all derivatives were tested via MTT assay, and their structure–activity relationships (SARs) were summarized. As the lead compound with the most potential, 7h was selected for further investigation of its anticancer mechanisms.
2. Results and Discussion
2.1. Synthesis of Enmein-Type Diterpenoid Derivatives
The synthesis of enmein-type diterpenoids is outlined in Scheme 1. Oridonin, as the starting material, was oxidised by Johns’ reagent at 0 °C to synthesize the ent-Kaurene 2 [30]. Compound 2 was oxidized by lead tetraacetate and anhydrous sodium carbonate to produce the spironolactone-type diterpenoid 3 [30]; after that, through further advancing the reaction, compound 3 was converted to the enmein-type diterpene compound 4. The assumed mechanisms of conversion from compound 2 to spironolactone-type diterpenoid 3 and enmein-type diterpenoid 4 are shown in Figure 2. The chemical structure of compound 4 was confirmed by HRMS, 1H NMR, 13C NMR, and crystal data.
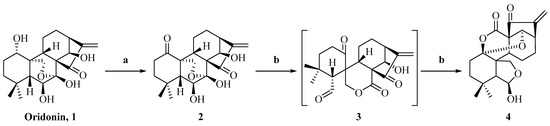
Scheme 1.
Synthesis of enmein-type diterpenoid derivative 4. Reagents and conditions: (a) Acetone, Jones Reagent, 0 °C, 1 h; (b) THF, anhydrous sodium carbonate, lead tetraacetate, 25 °C, 1 h.
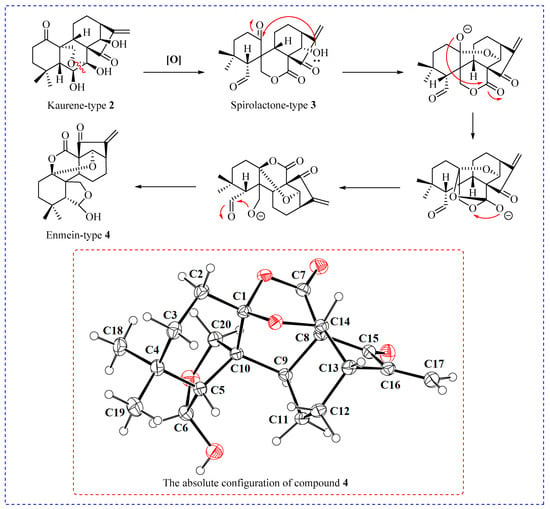
Figure 2.
The potential mechanism of conversion from compound 2 to enmein-type diterpenoid 4, and the absolute configuration of compound 4.
The cationic amino group, including carbamates, which are commonly used in the structural modification of natural products, can facilitate the binding between small molecules and protein receptors through electrostatic interactions, and further enhance the bio-activities of natural products [31,32]. We designed a series of novel derivatives, 5a–m, synthesized by introducing different amino acid esters at the C-6 position of compound 4 via acylation reaction (Scheme 2). The chemical structures of all derivatives, 5a–m, were determined by HRMS, 1H NMR, and 13C NMR spectra data.
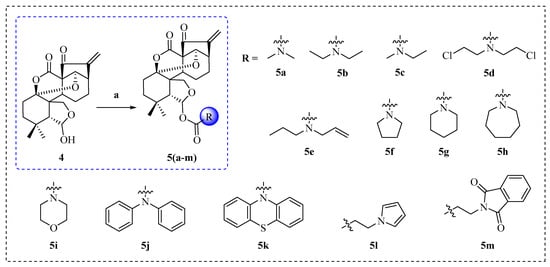
Scheme 2.
Synthesis of enmein-type diterpenoid derivatives 5a–m. Reagents and conditions: (a) Acyl chlorides, DCM, TEA, DMAP, 25 °C, 24 h.
To explore whether a flexible side chain containing aromatic groups could enhance the anticancer activities of enmein-type diterpenoids, a series of novel derivatives, 7a–h, were designed and synthesized (Scheme 3). Compound 4 and succinic anhydride were catalyzed by 4-dimethylaminopyridine (DMAP) to form intermediate compound 6 [33,34]. After that, compound 6 was acylated with different amines to obtain the target products, 7a–h. The chemical structures of all derivatives were determined by HRMS, 1H NMR, and 13C NMR spectral data.
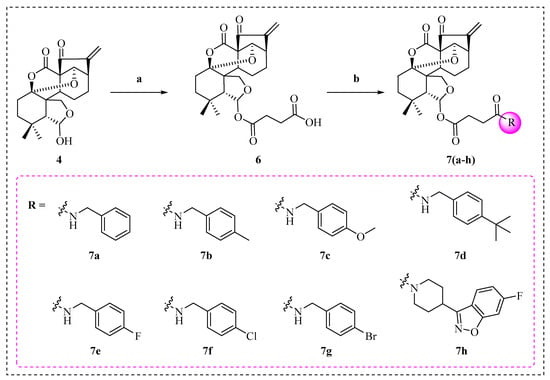
Scheme 3.
Synthesis of enmein-type diterpenoid derivatives 7a–h. Reagents and conditions: (a) Amines, DMAP, DCM, 25 °C, 4 h; (b) suitable substituent, DCM, PyBOP, TEA, 25 °C, 3 h.
2.2. In Vitro Anti-Proliferative Activity and SAR Analysis
A series of novel enmein-type diterpenoid derivatives, 5a–m, were synthesized by introducing a side chain containing a carbamate moiety at the C-6 position of compound 4. The anticancer activities of 5a–m were tested against three human cancer cell lines and one human normal cell line by MTT assay. The results are presented in Table 1.

Table 1.
In vitro anti-proliferative activities of 5a–m and 7a–h in different cell lines.
In A549 cells, all of the derivatives, 5a–m, exhibited better anticancer activities than their parental compound 4 (IC50 = 23.83 µM). Among them, compound 5m (IC50 = 6.85 µM), which contained an indole moiety, displayed the most remarkable anticancer activity, which was 3.48-fold better than that of compound 4. For the HepG2 cells and the MCF-7 cells, most derivatives showed better anticancer activities compared with their parental compound 4; however, overall, neither reached micro-molar levels (IC50 < 10.00 µM). It is noteworthy that the derivatives containing both longer flexible linker arms and aromatic groups in their side chains, such as 5l and 5m, exhibited better anticancer activities than other derivatives. However, the derivatives containing only aliphatic chain groups (5d, 5e) or aromatic groups (5j, 5k) in their side chains failed to exhibit anticancer activities at the low micromolar level. We assumed that the flexible linker arms and aromatic groups in the side chains are indispensable for the derivatives’ enhanced anticancer activities.
To verify the above conjecture, a series of novel derivatives, 7a–h, were synthesized using coupling compound 4 and aromatic groups, using succinic acid as a linker. The anticancer activities of 7a–h were tested against three human cancer cell lines and one human normal cell line by MTT assay. The results are also presented in Table 1.
In A549 cells, all derivatives, 7a–h, exhibited better anticancer activities than compound 4, and all attained low micro-molar levels (IC50 < 10.00 µM). Among them, derivative 7h exhibited the best anticancer activity (IC50 = 2.16 µM), which was 11.03-fold better than compound 4, and which was better than the positive control taxol. Furthermore, 7h also exhibited a relatively weak cytotoxic activity (IC50 > 50 µM) against human normal L-02 cell lines, indicating an excellent cancer cell-targeting property. For HepG2 cells, compounds 7f, 7g, and 7h showed low micro-molar-level anticancer activities. For MCF-7 cells, all compounds except compounds 7a, 7b, and 7g showed low micro-molar-level anticancer activities. These data suggest that the introduction of a suitable linking arm between the diterpenoid parental nucleus and the active moiety can remarkably improve the anticancer activities of derivatives.
Taken together with a suitable linking arm, derivative 7h, which was successively substituted with succinic acid and 6-fluoro-3-(piperidin-4-yl) benzo[d]isoxazole groups, exhibited low micro-molar-level inhibitory effects against all tested cancer cells, suggesting that it is a potential lead compound for clinical anticancer applications. Therefore, compound 7h was selected for further anticancer-mechanism exploration in A549 cells.
2.3. Cell Cycle Analysis
Cell cycle regulation is an essential process in maintaining normal bodily functions and is often disturbed in cancer cells. The inducement of cell cycle arrest is considered a key avenue for targeted cancer therapy [35]. As a novel enmein-type diterpenoid derivative with remarkable anti-proliferative effects against A549 cells, 7h may have the ability to induce cell cycle arrest. Therefore, A549 cells were treated with 7h, stained with PI/RNase solution, and analyzed for their cell cycle distribution by flow cytometry. The experimental results are shown in Figure 3. The detailed data revealed that, under the treatments with different concentrations (0, 2, 4, and 8 µM) of 7h, 56.37%, 62.99%, 71.04%, and 89.87% of A549 cells, respectively, were found to be in the G0/G1 phase. The above-mentioned results demonstrate that 7h effectively inhibited the proliferation of A549 cells by arresting the cell cycle at the G0/G1 phase, and this effect also showed a marked dose-dependent trend.
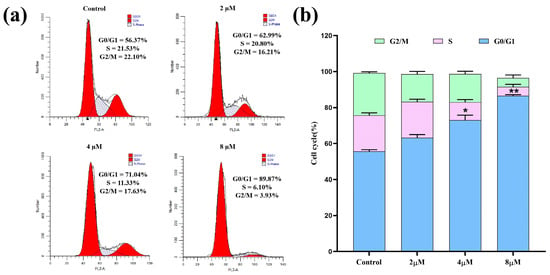
Figure 3.
Compound 7h induced cell cycle arrest in A549 cells. (a) A549 cells were treated with 7h (2, 4, and 8 µM) and 0.1% DMSO (the blank control group), and stained by PI/RNase. (b) The percentage of cell cycle arrest in different experimental groups (*: p < 0.05, **: p < 0.01, compared to the blank control group). All data are expressed as mean ± standard deviation (n = 3).
2.4. Apoptosis Analysis
To explore whether compound 7h could induce apoptosis in cancer cells, A549 cells were treated with different concentrations (0, 2, 4, and 8 µM) of 7h and then stained with AO/EB double staining. The morphological variations in the cells were observed using fluorescence microscopy, and the results are shown in Figure 4a. In the blank control group, normal cells were stained green with AO staining solution with intact nuclei. With the increased concentration of 7h, the cells gradually produced typical morphological transformations, such as nuclear chromatin condensation, cytoplasmic condensation, cell fragmentation, etc., and generated large amounts of apoptotic cells (EB staining in red).
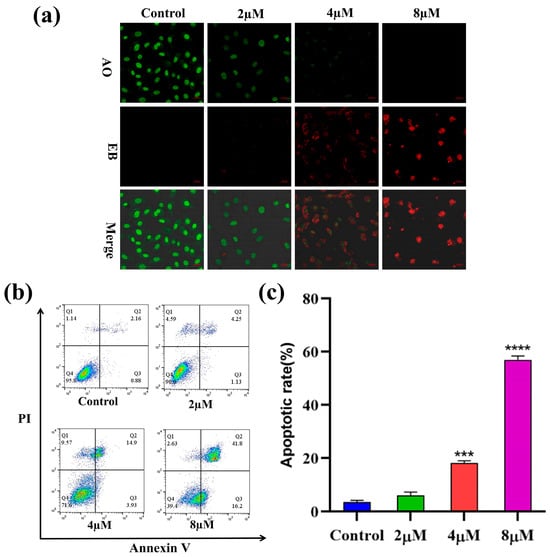
Figure 4.
Compound 7h induced A549 cell apoptosis. (a) A549 cells were treated with 7h (2, 4, and 8 µM) and 0.1% DMSO (blank control), and then stained by AO/EB. (b) A549 cells were treated with 7h (2, 4, and 8 µM) and 0.1% DMSO (the blank control group), and then stained by Annexin V-FITC/PI. (c) The percentage of apoptosis in different experimental groups (***: p < 0.001, ****: p < 0.0001, compared to the blank control group). All data are expressed as mean ± standard deviation (n = 3).
To further confirm the capacity of 7h to induce apoptosis in cancer cells, A549 cells were treated with 7h, detected using Annexin V-FITC/PI staining, and the apoptosis rate was detected by flow cytometry. The results showed that the percentage of total apoptosis in A549 cells treated with different concentrations of 7h increased from 5.38% to 58.00%, as opposed to an increase of 3.04% in the control group (Figure 4b,c). Among the types of apoptosis which occurred, late apoptosis was the main form in all apoptosis cells, increasing from 2.16% to 41.8%. Notably, the percentage of apoptosis in the A549 cells was correlated with 7h in a positively dose-dependent manner. The above results indicated that compound 7h could significantly induce apoptosis in A549 cells.
