A Combined Computational and Experimental Approach to Studying Tropomyosin Kinase Receptor B Binders for Potential Treatment of Neurodegenerative Diseases
Abstract
1. Introduction
2. Results
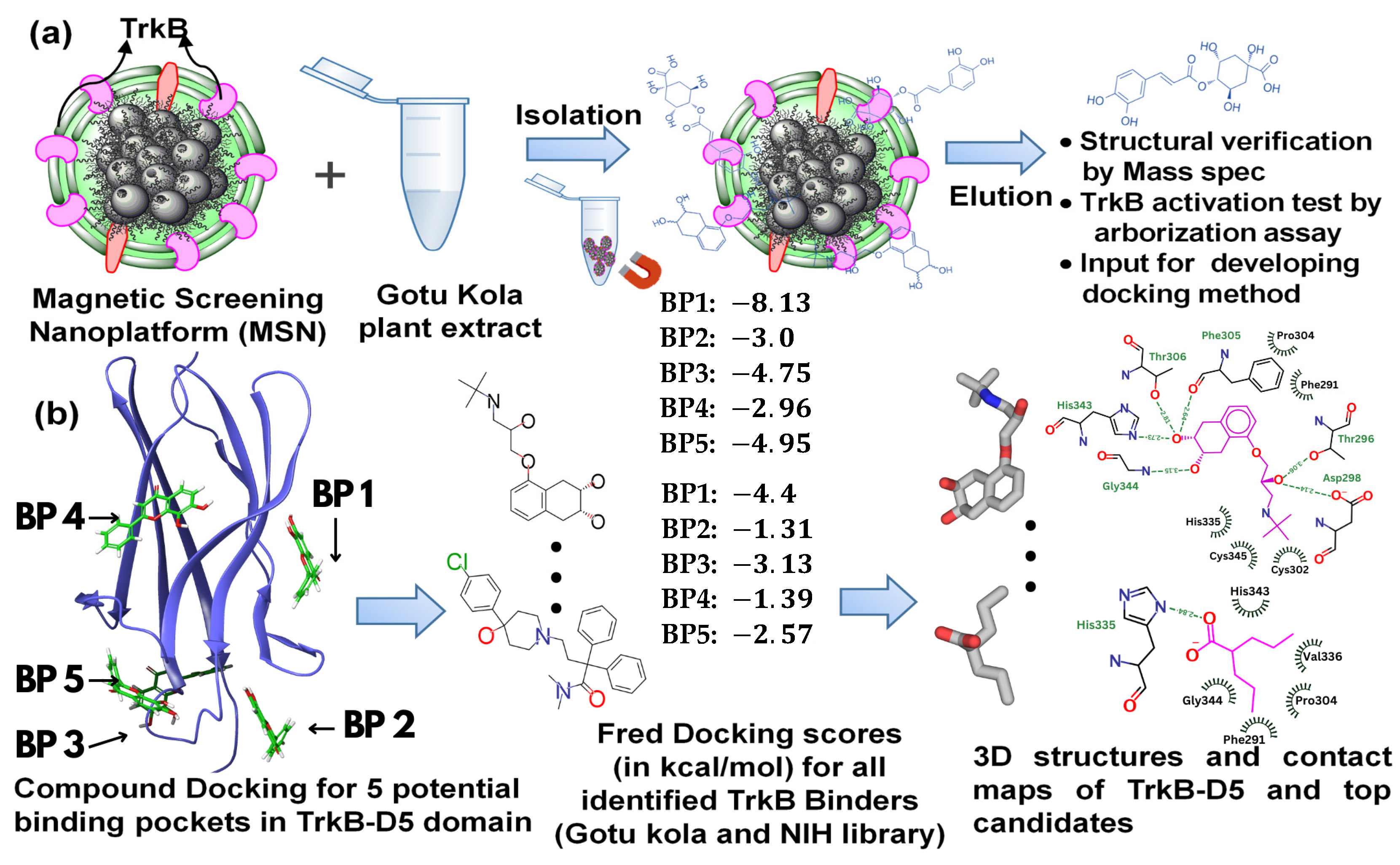
2.1. Magnetic Drug Screening Nanoplatform
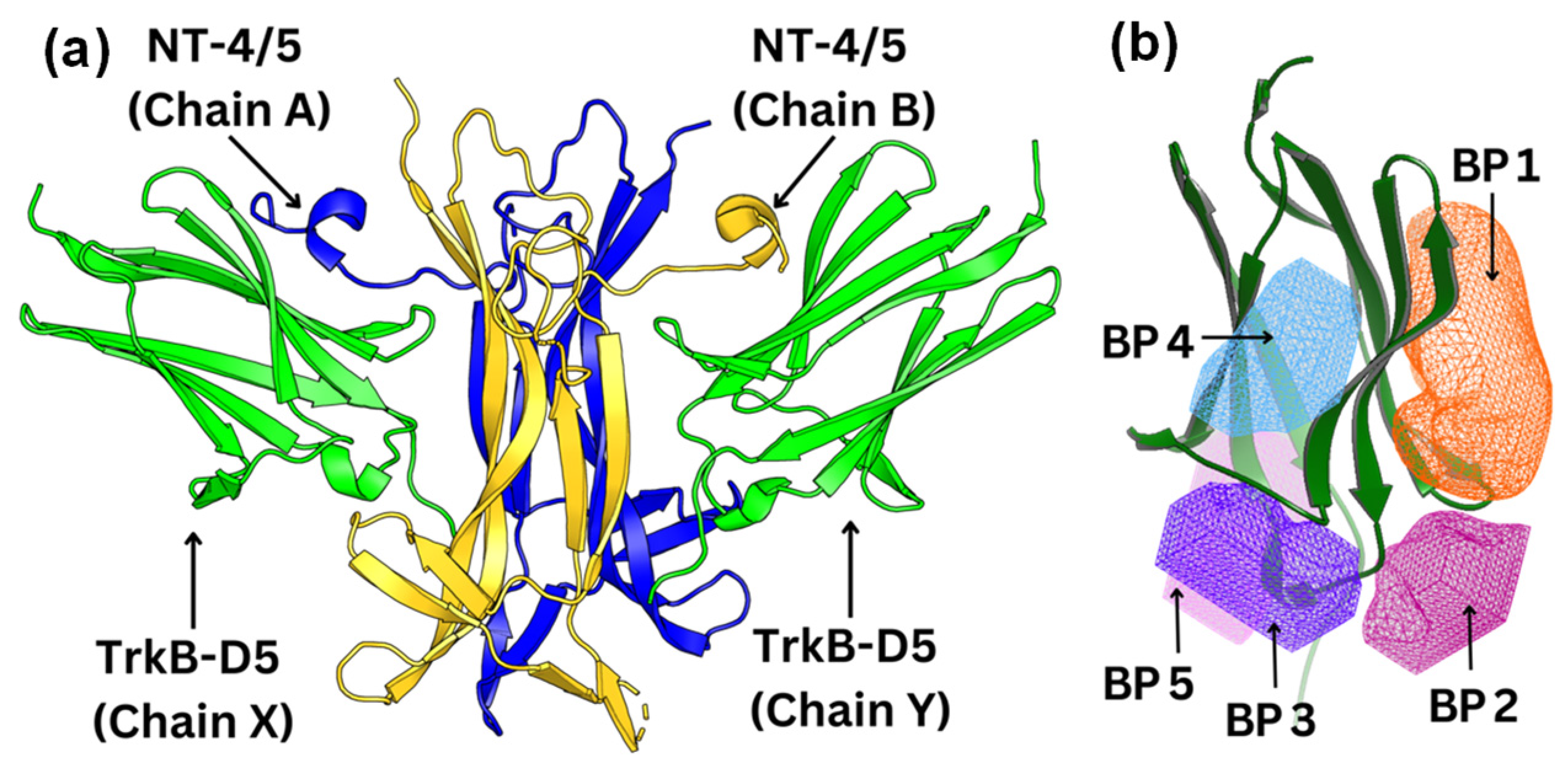
2.2. Docking of TrkB Binding Compounds from Gotu Kola Plant Extract
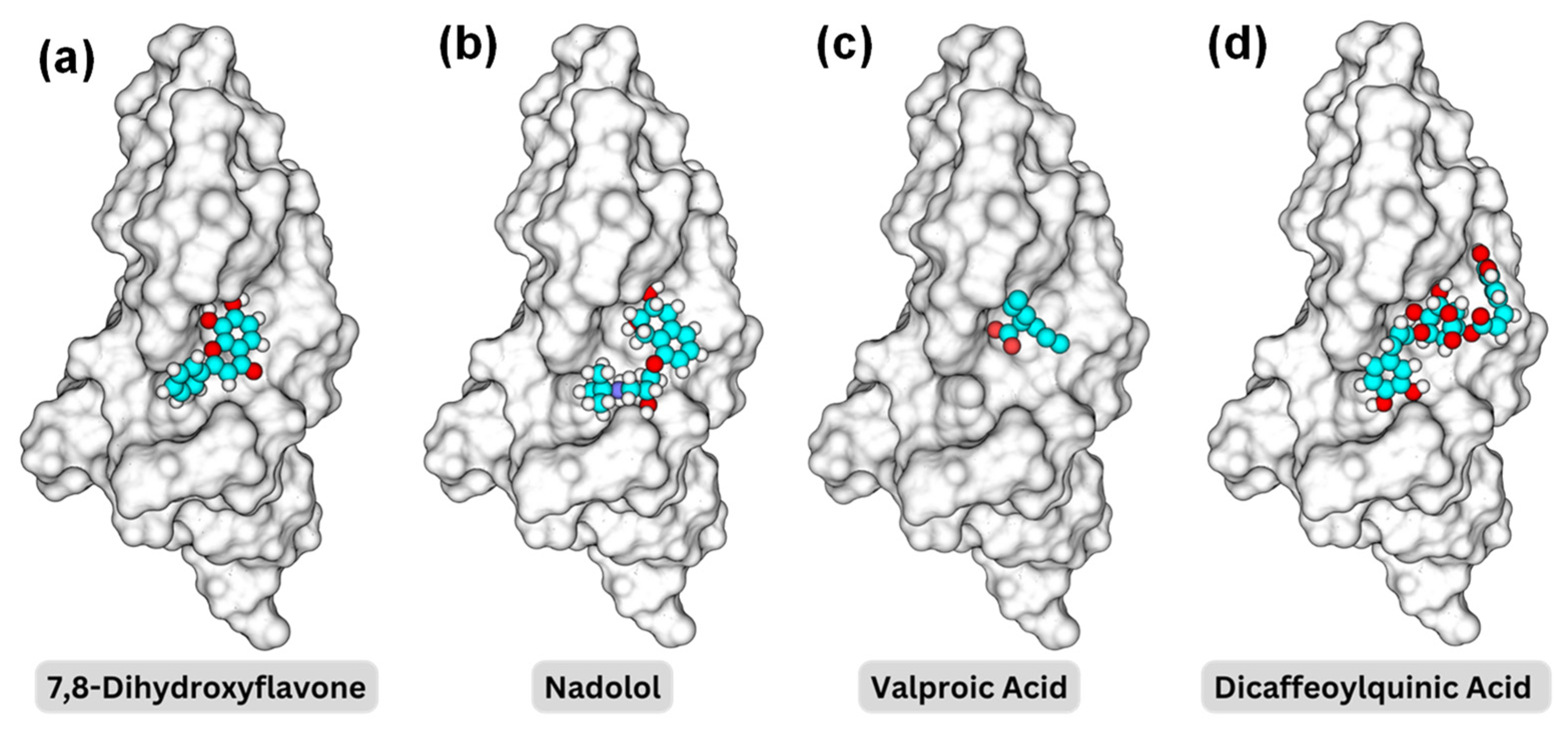
2.3. Screening of the NIH Clinical Collection Library
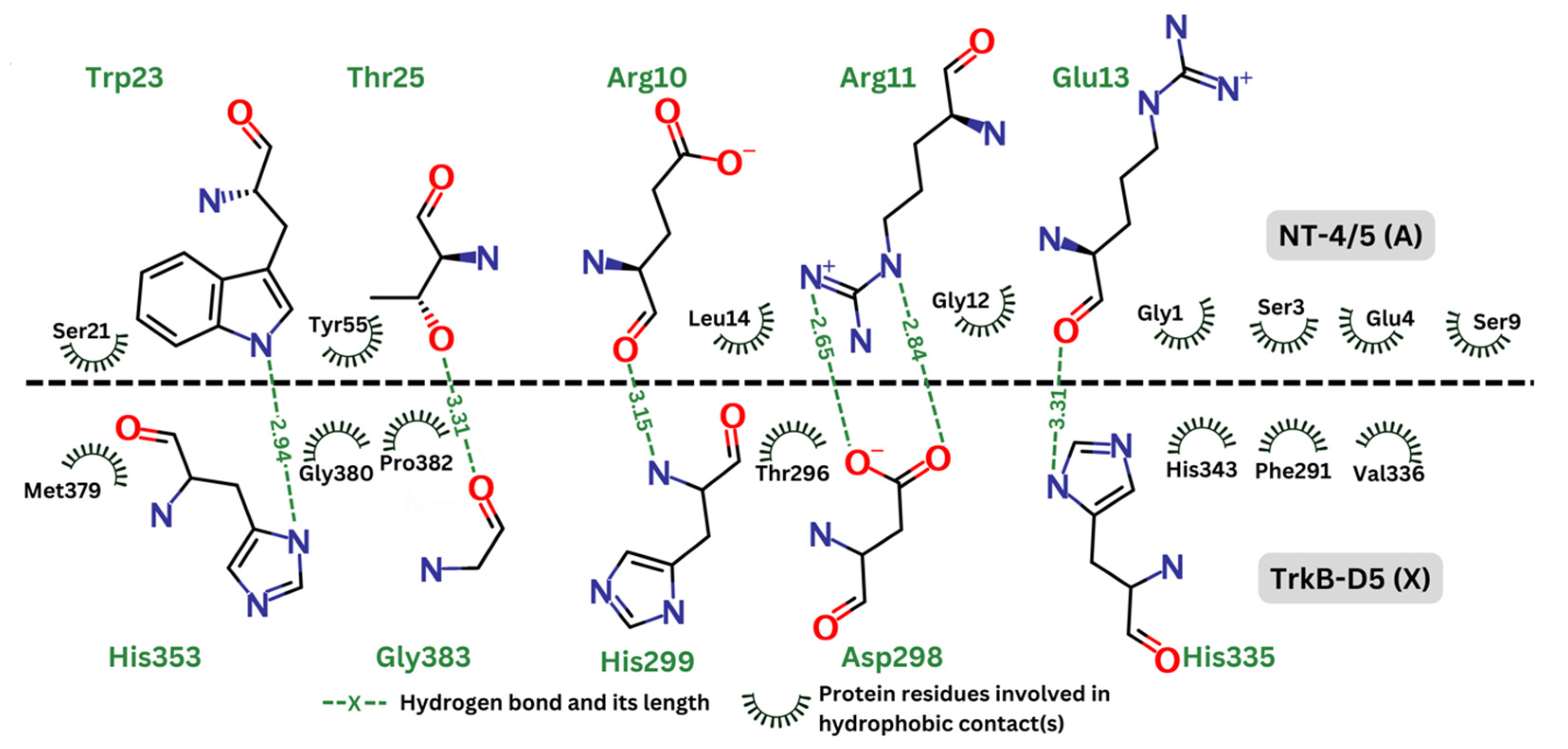
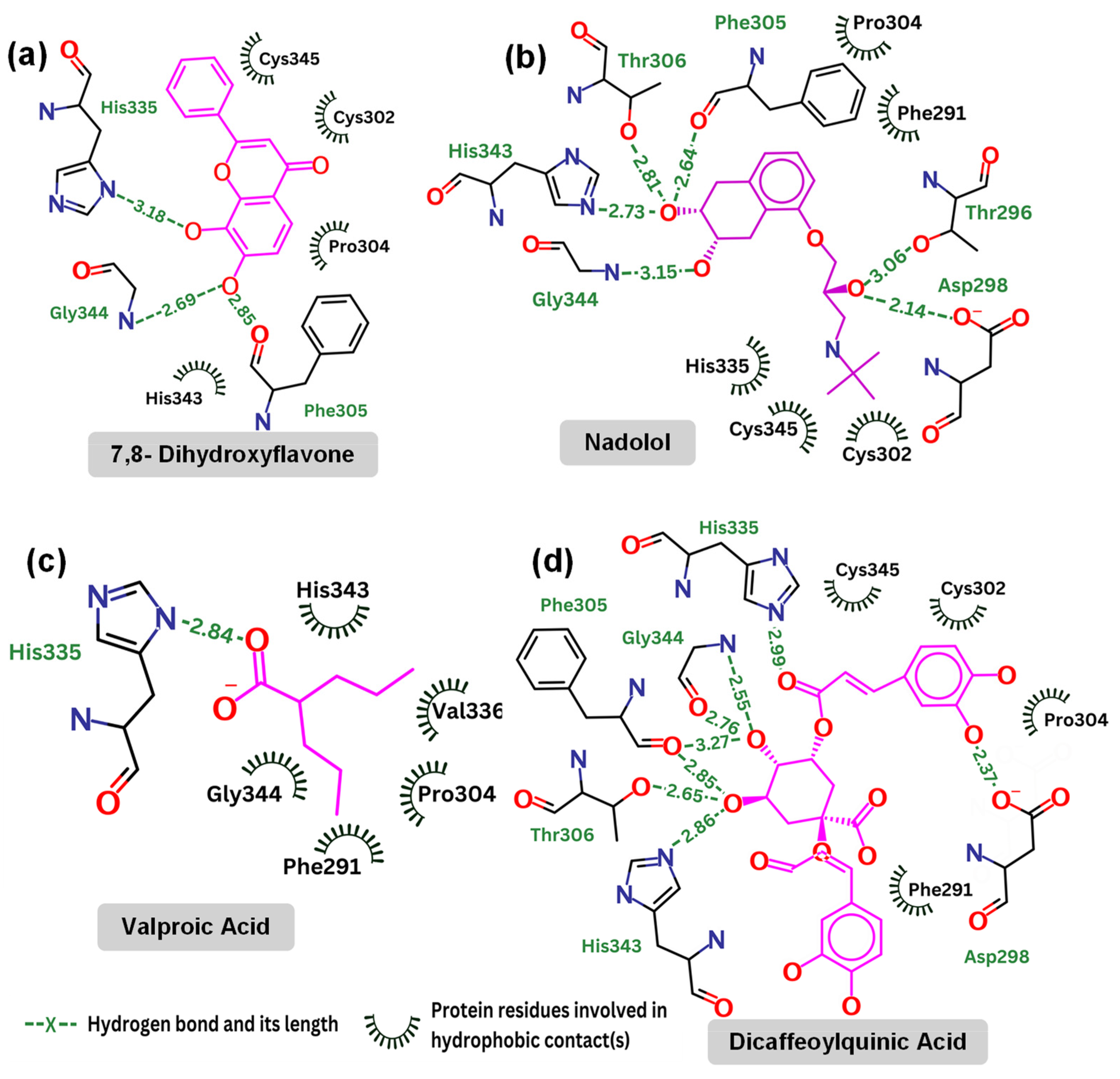
2.4. Detailed Analysis of TrkB and Top Candidate Interactions
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Magnetic Screening Nanoplatform (MSN)
4.3. Screening of NIH Library
4.4. Docking Computation of TrkB and Its Agonists
4.5. Structures of Docked Compounds
4.6. Biological Activity Evaluation of Top Compounds