2.5. Reactive Oxygen Species (ROS) Analysis
Reactive oxygen species (ROS) are secondary metabolites of cells and are closely linked to the regulation of multiple biological activities. Elevated intracellular ROS levels are thought to be a hallmark of apoptosis [35]. To determine whether 7h could affect the ROS content in cancer cells, the A549 cells were treated with different concentrations (0, 2, 4, and 8 µM) of 7h and then stained with H2DCFDA. The results of the laser scanning confocal microscopy observations that we conducted on the stained cells are shown in Figure 5a. Compared with the blank control group, all experimental groups treated with 7h were observed to exhibit remarkable green fluorescence, and the fluorescence intensity was positively correlated with the concentration of 7h in a dose-dependent manner.
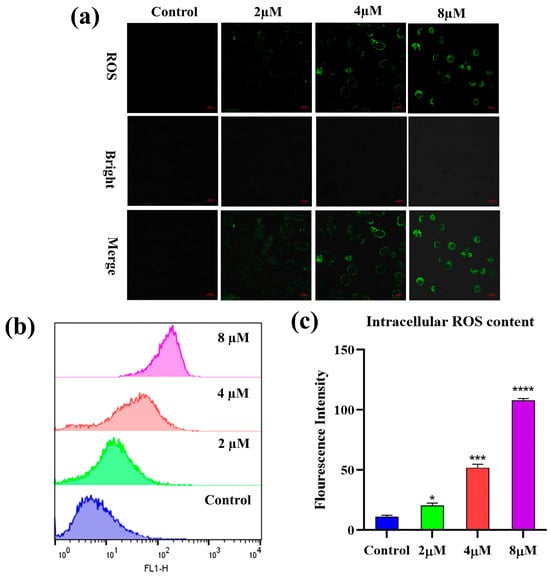
Figure 5.
Compound 7h induced ROS generation in A549 cells. (a,b) A549 cells were treated with 7h (2, 4, and 8 µM) and 0.1% DMSO (blank control), and then stained by H2DCFDA. (a) The morphological detection of A549 cells. (b) The determination of ROS content in A549 cells. (c) The percentage of ROS content in different experimental groups (*: p < 0.05, ***: p < 0.001, ****: p < 0.0001, compared to the blank control group). All data are expressed as mean ± standard deviation (n = 3).
Flow cytometry was used to detect changes in intracellular reactive oxygen species content, and the results are shown in Figure 5b,c. The intracellular ROS content of the 7h-treated A549 cells was increased significantly in a dose-dependent manner compared to the control group. The above results suggest that 7h-induced apoptosis in A549 cells may be involved in the notable induction of intracellular ROS production.
2.6. Mitochondrial Membrane Potential (MMP) Analysis
The depolarization of MMP is reckoned to be a hallmark of apoptosis [36]. To investigate the impact of 7h on the depolarization of MMP (δ:Ψm) in cancer cells, A549 cells were treated with different concentrations (0, 2, 4, and 8 µM) of 7h and then stained with JC-1. The laser confocal microscopy observations are shown in Figure 6a. In the blank control group, most cells were stained red with JC-1, indicating a high mitochondrial membrane potential. In contrast, in the 7h-treated experimental groups, the number of red cells decreased while the number of green cells increased, indicating a significant reduction in intracellular MMP. The results of the flow cytometry assay are illustrated in Figure 6b,c.
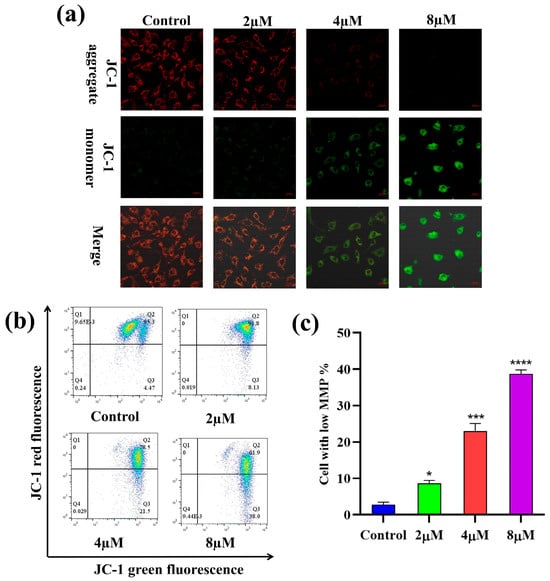
Figure 6.
Compound 7h induced MMP in A549 cells. (a,b) A549 cells were treated with 7h (2, 4, and 8 µM) and 0.1% DMSO (blank control), and then stained by JC-1. (a) The morphological detection of A549 cells. (b) The determination of MMP depolarization rate in A549 cells. (c) The percentage of MMP depolarization rate in different experimental groups (*: p < 0.05, ***: p < 0.001, ****: p < 0.0001, compared to blank the control group). All data are expressed as mean ± standard deviation (n = 3).
The results showed that the percentage of cells with depolarized MMP increased in a dose-dependent manner after the A549 cells were treated with different concentrations (0, 2, 4, and 8 µM) of 7h, from 3.24% to 15.21%, 25.6%, and 59.6%, respectively. Altogether, these findings suggest that 7h-induced cell cycle arrest and apoptosis in A549 cells may be linked to the depolarization of intracellular MMP.
2.7. The Predicted Potential Targets and Pathways of 7h
To further explore the anticancer mechanism of compound 7h, we predicted potential targets by molecular docking in the PharmMapper platform [37,38]. A total of 106 target proteins (Z-score > 0.5) were selected (Table S1). The Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment results were analyzed using the David platform [39,40].
GO enrichment analysis was performed against 7h to predict its potential biological processes (BP), cellular composition (CC), and molecular functions (MF). The top 20 functional categories (the minimum of the p-value) were screened separately. Compound 7h in BP mainly involves the hormone-mediated signaling pathway, phosphorylation, RNA polymerase II promoter protein phosphorylation, cell differentiation, signal transduction, and others (Table S2, Figure S1, and Figure 7a). Compound 7h in CC is mainly located in the cytosol, RNA polymerase II transcription factor, extracellular space, nucleoplasm, and others (Table S3, Figure S2 and Figure 7b). Compound 7h in MF mainly involves zinc ion binding, identical protein binding, ATP binding, protein homodimerization activity, protein tyrosine kinase activity, and others (Table S4, Figure S3 and Figure 7c). KEGG enrichment analysis was performed against 7h and the top 20 potential pathways were screened (Table S5, Figure S4 and Figure 7d). The prediction suggests that the main effects of compound 7h may be on cancer-related pathways and proteins in cells, potentially including the PI3K-Akt signaling pathway, the Ras signaling pathway, proteoglycans, the thyroid hormone signaling pathway, and others. The PI3K-Akt signaling pathway is proposed to be one of the potential major anticancer pathways of compound 7h. Taken together with compound 7h, the PI3K-Akt signaling pathway was selected for further studies to elucidate the exact anticancer mechanism of compound 7h.
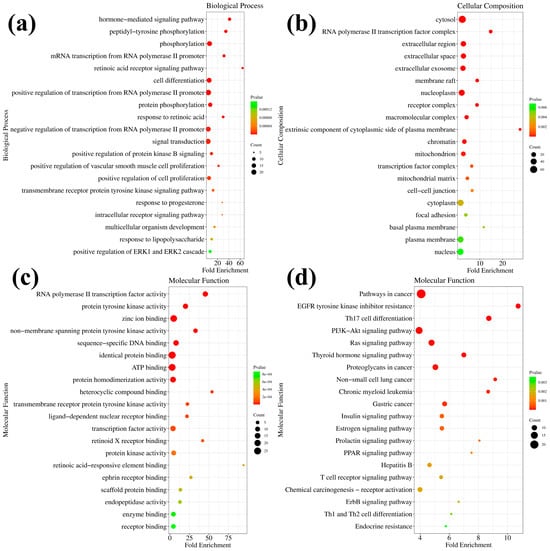
Figure 7.
GO and KEGG enrichment analysis of 7h. (a) The predicted molecular functions of 7h based on GO analysis. (b) The predicted biological processes of 7h based on GO analysis. (c) The predicted cellular compositions of 7h based on GO analysis. (d) The predicted pathways of 7h based on KEGG analysis.
2.8. Western Blot Analysis
The PI3K/Akt/mTOR pathway is a critical intracellular signaling pathway during cancer development that regulates cellular processes such as proliferation, differentiation, apoptosis, and autophagy. Inhibiting the aberrant activation of the PI3K/Akt/mTOR signaling pathway is envisaged as a promising anticancer strategy [41,42,43]. Based on KEGG analysis and researching the literature, the PI3K/Akt/mTOR signaling pathway is predicted to be one of the major pathways for 7h to exert its anticancer effects. Therefore, a western blot assay was performed to determine the expression of the main proteins, PI3K, p-PI3K, Akt, p-Akt, mTOR, and p-mTOR, to examine whether the anticancer ability of 7h against A549 cells is related to its modulation of the PI3K/Akt/mTOR pathway. The results demonstrated that 7h can effectively reduce the phosphorescent expression levels of p-PI3K and p-mTOR in A549 cells compared with the blank control in a dose-dependent manner (Figure 8). In addition, 7h also showed an effect on Akt protein phosphorylation expression. The above results suggest that 7h can exert significant anticancer effects by modulating the PI3K/Akt/mTOR signaling pathway in A549 cells.
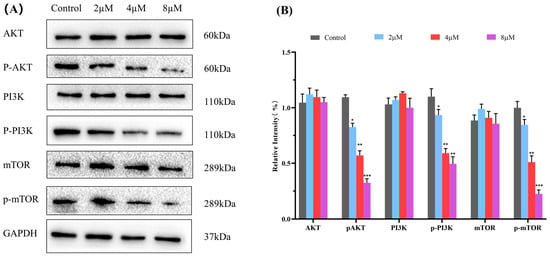
Figure 8.
Compound 7h inhibited the PI3K/AKT/mTOR pathway. (A) Western blot analysis of the expression levels of PI3K, p-PI3K, Akt, p-AKT, mTOR, and p-mTOR proteins in A549 cells treated with 7h (2, 4, and 8 µM) and 0.1% DMSO (the blank control group). (B) Statistical analysis. *: p < 0.05, **: p < 0.01, ***: p < 0.001, compared to blank the control group. All data are expressed as mean ± standard deviation (n = 3).
2.9. Molecular Docking Study
To explore the binding ability between ligand compounds and potential receptor proteins, compound 7h and its parental compound 4 were subjected to molecular docking studies with the target proteins PI3Kα (PDB ID: 6GVF), Akt1 (6HHF), and mTOR (4JT6), respectively [44,45]. The docking results are shown in Table 2. The docking study results showed that compound 7h exhibited superior binding ability to all three target proteins compared to its parental compound 4. Among them, compound 7h had the lowest docking binding energy (−11.6 kcal/mol) with the PI3Kα protein, which represented the highest binding affinity; therefore, PI3Kα might be one of the main targets of action of compound 7h.

Table 2.
The docking results of compounds 7h and 4.
After that, we visualized and analyzed the above docking results. The 3D action models of the PI3Kα protein with compounds 7h and 4, respectively, are shown in Figure 9a,c. The diterpene parent nucleus and 6-fluoro-3-(piperidin-4-yl) benzo[d]isoxazole moiety of compound 7h were embedded in the active pockets of the PI3Kα protein, respectively. Compared with compound 4, compound 7h showed a remarkable increase in the active pocket occupancy and binding stability of the PI3Kα protein. The 2D modelling of the interaction of the PI3Kα protein with compounds 7h and 4, respectively, is shown in Figure 9b,d. The 2D models of the interaction of the PI3Kα protein with compounds 7h and 4, respectively, are also shown in Figures S5 and S6 in the Supplementary Materials. Compound 7h has the ability to bind to amino acid residues in the PI3Kα protein via hydrogen bonds (LYS271, ARG818, ASN756, GLN815, and GLN630), pi-cation (LYS271 and ARG818), pi-anion (GLU172), alkyl (PHE666), pi-alkyl (LYS271 and ARG818) and halogen (ASN822). In addition, the van der Waals force field, which consists of equal amino acid residues (PRO835, GLY837, MET811, ILE633, SER629, and ASN170), is also crucial for compound 7h to be able to be immobilized in the active pocket of the PI3Kα protein.
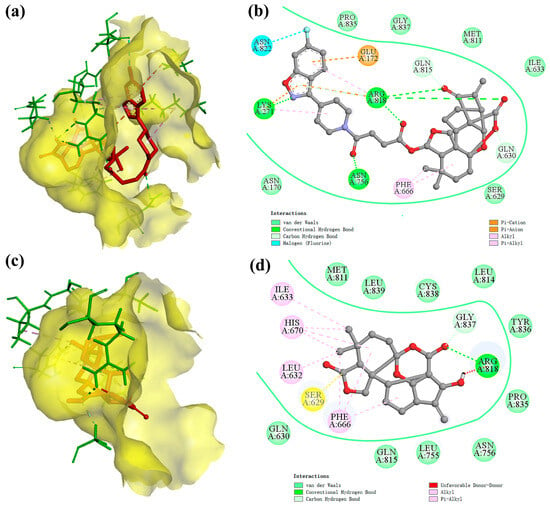
Figure 9.
The docking model of ligand molecules to receptor proteins. (a) The 3D interaction model of compound 7h with PI3Kα; (b) The 2D interaction model of compound 7h with PI3Kα; (c) The 3D interaction model of compound 4 with PI3Kα; (d) The 2D interaction model of compound 4 with PI3Kα.
Taken together, the introduction of a flexible side chain containing reactive groups at the C-6 position of the enmein-type diterpenoid 4 may enhance its PI3K inhibitory effect. Structural modifications that enhance the requisite capacity between the ligand molecule and its target protein may be a viable approach to improving the derivatives’ targeting abilities. The data and analysis of the above docking results provide critical clues for further structural modification or pharmacological research.
3. Conclusions
In conclusion, a novel 1,14-epoxy enmein-type diterpenoid and a library containing 21 novel enmein-type diterpenoid derivatives were constructed in this project. All the derivatives were evaluated for their anti-proliferative activities. Most derivatives displayed superior anti-proliferative activities against three cancer cell lines as compared with their parental compound 4. Derivative 7h, which was modified using succinic acid and an oxazole moiety, exhibited the most remarkable anti-proliferative activity against A549 cells, which was 11.03-fold higher than that of 4. Additionally, 7h exhibited a weak cytotoxicity against human normal L-02 cells and demonstrated a significant cancer cell-targeting property. The SARs analysis showed that the derivatives 6l, 6m, and 7a–h, which introduced a flexible linker arm between the diterpenoid parental core and the active moiety, exhibited more significant anticancer activity than derivatives 4a–k, which lacked the flexible linker arm. The biological studies revealed that 7h affected A549 cells by up-regulating their intracellular ROS levels, down-regulating their MMP levels, and inducing cell cycle arrest and apoptosis in a dose-dependent manner. Further mechanism studies showed that 7h exerted its anticancer effects through inhibition of the PI3K/Akt/mTOR pathway. Docking studies indicated that 7h exhibited the best binding effect with PI3K, suggesting its potential as a novel PI3K inhibitor through further structural modification. In conclusion, the above studies provide some leads for the exploration of enmein-type diterpene derivatives as potential anticancer agents. Meanwhile, it is hoped that this work inspires more research teams to focus on the study of enmein-type diterpenoids in cancer therapy.
4. Materials and Methods
4.1. Chemistry
4.1.1. Reagents and Apparatus
The starting material, Oridonin, was purchased from Chengdu Yirui Biotechnology Co., Ltd. (Chengdu, China), and the other reagents and solvents were purchased from Anegi (Shanghai, China) Medicinal Chemicals Co. All reagents were used directly after purchase and required no extra preparation. The chemical reactions were monitored using thin-layer chromatography, and the crude target products were purified using silica gel column chromatography. The silica gel preforms (model: HSGF254) and silica gel (100–200 mesh) were purchased from Yantai Yinlong Silica Gel Co (Yantai, China). The HRMS data were recorded on a Waters XEVO G2-XS QTOF high-resolution mass spectrometer. The NMR spectra were recorded on a Bruker AVANCE NEO 600 spectrometer, Fällanden, Switzerland, with CDCl3 or DMSO as solvent and tetramethylsilane (TMS) as internal standard. Coupling constants and chemical shift values are expressed in J (Hz) and d (ppm), respectively. The structures of all reported derivatives were consistent with the data from HRMS, 1H NMR, and 13C NMR spectra. The spectra of all derivatives are contained in Supplementary Materials.
4.1.2. Synthesis of Ent-6β,7β,14β-trihydroxy-1,15-dioxo-7,20-epoxy-16-kaurene (2)
Oridonin (1, 1.00 g, 2.74 mmol) was dissolved in ice-cold acetone (100 mL) and then Jones reagent was dropped slowly into the system at 0 °C and stirred for 1 h. The reaction was monitored throughout the experiment via thin-layer chromatography (TLC). After the reaction, the solution was removed by spinning under reduced pressure, redissolved by dichloromethane (DCM, 100 mL), and then extracted sequentially with 5% sodium bicarbonate solution, distilled water, and saturated saline. The organic phase was separated and dried with anhydrous sodium sulfate. The organic solvent was removed by decompression evaporation, and the crude mixture was purified by flash column chromatography (DCM/MeOH, 25/1, v/v), resulting in the target intermediate compound 2 (0.90 g, Yield: 90%) as a white granule [30].
4.1.3. Synthesis of Ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene (4)
Compound 2 (0.36 g, 1.00 mmol) and anhydrous sodium carbonate (1.06 g, 10.00 mmol) were co-dissolved in anhydrous tetrahydrofuran (100 mL). The mixture was reacted at 25° C and the lead tetraacetate (2.00 g, 4.5 mmol) was added in batches (0.5 g/per/5 min). After the reaction, the insoluble impurities were filtered off, and the solution was removed by spinning under reduced pressure, redissolved by DCM (100 mL), and then extracted sequentially with 5% sodium bicarbonate solution, distilled water, and saturated saline. The organic phase was separated and dried with anhydrous sodium sulfate. The organic solvent was removed by decompression evaporation, and the crude mixture was purified by flash column chromatography (DCM/MeOH, 50/1, v/v), resulting in the target compound 4 (0.31 g, Yield: 85%) as a white granule. 1H NMR (600 MHz, CDCl3) δ values were as follows: 6.28 (1H, s, CH2-17), 5.54 (1H, s, CH2-17), 5.43 (1H, brs, H-6), 4.26 (1H, d, J = 5.7 Hz, H-14), 3.99 (1H, d, J = 9.7 Hz, CH2-20), 3.73 (1H, d, J = 9.7 Hz, CH2-20), 3.25 (1H, m, H-13), 2.53 (1H, m, CH2-12), 2.51 (1H, m, CH2-2), 2.16 (1H, m, H-9), 2.08 (2H, m, CH2-2, CH2-12), 1.99 (1H, d, J = 13.8 Hz, CH-3), 1.80 (1H, m, CH2-11), 1.60 (2H, m, CH2-3, H-5), 1.47 (1H, d, J = 13.8 Hz, CH2-11), 1.12 (3H, s, CH3-18), 1.01 (3H, s, CH3-19). 13C NMR (151 MHz, CDCl3) δ 194.53 (C15), 168.84 (C7), 146.52 (C16), 121.39 (C17), 107.43 (C1), 102.01 (C6), 79.23 (C14), 72.71 (C20), 54.43 (C8), 53.54 (C5), 53.35 (C10), 46.82 (C13), 39.63 (C9), 36.11 (C3), 33.29 (C4), 30.69 (C2), 27.76 (C12), 23.66 (C18), 23.37 (C19), and 19.63 (C11). HRMS (ESI) m/z: calculated for C20H24O6 [M + Na]+, 383.1471, found, 383.1468. The single crystal data for compound 4 is detailed in Supplemental Materials.
4.1.4. General Procedure for the Synthesis of Target Derivatives 5a–m