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adkins-Jackson, P.B.; Belsky, D.W. Alzheimer’s disease risk biomarkers: Progress and challenges. Lancet Healthy Longev. 2022, 3, e575–e576. [Google Scholar] [CrossRef] [PubMed]
- Yiannopoulou, K.G.; Anastasiou, A.I.; Zachariou, V.; Pelidou, S.H. Reasons for Failed Trials of Disease-Modifying Treatments for Alzheimer Disease and Their Contribution in Recent Research. Biomedicines 2019, 7, 97. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.; Zhou, Y.; Lee, G.; Zhong, K.; Fonseca, J.; Cheng, F. Alzheimer’s disease drug development pipeline: 2024. Alzheimer's Dement. Transl. Res. Clin. Interv. 2024, 10, e12465. [Google Scholar] [CrossRef] [PubMed]
- Mullane, K.; Williams, M. Alzheimer’s disease beyond amyloid: Can the repetitive failures of amyloid-targeted therapeutics inform future approaches to dementia drug discovery? Biochem. Pharmacol. 2020, 177, 113945. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, I.; Marin, C.; Rey, M.J.; Ribalta, T.; Goutan, E.; Blanco, R.; Tolosa, E.; Marti, E. BDNF and full-length and truncated TrkB expression in Alzheimer disease. Implications in therapeutic strategies. J. Neuropathol. Exp. Neurol. 1999, 58, 729–739. [Google Scholar] [CrossRef]
- Numakawa, T.; Odaka, H.; Adachi, N. Actions of Brain-Derived Neurotrophin Factor in the Neurogenesis and Neuronal Function, and Its Involvement in the Pathophysiology of Brain Diseases. Int. J. Mol. Sci. 2018, 19, 3650. [Google Scholar] [CrossRef]
- Lima Giacobbo, B.; Doorduin, J.; Klein, H.C.; Dierckx, R.; Bromberg, E.; de Vries, E.F.J. Brain-Derived Neurotrophic Factor in Brain Disorders: Focus on Neuroinflammation. Mol. Neurobiol. 2019, 56, 3295–3312. [Google Scholar] [CrossRef]