Compound 3 (1.00 mmol) and appropriate acyl chloride (1.50 mmol) were co-dissolved in anhydrous DCM (20 mL), DMAP (0.1 mmol) and triethylamine (1.50 mmol) were added as catalysts, and the mixture was stirred for 24 h at 25° C. After the reaction, the solution was removed by spinning under reduced pressure, redissolved by DCM (100 mL), and then extracted sequentially with 5% sodium bicarbonate solution, distilled water, and saturated saline. The organic solvent was removed by decompression evaporation, and the crude mixture was purified by flash column chromatography. The solid was precipitated from n-hexane to give the target derivatives 5a–m.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl dimethylcarbamate (5a). Yield: 85%, white granule. 1H NMR (600 MHz, Chloroform-d) values were as follows: δ 6.31 (1H, s, CH2-17), 6.23 (1H, s, CH2-17), 5.57 (1H, brs, H-6), 4.29 (1H, d, J = 5.7 Hz, H-14), 4.04 (1H, d, J = 9.9 Hz, CH2-20), 3.77 (1H, d, J = 9.9 Hz, CH2-20), 3.29 (1H, m, H-13), 2.91 (3H, s, CH3-1′), 2.78 (3H, s, CH3-2′), 2.51 (1H, d, J = 5.6 Hz, H-9), 2.23 (1H, s), 2.18 (1H, d, J = 9.8 Hz), 2.10 (2H, m), 2.02 (1H, d, J = 14.0 Hz), 1.81 (1H, m), 1.67 (1H, m), 1.63 (1H, m), 1.51 (1H, d, J = 13.8 Hz), 1.19 (3H, s, CH3-18), 1.08 (3H, s, CH3-19). 13C NMR (151 MHz, CDCl3) δ 194.13 (C15), 168.25 (C7), 155.12 (C1′), 146.23 (C16), 121.82 (C17), 107.23 (C1), 103.22 (C6), 79.70 (C14), 72.71 (C20), 54.10 (C8), 53.24 (C5), 52.76 (C10), 46.21 (C13), 39.39 (C9), 36.64 (C1′), 36.09 (C3), 35.93 (C2′), 33.18 (C4), 31.15 (C2), 27.68 (C12), 23.65 (C18), 23.30 (C19), and 19.71 (C11). HRMS (ESI) m/z: calculated for C23H29NO7 [M + Na]+, 454.1842, found, 454.1847.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl diethylcarbamate (5b). Yield: 82%, white granule. 1H NMR (600 MHz, Chloroform-d) values were as follows: δ 6.30 (1H, s, CH2-17), 6.23 (1H, s, CH2-17), 5.57 (1H, brs, H-6), 4.28 (1H, d, J = 5.7 Hz, H-14), 4.03 (1H, d, J = 9.9 Hz, CH2-20), 3.75 (1H, d, J = 9.9 Hz, CH2-20), 3.28 (2H, m, CH2-2′), 3.25 (1H, m, H-13) 3.13 (1H, m, H-3′), 3.07 (1H, m, H-3′), 2.48 (1H, d, J = 5.4 Hz, H-9), 2.24 (1H, s), 2.17 (1H, d, J = 9.8 Hz), 2.10 (2H, m), 2.01 (1H, d, J = 14.0 Hz), 1.81 (1H, m), 1.60 (2H, m), 1.50 (1H, d, J = 13.8 Hz), 1.18 (3H, s, CH3-18), 1.11 (3H, m, CH3-4′), 1.07 (3H, s, CH3-19), 1.03 (3H, m, CH3-5′). 13C NMR (150 MHz, CDCl3) δ: 194.14 (C15), 168.26 (C7), 154.53 (C1′), 146.19 (C16), 121.86 (C17), 107.27 (C1), 103.09 (C6), 79.67 (C14), 72.56 (C20), 54.14 (C8), 53.24 (C5), 52.69 (C10), 46.27 (C13), 41.91 (C2′), 41.06 (C3′), 39.41 (C9), 36.13 (C3), 33.21 (C4), 31.21 (C2), 27.69 (C12), 23.67 (C18), 23.29 (C19), 19.61 (C11), 14.24 (C4′), and 13.41 (C5′). HRMS (ESI) m/z: calculated for C25H33NO7 [M + Na]+, 482.2155, found, 482.2154.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl ethyl(methyl)carbamate (5c). Yield: 86%, white granule. 1H NMR (600 MHz, Chloroform-d) values were as follows: δ 6.31 (1H, s, CH2-17), 6.23 (1H, s, CH2-17), 5.58 (1H, brs, H-6), 4.29 (1H, d, J = 5.7 Hz, H-14), 4.04 (1H, d, J = 9.9 Hz, CH2-20), 3.77 (1H, d, J = 9.9 Hz, CH2-20), 3.29 (2H, m, CH2-2′), 3.14 (1H, d, J = 46.9 Hz, H-13), 2.81 (3H, d, J = 79.5 Hz, CH3-3′), 2.50 (1H, d, J = 13.2 Hz, H-9), 2.24 (1H, s), 2.18 (1H, m), 2.10 (2H, m), 2.02 (1H, d, J = 13.9 Hz), 1.81 (1H, m), 1.66 (1H, m), 1.63 (1H, m), 1.51 (1H, d, J = 13.9 Hz), 1.19 (3H, s, CH3-18), 1.11-1.03 (6H, m, CH3-19, CH3-3′). 13C NMR (151 MHz, CDCl3) δ 194.02 (C15), 168.14 (C7), 154.57 (C1′), 146.1 (C16), 121.69 (C17), 107.14 (C1), 102.98 (C6), 79.58 (C14), 72.47 (C20), 54.01 (C8), 53.14 (C5), 52.62 (C10), 46.13 (C13), 43.89 (C2′), 39.29 (C9), 36.00 (C3), 33.08 (C4), 31.04 (C2), 29.70 (C3′), 27.58 (C12), 23.55 (C18), 23.20 (C19), 19.60 (C11) and 12.43 (C4′). HRMS (ESI) m/z: calculated for C24H31NO7 [M + Na]+, 468.1998, found, 468.1993.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl bis(2-chloroethyl) carbamate (5d). Yield: 80%, white granule. 1H NMR (600 MHz, CDCl3) values were as follows: δ 6.31 (1H, s, CH2-17), 6.20 (1H, s, CH2-17), 5.58 (1H, brs, H-6), 4.28 (1H, d, J = 5.7 Hz, H-14), 4.04 (1H, d, J = 9.9 Hz, CH2-20), 3.76 (1H, d, J = 9.9 Hz, CH2-20), 3.70 (2H, m, CH2-2′), 3.65 (2H, m, CH2-2′), 3.56 (2H, m, CH2-2′), 3.49 (2H, m, CH2-2′), 3.28 (1H, m, H-13), 2.45 (1H, m, H-9), 2.26 (1H, s), 2.10 (2H, m), 2.07 (1H, m), 2.08 (1H, d, J = 13.8 Hz), 2.02 (1H, d, J = 13.9 Hz), 1.82 (1H, m), 1.73 (1H, m), 1.63 (1H, m), 1.51 (1H, d, J = 14.0 Hz), 1.18 (3H, s, CH3-18), 1.07 (3H, s, CH3-19). 13C NMR (151 MHz, CDCl3) δ 193.80 (C15), 168.10 (C7), 154.65 (C1′), 146.01 (C16), 122.05 (C17), 107.03 (C1), 103.96 (C6), 79.88 (C14), 72.55 (C20), 54.02 (C8), 53.10 (C5), 52.71 (C10), 50.85 (C2′), 50.11 (C3′), 46.02 (C13), 41.85 (C4′), 41.32 (C5′), 39.32 (C9), 36.04 (C3), 33.21 (C4), 31.25 (C2), 27.65 (C12), 23.63 (C18), 23.18 (C19), and 19.68 (C11). HRMS (ESI) m/z: calculated for C25H31Cl2NO7 [M + Na]+, 550.1376, found, 550.1375.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl diallylcarbamate (5e). Yield: 85%, white granule. 1H NMR (600 MHz, DMSO) values were as follows: δ 6.13 (2H, s, CH2-17), 5.74 (1H, m, CH-4′), 5.69 (1H, brs, H-6), 5.66 (1H, m, CH-5′), 5.12 (2H, d, J = 10.7 Hz, CH2-7′), 5.07 (1H, d, J = 10.4 Hz, CH2-6′), 5.01 (1H, d, J = 10.4 Hz, CH2-6′), 4.41 (1H, d, J = 4.5 Hz, H-14), 4.04 (1H, d, J = 9.6 Hz, CH2-20), 3.81 (1H, m, CH2-3′), 3.73 (2H, d, J = 15.7 Hz, CH2-2′), 3.64 (1H, m, CH2-3′), 3.51 (1H, d, J = 9.6 Hz, CH2-20), 3.30 (1H, m, H-13), 2.32 (1H, d, J = 5.6 Hz), 2.19 (1H, s), 2.16 (1H, d, J = 10.2 Hz), 2.06 (1H, d, J = 10.2 Hz), 1.99 (1H, m), 1.86 (1H, d, J = 13.8 Hz), 1.70 (1H, m), 1.52 (1H, m), 1.46 (2H, d, J = 13.8 Hz), 1.23 (1H, s), 1.09 (3H, s, CH3-18), 0.99 (3H, s, CH3-19). 13C NMR (151 MHz, DMSO) δ 193.72 (C15), 167.64 (C7), 154.06 (C1′) 146.30 (C16), 133.64 (C4′), 133.38 (C5′), 121.31 (C17), 117.16 (C6′), 115.79 (C7′), 106.62 (C1), 102.26 (C6), 78.84 (C14), 71.41 (C20), 53.29 (C8), 52.49 (C5), 51.78 (C10), 48.85 (C2′), 47.76 (C3′), 45.47 (C13), 38.27 (C9), 35.36 (C3), 32.59 (C4), 30.69 (C2), 27.03 (C12), 23.22 (C18), 22.62 (C19), and 19.30 (C11). HRMS (ESI) m/z: calculated for C27H33NO7 [M + Na]+, 506.2155, found, 506.2152.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl pyrrolidine-1-carboxylate (5f). Yield: 80%, white granule. 1H NMR (600 MHz, DMSO) values were as follows: δ 6.12 (2H, d, J = 6.0 Hz, CH2-17), 5.70 (1H, brs, H-6), 4.41 (1H, d, J = 4.5 Hz, H-14), 4.02 (1H, d, J = 9.6 Hz, CH2-20), 3.53 (1H, d, J = 9.6 Hz, CH2-20), 3.30 (1H, m, H-13), 3.22 (2H, m, CH2-2′), 3.15 (2H, m, CH2-5′), 2.35 (1H, d, J = 5.4 Hz, H-9), 2.23 (1H, d, J = 10.1 Hz), 2.17 (2H, m), 1.99 (1H, m), 1.86 (1H, d, J = 13.9 Hz), 1.77 (2H, m, CH2-3′), 1.74 (2H, m, CH2-4′), 1.70 (1H, m), 1.55 (2H, m), 1.46 (1H, d, J = 13.7 Hz), 1.08 (3H, s, CH3-18), 0.99 (3H, s, CH3-19). 13C NMR (151 MHz, DMSO) δ 193.81 (C15), 167.69 (C7), 152.56 (C1′) 146.36 (C16), 121.28 (C17), 106.65 (C1), 101.13 (C6), 78.71 (C14), 71.48 (C20), 53.41 (C8), 52.57 (C5), 51.85 (C10), 46.11 (C13), 45.69 (C2′), 45.23 (C5′), 38.30 (C9), 35.34 (C3), 32.67 (C4), 30.64 (C2), 27.08 (C12), 25.25 (C3′), 24.26 (C4′), 23.25 (C18), 22.70 (C19), and 19.42 (C11). HRMS (ESI) m/z: calculated for C25H31NO7 [M + Na]+, 480.1998, found, 480.2003.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl piperidine-1-carboxylate (5g). Yield: 76%, white granule. 1H NMR (600 MHz, DMSO) values were as follows: δ 6.14 (2H, d, J = 4.5 Hz, CH2-17), 5.70 (1H, brs, H-6), 4.41 (1H, d, J = 4.4 Hz, H-14), 4.04 (1H, d, J = 9.6 Hz, CH2-20), 3.56 (1H, d, J = 9.6 Hz, CH2-20), 3.31 (1H, m, H-13), 3.30-3.14 (4H, m, CH2-2′, CH2-6′) 2.34 (1H, d, J = 5.6 Hz, H-9), 2.16 (2H, m), 2.02 (2H, m), 1.86 (1H, d, J = 13.9 Hz), 1.70 (1H, m), 1.55 (1H, d, J = 10.4 Hz), 1.50 (2H, m, CH2-4′), 1.47 (1H, m), 1.44 (1H, m), 1.42 (2H, m, CH2-3′), 1.37 (2H, m, CH2-5′), 1.10 (3H, s, CH3-18), 0.99 (3H, s, CH3-19). 13C NMR (151 MHz, DMSO) δ 194.25 (C15), 168.08 (C7), 153.36 (C1′), 146.73 (C16), 121.86 (C17), 107.11 (C1), 102.74 (C6), 79.29 (C14), 71.88 (C20), 53.74 (C8), 52.97 (C5), 52.56 (C10), 46.03 (C13), 44.51 (C2′, C6′), 38.71 (C9), 35.82 (C3), 33.05 (C4), 31.14 (C2), 27.52 (C12), 25.52 (C3′, C5′), 34.12 (C4′), 23.68 (C18), 23.11 (C19), and 19.70 (C11). HRMS (ESI) m/z: calculated for C26H33NO7 [M + Na]+, 494.2155, found, 494.2151.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl azepane-1-carboxylate (5h). Yield: 72%, white granule. 1H NMR (600 MHz, DMSO) values were as follows: δ 6.28 (2H, d, J = 3.9 Hz, CH2-17), 5.70 (1H, brs, H-6), 4.42 (1H, d, J = 4.5 Hz, H-14), 4.04 (1H, d, J = 9.6 Hz, CH2-20), 3.53 (1H, d, J = 9.6 Hz, CH2-20), 3.39 (1H, m, CH2-2′), 3.30 (1H, m, H-13), 3.26 (1H, m, CH2-3′), 3.23 (1H, m, CH2-2′), 3.13 (1H, m, CH2-3′), 2.31 (1H, d, J = 5.6 Hz, H-9), 2.19 (2H, m), 2.14 (1H, m), 2.01 (1H, m), 1.86 (1H, d, J = 13.8 Hz), 1.71 (1H, m), 1.59 (2H, d, J = 5.4 Hz), 1.56 (1H, m), 1.48 (4H, m, CH2-3′, CH2-6′), 1.44 (4H, m, CH2-4′, CH2-5′), 1.23 (1H, s), 1.09 (3H, s, CH3-18), 1.00 (3H, s, CH3-19). 13C NMR (151 MHz, DMSO) δ 193.84 (C15), 167.62 (C7), 154.03 (C1′), 146.28 (C16), 121.41 (C17), 106.66 (C1), 101.87 (C6), 78.79 (C14), 71.41 (C20), 53.37 (C8), 52.52 (C5), 51.85 (C10), 46.38 (C13), 45.70 (C2′), 45.65 (C7′), 38.26 (C9), 35.36 (C3), 32.61 (C4), 30.71 (C2), 28.20 (C12), 27.41 (C3′), 27.65 (C6′), 26.75 (C4′), 26.32 (C5′), 23.24 (C18), 23.65 (C19), and 19.27 (C11). HRMS (ESI) m/z: calculated for C27H35NO7 [M + Na]+, 508.2311, found, 508.2311.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl bmorpholine-4-carboxylate (5i). Yield: 65%, white granule. 1H NMR (600 MHz, CDCl3) values were as follows: δ 6.32 (1H, s, CH2-17), 6.27 (1H, s, CH2-17), 5.59 (1H, brs, H-6), 4.29 (1H, d, J = 4.5 Hz, H-14), 4.05 (1H, d, J = 9.9 Hz, CH2-20), 3.77 (1H, d, J = 9.9 Hz, CH2-20), 3.66 (2H, s, CH2-2′), 3.59 (2H, s, CH2-3′), 3.51 (1H, m, CH2-4′), 3.39 (1H, m, CH2-4′), 3.35 (1H, m, CH2-5′), 3.29 (1H, m, H-13), 3.24 (1H, m, CH2-4′), 2.46 (1H, d, J = 5.2 Hz, H-9), 2.24 (1H, s), 2.10 (2H, m), 2.04 (2H, m), 1.82 (1H, m), 1.65 (1H, m), 1.62 (2H, m), 1.52 (1H, d, J = 13.9 Hz), 1.20 (3H, s, CH3-18), 1.08 (3H, s, CH3-19). 13C NMR (151 MHz, CDCl3) δ 193.95 (C15), 168.02 (C7), 153.78 (C1′), 145.95 (C16), 121.95 (C17), 107.03 (C1), 103.64 (C6), 79.71 (C14), 72.43 (C20), 66.55 (C2′), 66.27 (C3′), 53.91 (C8), 53.05 (C5), 52.82 (C10), 46.00 (C13), 44.07 (C4′), 43.78 (C5′), 39.23 (C9), 35.95 (C3), 33.06 (C4), 31.08 (C2), 27.56 (C12), 23.52 (C18), 23.15 (C19), and 19.56 (C11). HRMS (ESI) m/z: calculated for C25H31NO8 [M + Na]+, 496.1947, found, 496.1942.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl diphenylcarbamate (5j). Yield: 72%, white granule. 1H NMR (600 MHz, DMSO) values were as follows: δ 7.31 (4H, t, J = 7.7 Hz, H-Ar), 7.26-7.24 (4H, m, H-Ar), 7.19 (2H, t, J = 7.3 Hz, H-Ar), 6.22 (1H, s, CH2-17), 6.09 (1H, s, CH2-17), 5.59 (1H, brs, H-6), 4.29 (1H, d, J = 5.7 Hz, H-14), 4.04 (1H, d, J = 9.6 Hz, CH2-20), 3.53 (1H, d, J = 9.5 Hz, CH2-20), 3.14 (1H, m, H-13), 2.13 (1H, m, H-9), 2.01 (1H, s), 1.80 (2H, m), 1.72 (1H, m), 1.63 (1H, m), 1.43 (1H, d, J = 13.7 Hz), 1.27 (1H, m), 1.24 (1H, m), 1.23 (1H, m), 1.08 (3H, s, CH3-18), 0.98 (3H, s, CH3-19). 13C NMR (151 MHz, DMSO) δ 193.24 (C15), 167.66 (C7), 153.10 (C1′), 146.54 (C16), 129.14 (Ar), 127.08 (Ar), 120.56 (C17), 106.44 (C1), 102.00 (C6), 79.01 (C14), 71.31 (C20), 53.04 (C8), 52.56 (C5), 51.48 (C10), 45.49 (C13), 38.17 (C9), 35.28 (C3), 32.63 (C4), 30.98 (C2), 26.93 (C12), 23.22 (C18), 22.08 (C19), and 16.93 (C11). HRMS (ESI) m/z: calculated for C33H33NO7 [M + Na]+, 578.2155, found, 578.2150.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl 10H-phenothiazine-10-carboxylate (5k). Yield: 88%, yellow granule. 1H NMR (600 MHz, DMSO) values were as follows: δ 7.32 (2H, s, H-Ar), 7.23 (2H, d, J = 7.8 Hz, H-Ar), 7.09 (2H, s, H-Ar), 6.99 (2H, s, H-Ar), 6.09 (1H, s, CH2-17), 5.85 (1H, s, CH2-17), 5.37 (1H, brs, H-6), 4.07 (1H, d, J = 5.7 Hz, H-14), 3.87 (1H, d, J = 9.6 Hz, 1H, CH2-20), 3.39 (1H, d, J = 9.6 Hz, CH2-20), 2.94 (1H, m, H-13), 1.92 (1H, s, H-9), 1.80 (1H, s), 1.64 (1H, s), 1.58 (1H, d, J = 13.9 Hz), 1.53 (1H, d, J = 15.1 Hz), 1.39 (1H, m), 1.19 (1H, d, J = 13.9 Hz), 0.99 (3H, s), 0.83 (3H, s, CH3-18), 0.76 (3H, s, CH3-19). 13C NMR (151 MHz, DMSO) δ 193.26 (C15), 167.59 (C7), 151.35 (C1′), 146.41 (C16), 137.40 (Ar), 131.52 (Ar×2), 129.67 (Ar), 127.48 (Ar×2), 127.36 (Ar×2), 127.13 (Ar×2), 126.89 (Ar×2), 120.78 (C17), 106.42 (C1), 103.37 (C6), 79.33 (C14), 71.32 (C20), 52.96 (C8), 52.26 (C5), 52.05 (C10), 45.31 (C13), 38.16 (C9), 35.19 (C3), 32.48 (C4), 30.56 (C2), 26.94 (C12), 23.15 (C18), 22.60 (C19), and 17.55 (C11). HRMS (ESI) m/z: calculated for C33H31NO7 [M + Na]+, 608.1719, found, 608.1718.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl 3-(1H-pyrrol-1-yl) propanoate (5l). Yield: 85%, white granule. 1H NMR (600 MHz, DMSO-d6) values were as follows: δ 6.66 (2H, s, H-Ar), 6.12 (1H, s, H-Ar), 6.09 (1H, s, H-Ar), 5.93 (2H, s, CH2-17), 5.68 (1H, brs, H-6), 4.39 (1H, d, J = 5.7 Hz), 4.07 (2H, t, J = 6.6 Hz, CH2-3′), 4.04 (1H, d, J = 9.6 Hz), 3.48 (1H, d, J = 9.6 Hz), 3.27 (1H, m), 2.72 (1H, m, CH2-2′), 2.67 (1H, m, CH2-2′), 2.27 (1H, d, J = 5.3 Hz), 2.15 (1H, m), 2.08 (1H, s), 1.95 (1H, d, J = 15.7 Hz), 1.90 (1H, m), 1.84 (1H, d, J = 13.8 Hz), 1.67 (1H, m), 1.49 (1H, m), 1.46 (1H, m), 1.44 (1H, m), 1.07 (3H, s, CH3-18), 0.98 (3H, s, CH3-19). 13C NMR (151 MHz, DMSO) δ 193.68 (C15), 170.10 (C1′), 167.72 (C7), 146.48 (C16), 121.03 (C17), 120.51 (Ar), 107.69 (Ar), 106.52 (C1), 100.67 (C6), 78.94 (C14), 71.49 (C20), 53.22 (C8), 52.54 (C5), 51.79 (C10), 45.44 (C13), 44.32 (C2′), 38.32 (C9), 36.46 (C3′), 35.34 (C3), 32.63 (C4), 30.50 (C2), 27.00 (C12), 23.07 (C18), 22.78 (C19), and 18.81 (C11). HRMS (ESI) m/z: calculated for C27H31NO7 [M + Na]+, 504.1998, found, 504.1997.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl 3-(1,3-dioxoisoindolin-2-yl) propanoate (5m). Yield: 90%, white granule. 1H NMR (600 MHz, DMSO) values were as follows: δ 7.87 (2H, d, J = 5.3 Hz, H-Ar), 7.84 (2H, d, J = 5.2 Hz, H-Ar), 6.13 (1H, s, CH2-17), 6.03 (1H, s, CH2-17), 5.68 (1H, brs, H-6), 4.37 (1H, d, J = 5.7 Hz, H-14), 3.93 (1H, d, J = 9.6 Hz, 1H, CH2-20), 3.82 (1H, m, CH2-2′), 3.75 (1H, m, CH2-2′), 3.26 (1H, m, H-9), 3.16 (1H, d, J = 9.6 Hz, CH2-20), 2.66 (1H, m, CH2-3′), 2.61 (1H, m, CH2-3′), 2.24 (1H, s), 2.10 (2H, m), 2.07 (1H, m), 1.86 (1H, m), 1.81 (1H, d, J = 14.0 Hz), 1.64 (1H, m), 1.49 (1H, m), 1.42 (2H, m), 1.03 (3H, s, CH3-18), 0.94 (3H, s, CH3-19). 13C NMR (151 MHz, DMSO) δ 193.67 (C15), 169.54 (C4′, C7′), 167.59 (C7, C1′), 146.41 (C16), 134.50 (Ar×2), 131.49 (Ar×2), 123.14 (Ar×2), 121.06 (C17), 106.42 (C1), 100.40 (C6), 78.74 (C14), 71.45 (C20), 53.13 (C8), 52.46 (C5), 51.67 (C10), 45.35 (C13), 38.26 (C9), 35.30 (C3), 33.50 (C4), 32.80 (C2′), 32.53 (C3′), 30.42 (C2), 26.93 (C12), 23.01 (C18), 22.64 (C19), and 18.85 (C11). HRMS (ESI) m/z: calculated for C31H31NO9 [M + Na]+, 584.1897, found, 584.1901.
4.1.5. Synthesis of 4-(Ent-6β,14α-dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemi-ketal-6,7-seco-16-kaurene-6β-yloxy)-4-oxobutanoic Acid (6)
Compound 4 (0.36 g, 1.00 mmol), succinic anhydride (0.15 g, 1.5 mmol), and DMAP (15mg, 0.12 mmol) were co-dissolved in anhydrous DCM (50 mL) and stirred for 4 h at 25 °C. After the reaction, the solution was removed by spinning under reduced pressure, redissolved by DCM (100 mL), and then extracted sequentially with 5% sodium bicarbonate solution, distilled water, and saturated saline. The organic solvent was removed by decompression evaporation, and the crude mixture was purified by flash column chromatography (DCM/MeOH, 50/3, v/v), resulting in the target intermediate compound 6 (0.37g, Yield: 80%) as a white granule. 1H NMR (600 MHz, DMSO) values were as follows: δ 12.25 (1H, s, OH-4′), 6.13 (1H, s, CH2-17), 6.11 (1H, s, CH2-17), 5.70 (1H, brs, H-6), 4.42 (1H, d, J = 5.7 Hz, H-14), 4.07 (1H, d, J = 9.6 Hz, CH2-20), 3.53 (1H, d, J = 9.6 Hz, CH2-20), 3.30 (1H, m, H-13), 2.47 (1H, m, H-9), 2.45 (1H, d, J = 5.4 Hz, CH2-3′), 2.42 (2H, d, J = 1.9 Hz, CH2-2′), 2.36 (1H, d, J = 5.1 Hz, CH2-3′), 2.16 (3H, m), 1.98 (1H, m), 1.86 (1H, d, J = 13.9 Hz), 1.70 (1H, m), 1.54 (2H, m), 1.46 (1H, d, J = 13.8 Hz), 1.23 (1H, s), 1.08 (3H, s, CH3-18), 0.99 (3H, s, CH3-19). 13C NMR (151 MHz, DMSO) δ 193.77 (C15), 173.35 (C4′), 171.14 (C1′), 167.73 (C7), 146.47 (C16), 121.13 (C17), 106.59 (C1), 100.84 (C6), 78.96 (C14), 71.50 (C20), 53.38 (C8), 52.56 (C5), 51.97 (C10), 45.67 (C13), 38.33 (C9), 35.37 (C3), 32.60 (C4), 30.56 (C2), 29.03 (C2′), 28.37 (C3′), 27.03 (C12), 23.11 (C18), 22.77 (C19), and 19.03 (C11). HRMS (ESI) m/z: calculated for C24H28O9 [M + Na]+, 483.1631, found, 483.1633.