- Wang, Z.H.; Xiang, J.; Liu, X.; Yu, S.P.; Manfredsson, F.P.; Sandoval, I.M.; Wu, S.; Wang, J.Z.; Ye, K. Deficiency in BDNF/TrkB Neurotrophic Activity Stimulates delta-Secretase by Upregulating C/EBPbeta in Alzheimer’s Disease. Cell Rep. 2019, 28, 655–669. [Google Scholar] [CrossRef]
- Devi, L.; Ohno, M. TrkB reduction exacerbates Alzheimer’s disease-like signaling aberrations and memory deficits without affecting beta-amyloidosis in 5XFAD mice. Transl. Psychiatry 2015, 5, e562. [Google Scholar] [CrossRef]
- Jiao, S.S.; Shen, L.L.; Zhu, C.; Bu, X.L.; Liu, Y.H.; Liu, C.H.; Yao, X.Q.; Zhang, L.L.; Zhou, H.D.; Walker, D.G.; et al. Brain-derived neurotrophic factor protects against tau-related neurodegeneration of Alzheimer’s disease. Transl. Psychiatry 2016, 6, e907. [Google Scholar] [CrossRef]
- Ng, T.K.S.; Ho, C.S.H.; Tam, W.W.S.; Kua, E.H.; Ho, R.C. Decreased Serum Brain-Derived Neurotrophic Factor (BDNF) Levels in Patients with Alzheimer’s Disease (AD): A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Amidfar, M.; de