4.1.6. General Procedure for the Synthesis of Target Derivatives 7a–h
Compound 6 (1.00 mmol), PyBOP (1.5 mmol), and triethylamine (1.50 mmol) were co-dissolved in anhydrous DCM (20 mL), activated for 1 h at 25 °C, and then amine (1.50 mmol) was added and stirred for 2 h. After the reaction, the solution was removed by spinning under reduced pressure, redissolved by DCM (100 mL), and then extracted sequentially with 5% sodium bicarbonate solution, distilled water, and saturated saline. The organic solvent was removed by decompression evaporation, and the crude mixture was purified by flash column chromatography. The solid was precipitated from n-hexane to give the target derivatives 7a–h.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl 4-(benzylamino)-4-oxobutanoate (7a). Yield: 75%, white granule. 1H NMR (600 MHz, DMSO) values were as follows: δ 8.37 (1H, s, NH), 7.30 (2H, t, J = 7.6 Hz, H-Ar), 7.24 (2H, t, J = 14.2 Hz, H-Ar), 7.22 (1H, s, H-Ar), 6.12 (2H, J = 14.1 Hz, CH2-17), 5.70 (1H, brs, H-6), 4.42 (1H, d, J = 4.4 Hz, H-14), 4.24 (2H, d, J = 3.4 Hz, CH2-5′), 4.07 (1H, d, J = 9.6 Hz, CH2-20), 3.52 (1H, d, J = 9.6 Hz, CH2-20), 3.30 (1H, s, H-13), 2.48 (d, J = 6.5 Hz, CH2-2′), 2.39 (2H, t, J = 6.4 Hz, CH2-3′), 2.35 (1H, d, J = 6.5 Hz, CH2-2′), 2.16 (2H, s), 1.98 (2H, d, J = 5.8 Hz), 1.86 (1H, d, J = 6.8 Hz), 1.70 (1H, m), 1.53 (2H, d, J = 9.2 Hz), 1.46 (1H, d, J = 13.8 Hz), 1.23 (2H, s), 1.08 (3H, s, CH3-18), 1.00 (3H, s, CH3-19). 13C NMR (150 MHz, DMSO) δ: 193.80 (C15), 171.38 (C4′), 170.57 (C1′), 167.74 (C7), 146.47 (C16), 139.51 (Ar), 128.26 (Ar), 127.18 (Ar), 126.74 (Ar), 121.13 (C17), 106.60 (C1), 100.77 (C6), 78.93 (C14), 71.50 (C20), 53.29 (C8), 52.57 (C5), 51.94 (C10), 45.71 (C13), 42.04 (C5′), 38.33 (C9), 35.37 (C3), 32.60 (C4), 30.56 (C2), 29.43 (C2′), 29.28 (C3′), 27.04 (C12), 23.12 (C18), 22.76 (C19), and 19.03 (C11). HRMS (ESI) m/z: calculated for C31H35NO8 [M + Na]+, 572.2260, found, 572.2259.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl 4-((4-methylbenzyl) amino)-4-oxobutanoate (7b). Yield: 72%, white granule. 1H NMR (600 MHz, DMSO-d6) values were as follows: δ 8.30 (1H, s, NH), 7.11 (4H, s, Ar), 6.14 (1H, s, CH2-17), 6.11 (1H, s, CH2-17), 5.70 (1H, brs, H-6), 4.41 (1H, d, J = 5.8 Hz, H-14), 4.19 (2H, s, CH2-5′), 4.06 (1H, d, J = 9.3 Hz, CH2-20), 3.52 (1H, d, J = 9.9 Hz, CH2-20), 3.30 (1H, m), 2.48 (1H, m, CH2-2′), 2.37 (1H, m, CH2-2′), 2.36 (2H, m, CH2-3′), 2.27 (3H, m, CH3-6′), 2.16 (2H, m), 1.98 (1H, m), 1.86 (1H, d, J = 13.6 Hz), 1.70 (1H, m), 1.54 (2H, d, J = 13.3 Hz), 1.46 (1H, d, J = 13.5 Hz), 1.24 (2H, s), 1.08 (3H, s, CH3-18), 1.00 (3H, s, CH3-19). 13C NMR (150 MHz, DMSO) δ: 193.77 (C15), 171.35 (C4′), 170.46 (C1′), 167.72 (C7), 146.46 (C16), 136.45 (Ar), 135.74 (Ar), 128.78 (Ar), 127.18 (Ar), 121.09 (C17), 106.59 (C1), 100.74 (C6), 78.92 (C14), 71.51 (C20), 53.29 (C8), 52.57 (C5), 51.95 (C10), 45.71 (C13), 41.80 (C5′), 38.34 (C9), 35.36 (C3), 32.58 (C4), 30.56 (C2), 29.44 (C2′), 29.30 (C3′), 27.03 (C12), 23.10 (C18), 22.75 (C19), 20.65 (C6′), and 19.01 (C11). HRMS (ESI) m/z: calculated for C32H37NO8 [M + Na]+, 586.2417, found, 586.2420.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl 4-((4-meth0xybenzyl) amino)-4-oxobutanoate (7c). Yield: 80%, white granule. 1H NMR (600 MHz, DMSO) values were as follows: δ 8.28 (1H, s, NH), 7.15 (2H, d, J = 8.2 Hz, H-Ar), 6.86 (2H, d, J = 8.5 Hz, H-Ar), 6.12 (2H, J = 16.1 Hz, CH2-17), 5.70 (1H, brs, H-6), 4.41 (1H, d, J = 5.7 Hz, H-14), 4.16 (2H, d, J = 4.0 Hz, CH2-5′), 4.07 (1H, d, J = 9.5 Hz, CH2-20), 3.72 (3H, s, OCH3-6′), 3.52 (1H, d, J = 9.6 Hz, CH2-20), 3.30 (1H, m, H-13), 2.48 (2H, d, J = 6.1 Hz, CH2-2′), 2.35 (2H, d, J = 6.2 Hz, CH2-3′), 2.18(2H, m), 1.98 (1H, m), 1.86 (1H, d, J = 13.8 Hz), 1.70 (1H, m), 1.54 (2H, d, J = 9.8 Hz), 1.46 (1H, d, J = 13.7 Hz), 1.23 (2H, s), 1.08 (3H, s, CH3-18), 0.99 (3H, s, CH3-19). 13C NMR (150 MHz, DMSO) δ: 193.78 (C15), 171.37 (C4′), 170.39 (C1′), 167.73 (C7), 146.47 (C16), 131.43 (Ar), 128.53 (Ar×3), 121.11 (C17), 113.64 (Ar×2) 106.59 (C1), 100.74 (C6), 78.92 (C14), 71.50 (C20), 55.06 (C8), 53.29 (C6′), 52.57 (C5), 51.94 (C10), 45.71 (C13), 41.51 (C5′), 38.33 (C9), 35.37 (C3), 32.59 (C4), 30.55 (C2), 29.44 (C2′), 29.29 (C3′), 27.03 (C12), 23.11 (C18), 22.76 (C19), and 19.02 (C11). HRMS (ESI) m/z: calculated for C32H37NO9 [M + Na]+, 602.2366, found, 602.2366.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl 4-((4-(tert-butyl)benz-yl)amino)-4-oxobutanoate (7d). Yield: 70%, white granule. 1H NMR (600 MHz, DMSO) values were as follows: δ 8.31 (1H, s, NH), 7.31 (2H, d, J = 8.3 Hz, H-Ar), 7.15 (2H, d, J = 8.3 Hz, H-Ar), 6.12 (2H, d, J = 13.6 Hz, CH2-17), 5.69 (1H, brs, H-6), 4.41 (1H, d, J = 4.5 Hz, H-14), 4.19 (2H, dd, J = 5.3, 3.7 Hz, CH2-5′), 4.07 (1H, d, J = 9.6 Hz, CH2-20), 3.52 (1H, d, J = 9.6 Hz, CH2-20), 3.29 (1H, m, H-13), 2.48 (1H, d, J = 3.7 Hz, CH2-2′), 2.37 (1H, d, J = 6.9 Hz, CH2-2′), 2.35 (1H, d, J = 6.5 Hz, CH2-3′), 2.16 (2H, m), 1.95 (1H, m), 1.86 (1H, d, J = 13.8 Hz), 1.70 (1H, m), 1.53 (2H, m), 1.46 (1H, d, J = 13.8 Hz), 1.26 (9H, s, CH3-7′, CH3-8′, CH3-9′), 1.23 (2H, s), 1.08 (3H, s, CH3-18), 1.00 (3H, s, CH3-19). 13C NMR (150 MHz, DMSO) δ: 193.78 (C15), 171.38 (C4′), 170.44 (C1′), 167.72 (C7), 149.13 (Ar), 146.48 (C16), 136.47 (Ar), 127.07 (Ar), 124.99 (Ar), 121.10 (C17), 106.59 (C1), 100.78 (C6), 78.93 (C14), 71.50 (C20), 53.29 (C8), 52.57 (C5), 51.94 (C10), 45.71 (C13), 41.79 (C5′), 38.34 (C9), 35.37 (C3), 34.15 (C6′), 32.59 (C4), 31.18 (C7′, C8′, C9′), 30.56 (C2), 29.42 (C2′), 29.30 (C3′), 27.04 (C12), 23.11 (C18), 22.75 (C19), and 19.03 (C11). HRMS (ESI) m/z: calculated for C35H43NO8 [M + H]+, 628.2886, found, 628.2890.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl 4-((4-fluorobenzyl) amino)-4-oxobutanoate (7e). Yield: 65%, white granule. 1H NMR (600 MHz, DMSO) values were as follows: δ 8.38 (1H, s, NH), 7.26 (2H, s, H-Ar), 7.12 (2H, s, H-Ar), 6.12 (2H, J = 16.6 Hz, CH2-17), 5.70 (1H, brs, H-6), 4.42 (1H, s, H-14), 4.22 (2H, s, CH2-5′), 4.06 (1H, d, J = 9.7 Hz, CH2-20), 3.52 (1H, d, J = 9.8 Hz, CH2-20), 3.30 (1H, m, H-13), 2.47 (1H, d, J = 6.2 Hz, CH2-3′), 2.37 (2H, d, J = 6.5 Hz, CH2-2′), 2.35 (1H, s, CH2-3′), 2.15 (2H, m), 1.98 (1H, m), 1.86 (1H, d, J = 13.4 Hz), 1.69 (1H, m), 1.54 (2H, m), 1.47 (1H, m), 1.23 (2H, s), 1.08 (3H, s, CH3-18), 0.99 (3H, s, CH3-19). 13C NMR (150 MHz, DMSO) δ: 193.80 (C15), 171.36 (C4′), 170.60 (C1′), 167.74 (C7), 161.95 (Ar), 160.35 (Ar), 146.48 (C16), 135.73 (Ar), 129.11 (Ar), 121.13 (C17), 115.02 (Ar), 114.88 (Ar), 106.60 (C1), 100.77 (C6), 78.94 (C14), 71.51 (C20), 53.29 (C8), 52.57 (C5), 51.94 (C10), 45.72 (C13), 41.34 (C5′), 38.34 (C9), 35.37 (C3), 32.59 (C4), 30.55 (C2), 29.43 (C2′), 29.25 (C3′), 27.04 (C12), 23.11 (C18), 22.76 (C19), and 19.03 (C11). HRMS (ESI) m/z: calculated for C31H34NO8F [M + Na]+, 590.2166, found, 590.2166.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl 4-((4-chlorobenzyl) amino)-4-oxobutanoate (7f). Yield: 66%, white granule. 1H NMR (600 MHz, DMSO) values were as follows: δ 7.25-7.22 (2H, m, H-Ar), 7.00 (2H, t, J = 8.6 Hz, H-Ar), 6.30 (1H, s, CH2-17), 6.21 (1H, s, CH2-17), 5.92 (1H, s, NH), 5.57 (1H, brs, H-6), 4.39 (2H, dd, J = 14.8, 5.7 Hz, H-14, CH2-5′), 4.28 (1H, d, J = 5.7 Hz), 4.03 (1H, d, J = 9.8 Hz, CH2-20), 3.73 (1H, d, J = 9.8 Hz, CH2-20), 3.27 (1H, m, H-13), 2.71 (1H, m), 2.51 (1H, d, J = 10.6 Hz, CH2-3′), 2.47 (1H, d, J = 5.6 Hz, CH2-2′), 2.41 (1H, d, J = 9.3 Hz, CH2-3′), 2.25 (1H, d, J = 10.7 Hz, CH2-2′), 2.20 (1H, s), 2.08 (2H, d, J = 10.7 Hz), 2.02 (1H, d, J = 14.0 Hz), 1.81 (1H, m), 1.62 (1H, d, J = 13.0 Hz), 1.50 (1H, d, J = 13.9 Hz), 1.25 (2H, s), 1.14 (3H, s, CH3-18), 1.06 (3H, s, CH3-19). 13C NMR (150 MHz, DMSO) δ: 194.03 (C15), 171.81 (C4′), 171.04 (C1′), 168.28 (C7), 161.50 (Ar), 146.23 (C16), 134.02 (Ar), 129.58 (Ar), 129.52 (Ar), 121.83 (C17), 115.75 (Ar), 115.61 (Ar), 107.01 (C1), 101.61 (C6), 79.88 (C14), 72.67 (C20), 54.07 (C8), 53.24 (C5), 52.64 (C10), 46.31 (C13), 43.12 (C5′), 39.44 (C9), 35.97 (C3), 33.27 (C4), 31.03 (C2), 30.58 (C2′), 29.79 (C3′), 27.66 (C12), 23.61 (C18), 23.32 (C19), and 19.44 (C11). HRMS (ESI) m/z: calculated for C31H34NO8Cl [M + Na]+, 606.1871, found, 606.1871.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl 4-((4-bromobenzyl) amino)-4-oxobutanoate (7g). Yield: 60%, white granule. 1H NMR (600 MHz, DMSO) values were as follows: δ 8.41 (1H, s, NH), 7.49 (2H, d, J = 8.4 Hz, H-Ar), 7.19 (2H, d, J = 8.4 Hz, H-Ar), 6.13 (1H, s, CH2-17), 6.10 (1H, s, CH2-17), 5.70 (1H, brs, H-6), 4.41 (1H, d, J = 4.4 Hz, H-14), 4.20 (2H, d, J = 3.5 Hz, CH2-5′), 4.06 (1H, d, J = 9.6 Hz, CH2-20), 3.51 (1H, d, J = 9.6 Hz, CH2-20), 3.30 (1H, m, H-13), 2.48 (1H, d, J = 5.2 Hz, CH2-3′), 2.37 (2H, d, J = 6.8 Hz, CH2-2′), 2.34 (1H, d, J = 5.2 Hz, CH2-3′), 2.15 (2H, m), 1.97 (1H, m), 1.85 (1H, d, J = 9.9 Hz), 1.69 (1H, m), 1.53 (2H, d, J = 12.6 Hz), 1.46 (1H, d, J = 13.8 Hz), 1.23 (2H, s), 1.07 (3H, s, CH3-18), 0.99 (3H, s, CH3-19). 13C NMR (150 MHz, DMSO) δ: 193.83 (C15), 171.37 (C4′), 170.72 (C1′), 167.76 (C7), 146.48 (C16), 139.09 (Ar), 131.11 (Ar), 129.43 (Ar), 121.16 (C17), 119.37 (Ar), 106.62 (C1), 100.78 (C6), 78.95 (C14), 71.51 (C20), 53.30 (C8), 52.59 (C5), 51.95 (C10), 45.73 (C13), 41.45 (C5′), 38.35 (C9), 35.38 (C3), 32.60 (C4), 30.56 (C2), 29.43 (C2′), 29.26 (C3′), 27.05 (C12), 23.12 (C18), 22.77 (C19), and 19.04 (C11). HRMS (ESI) m/z: calculated for C31H34NO8Br [M + Na]+, 650.1365, found, 650.1369.
ent-6β,14α-Dihydroxy-(1,7–1,14)-epoxy-7,15-dioxo-6,20-hemiketal-6,7-seco-16-kaurene-6β-yl 4-(4-(6-fluorobenzo[d] isoxazol-3-yl) piperidin-1-yl)-4-oxobutanoate (7h). Yield: 85%, white granule. 1H NMR (600 MHz, DMSO-d6) values were as follows: δ 8.05 (1H, dd, J = 8.7, 5.3 Hz, H-Ar), 7.70 (1H, d, J = 9.0 Hz, H-Ar), 7.29 (1H, t, J = 9.0 Hz, H-Ar), 6.12 (2H, s, CH2-17), 5.68 (1H, d, J = 11.3 Hz, H-6), 4.41 (2H, t, J = 8.7 Hz), 4.08 (1H, s, H-14), 3.96 (1H, d, J = 12.1 Hz, H-7′), 3.50 (2H, m, CH2-5′), 3.31 (1H, m), 3.20 (2H, m, CH2-9′), 2.80 (1H, s), 2.59 (2H, m, CH2-2′), 2.47 (1H, m, CH2-3′), 2.36 (1H, m, CH2-3′), 2.17 (2H, m, CH2-6′), 2.04 (2H, d, J = 14.8 Hz, CH2-8′), 2.00 (1H, m), 1.87 (1H, d, J = 13.9 Hz, 1H), 1.78 (1H, m), 1.70 (1H, m), 1.61 (1H, m), 1.55 (1H, d, J = 9.7 Hz), 1.46 (1H, d, J = 14.2 Hz), 1.23 (2H, s), 1.09 (3H, s, CH3-18), 1.00 (3H, s, CH3-19). 13C NMR (150 MHz, DMSO) δ: 193.83 (C15), 171.54 (C4′), 169.09 (C1′), 167.71 (C7), 163.11 (Ar), 162.89 (Ar), 160.94 (C10′), 146.48 (C16), 123.89 (Ar), 121.12 (C17), 117.15 (Ar), 112.67 (Ar), 106.60 (C1), 100.66 (C6), 97.50 (Ar), 78.91 (C14), 71.49 (C20), 53.30 (C8), 52.57 (C5), 51.92 (C10), 45.71 (C13), 44.51 (C5′), 41.03 (C9′), 38.32 (C9), 35.36 (C3), 33.23 (C7′), 32.61 (C4), 30.54 (C2), 30.17 (C6′), 29.73 (C8′), 29.38 (C12), 27.09 (C2′), 27.03 (C3′), 23.11 (C18), 22.80 (C19), and 19.09 (C11). HRMS (ESI) m/z: calculated for C36H39N2O9F [M + Na]+, 685.2537, found, 685.2542.
4.2. Biological Testing
4.2.1. Cell Culture
Human non-small cell lung cancer cell lines (A549), human liver cancer cell lines (HepG2), human breast cancer cell lines (MCF-7), and human normal cell lines (L-02) were purchased from the cell bank of the Chinese Academy of Sciences. All cells were placed in RPMI-1640 medium (added 10% fetal bovine serum, 100 units/mL streptomycin, and 100 µg/mL penicillin) and cultured in an aseptic environment with 5% CO2 at 37 °C.
4.2.2. In Vitro Anti-Proliferative Activity
All derivatives were evaluated for their anti-proliferative activities against three cancer cell lines and one normal cell line by MTT assay. The cells in the logarithmic growth phase were inoculated into 96-well plates (3 × 103 cells/well) and incubated for 48 h in a sterile environment with 5% CO2 at 37 °C to make them attach via cell adhesion; then, the cells were co-incubated with target compounds for 48 h. After that, MTT solution (10 µL/well) was added to the cell culture medium to continue incubation for 4 h, following which the medium was replaced with DMSO (150 µL/well). The absorbance values at 490 nm were determined using an enzyme meter (Safire 2, Tecan, Männedorf, Switzerland) and further converted to half-inhibitory concentration values of these compounds.
4.2.3. Experimental Method for Cell Cycle Analysis
A549 cells were inoculated into 24-well plates and attached via cell adhesion. Then, gradient concentrations (0, 2, 4, and 8 µM) of 7h were added and the cells were co-incubated for 48 h. The cells were cleaned with ice-cold phosphate buffer solution (PBS), digested with trypsin, collected, and prepared as a cell suspension. The cell suspension was treated with PI/RNase staining solution, the data were gathered using a flow cytometer (FACS Calibur, BD, Milpitas, CA, USA), and the cell cycle distribution was analyzed using Flow Jo software. Three attempts were performed for each treatment to eliminate experimental bias.
4.2.4. Experimental Methods for Apoptosis Analysis
Cell Apoptosis Morphological Detection
A549 cells were inoculated into 24-well plates and attached via cell adhesion. Then, gradient concentrations (0, 2, 4, and 8 µM) of 7h were added and co-incubated for 48 h. The cells were cleaned with ice-cold PBS, stained with AO/EB, mixed, and then incubated in cells for 5 min in darkness at 25 °C. The apoptosis morphology of the cells in each group was observed and photographed using a laser confocal microscope (LSM710, Zeiss, Oberkochen, Germany).
Cell Apoptosis Rate Detection
A549 cells were inoculated into 6-well plates and attached via cell adhesion. Then, gradient concentrations (0, 2, 4, and 8 µM) of 7h was added and co-incubated for 48 h. The cells were gathered and prepared as cell suspensions according to the method described in Section 4.2.3. The cell suspension was treated with Annexin V-FITC/PI staining solution, and the data were gathered using a flow cytometer (FACS Calibur, BD, Milpitas, CA, USA) and analyzed using Flow Jo software. Three attempts were performed for each treatment to eliminate experimental bias.
4.2.5. Experimental Methods for ROS Measurement
Cell Morphological Detection
A549 cells were inoculated into 24-well plates and attached via cell adhesion. Then, gradient concentrations (0, 2, 4, and 8 µM) of 7h were added and the cells were co-incubated for 48 h. The cells were cleaned with ice-cold PBS, stained with H2DCFDA, mixed, and incubated in cells for 20 min in darkness at 25 °C. The apoptosis morphology of the cells in each group was observed and photographed using a laser confocal microscope (LSM710, Zeiss, Oberkochen, Germany).
ROS Content Determination
A549 cells were inoculated into 6-well plates and attached via cell adhesion. Then, gradient concentrations (0, 2, 4, and 8 µM) of 7h were added and the cells were co-incubated for 48 h. The cells were gathered and prepared as cell suspensions according to the method described in Section 4.2.3. The cell suspensions were treated with H2DCFDA staining solution in darkness for 20 min at 25 °C and then cleaned with PBS to remove residual stain. The experimental data were gathered using flow cytometry (FACS Calibur, BD, Milpitas, CA, USA) and analyzed using Flow Jo software. Three attempts were performed for each treatment to eliminate experimental bias.
4.2.6. Experimental Method for MMP Measurement
Cell Morphological Detection
A549 cells were inoculated into 24-well plates and attached via cell adhesion. Then, gradient concentrations (0, 2, 4, and 8 µM) of compound 7h were added and the cells were co-incubated for 48 h. The cells were cleaned with ice-cold PBS, stained with JC-1, mixed, and then incubated in cells for 20 min in darkness at 25 °C. The apoptotic morphology of the cells in each group was observed and photographed using a laser confocal microscopy (LSM710, Zeiss, Oberkochen, Germany).
Cell Morphological Detection
A549 cells were inoculated into 6-well plates and attached via cell adhesion. Then, gradient concentrations (0, 2, 4, and 8 µM) of 7h were added and the cells were co-incubated for 48 h. The cells were collected and prepared as cell suspensions according to the method described in Section 4.2.3. Cell suspensions were treated with JC-1 staining solution in darkness for 20 min at 25 °C, and then washed with PBS buffer to remove residual stain. The experimental data were gathered using flow cytometry (FACS Calibur, BD, Milpitas, CA, USA) and analyzed by Flow Jo software. Three attempts were performed for each treatment to eliminate experimental bias.
4.2.7. Experimental Methods for GO and KEGG Analysis
The potential targets of compound 7h were obtained from the PharmMapper online platform (http://www.lilab-ecust.cn/pharmmapper (accessed on 27 July 2024)). Firstly, the chemical structure of 7h was constructed using ChemBio3D Ultra 14.0 software, and the energy minimum conformation was obtained using the “Calculations-MM2-Minemize Energy” module and saved in MOL2 format. The 3D structure was imported into the PharmMapper server, and the following parameters were qualified: generate conformers: yes; maximum generated conformations: 300; select target set: human protein target only; number of reserved matched targets: 300. The proteins (Z-score > 0.5) were screened as candidate targets for 7h. The GO and KEGG analyses were performed using the DAVID platform (https://david.ncifcrf.gov (accessed on 27 July 2024)). R (4.4.1) software was used to analyze and visualize the results.
4.2.8. Experimental Method of Western Blot Analysis
Western blot was used to determine the effect of 7h against the related protein expression in A549 cells. A549 cells were inoculated in 6-well plates and treated with a grdient concentrations (0, 2, 4, and 8 µM) of 7h for 48 h. The cells were collected, lysed in RIPA buffer, and heated at 100 °C for 10 min. The samples were processed via 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% SDS-PAGE) and then transferred to a poly vinylidene difluoride (PVDE) membrane. The membrane was immersed in tris-buffered saline and tween (TBST) containing 5% skimmed milk and blocked for 2 h at 25 °C, and then incubated overnight at 4 °C with the addition of the internal reference protein primary antibody (GAPDH; dilution ratio 1:5000) and the target protein primary antibody (PI3K, p-PI3K, Akt, p-Akt, mTOR, and p-mTOR; dilution ratio 1:1000). Following this, the membrane was incubated with an HRP-labeled (horseradish peroxidase) secondary antibody for 2 h at 25 °C and positive bands were shown on X-ray film using an enhanced chemiluminescence system (Kodak, Rochester, NY, USA). The information on the primary antibodies is as follow: PI3K (Catalog: 4292, Cell Signaling Technology, Danvers, MA, USA), p-PI3K (Catalog: 4228, Cell Signaling Technology), Akt (Catalog: 9272, Cell Signaling Technology), p-Akt (Catalog: 4060, Cell Signaling Technology), mTOR (Catalog: 2972, Cell Signaling Technology), p-mTOR (Catalog: 5536, Cell Signaling Technology), and GAPDH (Catalog: 2118, Cell Signaling Technology).
4.2.9. Experimental Method of Molecular Docking Study
The 3D model of ligand compounds was constructed using the ChemBio3D Ultra 14.0 software. The 3D structural models of receptor proteins PI3Kα (PDB ID: 6GVF), Akt1 (PDB ID: 6HHF), and mTOR (PDB ID: 4JT6) were downloaded from the RCSB Protein Data Bank (https://www.rcsb.org/ (accessed on 27 July 2024)) and reprocessed using the Pymol (3.0) software. Molecular docking studies were performed using the Autodock Vina 1.1.2 software [44]. Visualization and interaction analysis of the docking results were obtained using the Discovery Studio 2019 Client (V19.10) software [45].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules29174066/s1. The supplementary material includes HRMS, 1H NMR, and 13C NMR spectral data of all derivatives, the single-crystal data and absolute configuration of compound 4, the GO and KEGG analyses of 7h, and the molecular docking results.