Oliveira, J.; Kucharska, E.; Budni, J.; Kim, Y.K. The role of CREB and BDNF in neurobiology and treatment of Alzheimer’s disease. Life Sci. 2020, 257, 118020. [Google Scholar] [CrossRef]
- Atasoy, I.L.; Dursun, E.; Gezen-Ak, D.; Metin-Armagan, D.; Ozturk, M.; Yilmazer, S. Both secreted and the cellular levels of BDNF attenuated due to tau hyperphosphorylation in primary cultures of cortical neurons. J. Chem. Neuroanat. 2017, 80, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Rosa, E.; Mahendram, S.; Ke, Y.D.; Ittner, L.M.; Ginsberg, S.D.; Fahnestock, M. Tau downregulates BDNF expression in animal and cellular models of Alzheimer’s disease. Neurobiol. Aging 2016, 48, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Wang, Z.H.; Ahn, E.H.; Liu, X.; Yu, S.P.; Manfredsson, F.P.; Sandoval, I.M.; Ju, G.; Wu, S.; Ye, K. Delta-secretase-cleaved Tau antagonizes TrkB neurotrophic signalings, mediating Alzheimer’s disease pathologies. Proc. Natl. Acad. Sci. USA 2019, 116, 9094–9102. [Google Scholar] [CrossRef]