Author Contributions
Conceptualization, J.W. (Jiafeng Wang) and M.B.; Data curation, J.W. (Jiafeng Wang) and L.W.; Formal analysis, J.W. (Jiafeng Wang) and L.W.; Methodology, Y.L. and M.B.; Software, Y.Z. and J.W. (Jiale Wu); Validation, J.W. (Jiafeng Wang), Y.Z., L.W., S.P. and Y.L.; Writing—original draft, J.W. (Jiafeng Wang); Writing—review & editing, M.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Fundamental Research Funds for Education Department of Heilongjiang Province (Grant No. 2022-KYYWF-0787) and the Natural Science Foundation of Heilongjiang Province of China (Grant No. LH2022H113).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data will be provided upon request.
Conflicts of Interest
The authors reported no potential conflicts of interest.
References
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Gaidai, O.; Yan, P.; Xing, Y.H. Future world cancer death rate prediction. Sci. Rep. 2023, 13, 303. [Google Scholar] [CrossRef]
- Herrero Álvarez, N.; Bauer, D.; Hernández-Gil, J.; Lewis, J.S. Recent Advances in Radiometals for Combined Imaging and Therapy in Cancer. ChemMedChem 2021, 16, 2909–2941. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Liu, X. Radionanomedicine: Advanced Strategy for Precision Theranostics of Breast Cancer. J. Biomed. Nanotechnol. 2022, 18, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Bian, X.; Li, P.; Huang, Y.; Li, C. Carrier-Free Nanomedicine for Cancer Immunotherapy. J. Biomed. Nanotechnol. 2022, 18, 939–956. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.C.; Ni, J.J.; Cui, W.Y.; Wang, B.Y.; Zhuo, W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res. 2019, 9, 1354–1366. [Google Scholar] [PubMed]
- Fehrenbacher, J.C. Chemotherapy-induced peripheral neuropathy. Prog. Mol. Biol. Transl. Sci. 2015, 131, 471–508. [Google Scholar] [PubMed]
- Zhang, J.; Chai, H. Recent Advances in Drug Discovery and Cancer Diagnoses. Curr. Top. Med. Chem. 2020, 20, 1855–1857. [Google Scholar] [CrossRef]
- Barot, K.P.; Nikolova, S.; Ivanov, I.; Ghate, M.D. Novel anticancer agents and targets: Recent advances and future perspectives. Mini Rev. Med. Chem. 2013, 13, 1239–1255. [Google Scholar] [CrossRef]
- Zhang, L.; Song, J.; Kong, L.; Yuan, T.; Li, W.; Zhang, W.; Hou, B.; Lu, Y.; Du, G. The strategies and techniques of drug discovery from natural products. Pharmacol. Ther. 2020, 216, 107686. [Google Scholar] [CrossRef]
- Chan, W.J.; Adiwidjaja, J.; McLachlan, A.J.; Boddy, A.V.; Harnett, J.E. Interactions between natural products and cancer treatments: Underlying mechanisms and clinical importance. Cancer Chemother. Pharmacol. 2023, 91, 103–119. [Google Scholar] [CrossRef]
- Hashem, S.; Ali, T.A.; Akhtar, S.; Nisar, S.; Sageena, G.; Ali, S.; Al-Mannai, S.; Therachiyil, L.; Mir, R.; Elfaki, I.; et al. Targeting cancer signaling pathways by natural products: Exploring promising anticancer agents. Biomed. Pharmacother. 2022, 150, 113054. [Google Scholar] [CrossRef] [PubMed]
- Shaik, B.B.; Katari, N.K.; Jonnalagadda, S.B. Role of Natural Products in Developing Novel Anticancer Agents: A Perspective. Chem. Biodivers. 2022, 19, e202200535. [Google Scholar] [CrossRef] [PubMed]
- Najmi, A.; Javed, S.A.; Al Bratty, M.; Alhazmi, H.A. Modern Approaches in the Discovery and Development of Plant-Based Natural Products and Their Analogues as Potential Therapeutic Agents. Molecules 2022, 27, 349. [Google Scholar] [CrossRef] [PubMed]
- Cabral, C.; Efferth, T.; Pires, I.M.; Severino, P.; Lemos, M.F.L. Natural Products as a Source for New Leads in Cancer Research and Treatment. Evid.-Based Complement. Altern. Med. 2018, 2018, 8243680. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Naeem, A.; Hu, P.; Yang, M.; Zhang, J.; Liu, Y.; Zhu, W.; Zheng, Q. Natural Products as Anticancer Agents: Current Status and Future Perspectives. Molecules 2022, 27, 8367. [Google Scholar] [CrossRef] [PubMed]
- Varghese, R.; Dalvi, Y.B. Natural Products as Anticancer Agents. Curr. Drug Targets 2021, 22, 1272–1287. [Google Scholar] [CrossRef]
- Ibrahim, T.S.; Khongorzul, P.; Muyaba, M.; Alolga, R.N. Ent-kaurane diterpenoids from the Annonaceae family: A review of research progress and call for further research. Front. Pharmacol. 2023, 14, 1227574. [Google Scholar] [CrossRef]
- Sarwar, M.S.; Xia, Y.X.; Liang, Z.M.; Tsang, S.W.; Zhang, H.J. Mechanistic Pathways and Molecular Targets of Plant-Derived Anticancer ent-Kaurane Diterpenes. Biomolecules 2020, 10, 144. [Google Scholar] [CrossRef]
- Luo, H.; Vong, C.T.; Chen, H.; Gao, Y.; Lyu, P.; Qiu, L.; Zhao, M.; Liu, Q.; Cheng, Z.; Zou, J.; et al. Naturally occurring anticancer compounds: Shining from Chinese herbal medicine. Chin. Med. 2019, 14, 48. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, C.T.; Ma, W.; Xie, X.; Huang, Q. Oridonin: A Review of Its Pharmacology, Pharmacokinetics and Toxicity. Front. Pharmacol. 2021, 12, 645824. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Chen, X.; Qu, S.; Yao, B.; Xu, Y.; Wu, J.; Jin, Y.; Ma, C. Oridonin induces G2/M cell cycle arrest and apoptosis via the PI3K/Akt signaling pathway in hormone-independent prostate cancer cells. Oncol. Lett. 2017, 13, 2838–2846. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.H.; Pi, J.; Jin, H.; Cai, J.Y. Oridonin-induced mitochondria-dependent apoptosis in esophageal cancer cells by inhibiting PI3K/AKT/mTOR and Ras/Raf pathways. J. Cell. Biochem. 2019, 120, 3736–3746. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.X.; He, Z.A.; Chen, Z.Y.; Wang, Y.X.; Bai, S.P.; Sun, H.D. Cytotoxic ent-kaurane diterpenoids from Isodon macrophyllus. J. Asian Nat. Prod. Res. 2009, 11, 693–697. [Google Scholar] [CrossRef]
- Yang, J.; Wang, W.G.; Wu, H.Y.; Du, X.; Li, X.N.; Li, Y.; Pu, J.X.; Sun, H.D. Bioactive Enmein-Type ent-Kaurane Diterpenoids from Isodon phyllostachys. J. Nat. Prod. 2016, 79, 132–140. [Google Scholar] [CrossRef]
- Li, L.M.; Li, G.Y.; Ding, L.S.; Lei, C.; Yang, L.B.; Zhao, Y.; Weng, Z.Y.; Li, S.H.; Huang, S.X.; Xiao, W.L.; et al. Sculponins A-C, three new 6,7-seco-ent-kauranoids from Isodon sculponeatus. Tetrahedron Lett. 2007, 48, 9100–9103. [Google Scholar] [CrossRef]
- Yan, F.L.; Guo, L.Q.; Wang, C.M.; Zhang, J.X. Chemical constituents of Isodon nervosus and their cytotoxicity. J. Asian Nat. Prod. Res. 2009, 11, 326–331. [Google Scholar] [CrossRef]
- Wang, L.; Li, D.; Xu, S.; Cai, H.; Yao, H.; Zhang, Y.; Jiang, J.; Xu, J. The conversion of oridonin to spirolactone-type or enmein-type diterpenoid: Synthesis and biological evaluation of ent-6,7-seco-oridonin derivatives as novel potential anticancer agents. Eur. J. Med. Chem. 2012, 52, 242–250. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Brindisi, M. Organic carbamates in drug design and medicinal chemistry. J. Med. Chem. 2015, 58, 2895–2940. [Google Scholar] [CrossRef] [PubMed]
- Bu, M.; Cao, T.; Li, H.; Guo, M.; Yang, B.B.; Zeng, C.; Hu, L. Synthesis of 5α,8α-Ergosterol Peroxide 3-Carbamate Derivatives and a Fluorescent Mitochondria-Targeting Conjugate for Enhanced Anticancer Activities. ChemMedChem 2017, 12, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.X.; Chen, C.; Liu, X.Q.; Li, Y.; Lin, Y.L.; Wu, X.T.; Kong, L.Y.; Luo, J.G. Discovery and optimization of withangulatin A derivatives as novel glutaminase 1 inhibitors for the treatment of triple-negative breast cancer. Eur. J. Med. Chem. 2021, 210, 112980. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.J.; Liu, M.C.; Xiang, H.M.; Zhao, Q.; Xue, W.; Yang, S. Synthesis and in vitro antitumor evaluation of betulin acid ester derivatives as novel apoptosis inducers. Eur. J. Med. Chem. 2015, 102, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Hameed, R.H.; Mohamed, M.S.; Awad, S.M.; Hassan, B.B.; Khodair, M.A.E.; Mansour, Y.E. Novel benzo chromene derivatives: Design, synthesis, molecular docking, cell cycle arrest, and apoptosis induction in human acute myeloid leukemia HL-60 cells. J. Enzym. Inhib. Med. Chem. 2023, 38, 405–422. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Zheng, Z.; Wei, X.; Liu, Y.; Li, W.; Fang, B.; Yun, D.; Dong, Z.; Yi, B.; Li, W.; et al. Rational design, synthesis, and pharmacological characterisation of dicarbonyl curcuminoid analogues with improved stability against lung cancer via ROS and ER stress mediated cell apoptosis and pyroptosis. J. Enzym. Inhib. Med. Chem. 2022, 37, 2357–2369. [Google Scholar] [CrossRef]
- Wang, X.; Pan, C.X.; Gong, J.Y.; Liu, X.F.; Li, H.L. Enhancing the Enrichment of Pharmacophore-Based Target Prediction for the Polypharmacological Profiles of Drugs. J. Chem. Inf. Model. 2016, 56, 1175–1183. [Google Scholar] [CrossRef]
- Wang, X.; Shen, Y.; Wang, S.; Li, S.; Zhang, W.; Liu, X.; Lai, L.; Pei, J.; Li, H. PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017, 45, W356–W360. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.H.; Wang, S.; Zhang, Y.; Huang, T.; Cai, Y.D. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS ONE 2017, 12, e0184129. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.H.; Lu, G.; Huang, T.; Cai, Y.D. Analysis of cancer-related lncRNAs using gene ontology and KEGG pathways. Artif. Intell. Med. 2017, 76, 27–36. [Google Scholar] [CrossRef]
- Yu, L.; Wei, J.; Liu, P. Attacking the PI3K/Akt/mTOR signaling pathway for targeted therapeutic treatment in human cancer. Semin. Cancer Biol. 2022, 85, 69–94. [Google Scholar] [CrossRef] [PubMed]
- Tewari, D.; Patni, P.; Bishayee, A.; Sah, A.N.; Bishayee, A. Natural products targeting the PI3K-Akt-mTOR signaling pathway in cancer: A novel therapeutic strategy. Semin. Cancer Biol. 2022, 80, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.C. Targeting the PI3K/Akt/mTOR pathway in non-small cell lung cancer (NSCLC). Thorac. Cancer 2020, 11, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Yang, J.; Yu, W.; Li, H.; Cai, M.; Xu, J.L.; Xu, H.D.; Shi, Y.F.; Guan, X.; Cheng, X.D.; et al. Discovery of benzochalcone derivative as a potential antigastric cancer agent targeting signal transducer and activator of transcription 3 (STAT3). J. Enzym. Inhib. Med. Chem. 2022, 37, 2004–2016. [Google Scholar] [CrossRef]
- Ahmed, R.I.; Osman, E.E.; Awadallah, F.M.; El-Moghazy, S.M. Design, synthesis and molecular docking of novel diarylcyclohexenone and diarylindazole derivatives as tubulin polymerization inhibitors. J. Enzym. Inhib. Med. Chem. 2017, 32, 176–188. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).