- Giuffrida, M.L.; Copani, A.; Rizzarelli, E. A promising connection between BDNF and Alzheimer’s disease. Aging 2018, 10, 1791–1792. [Google Scholar] [CrossRef]
- Ando, S.; Kobayashi, S.; Waki, H.; Kon, K.; Fukui, F.; Tadenuma, T.; Iwamoto, M.; Takeda, Y.; Izumiyama, N.; Watanabe, K.; et al. Animal model of dementia induced by entorhinal synaptic damage and partial restoration of cognitive deficits by BDNF and carnitine. J. Neurosci. Res. 2002, 70, 519–527. [Google Scholar] [CrossRef]
- Fischer, D.L.; Sortwell, C.E. BDNF provides many routes toward STN DBS-mediated disease modification. Mov. Disord. 2019, 34, 22–34. [Google Scholar] [CrossRef]
- Zhang, F.; Kang, Z.; Li, W.; Xiao, Z.; Zhou, X. Roles of brain-derived neurotrophic factor/tropomyosin-related kinase B (BDNF/TrkB) signalling in Alzheimer’s disease. J. Clin. Neurosci. 2012, 19, 946–949. [Google Scholar] [CrossRef]
- Pilakka-Kanthikeel, S.; Atluri, V.S.; Sagar, V.; Saxena, S.K.; Nair, M. Targeted brain derived neurotropic factors (BDNF) delivery across the blood-brain barrier for neuro-protection using magnetic nano carriers: An in-vitro study. PLoS ONE 2013, 8, e62241. [Google Scholar] [CrossRef]
- Jang, S.W.; Liu, X.; Yepes, M.; Shepherd, K.R.; Miller, G.W.; Liu, Y.; Wilson, W.D.; Xiao, G.; Blanchi, B.; Sun, Y.E.; et al. A selective TrkB agonist with potent neurotrophic activities by 7,8-dihydroxyflavone. Proc. Natl. Acad. Sci. USA 2010, 107, 2687–2692. [Google Scholar] [CrossRef]
- Todd, D.; Gowers, I.; Dowler, S.J.; Wall, M.D.; McAllister, G.; Fischer, D.F.; Dijkstra, S.; Fratantoni, S.A.; van de Bospoort, R.; Veenman-Koepke, J.; et al. A Monoclonal Antibody TrkB Receptor Agonist as a Potential Therapeutic for Huntington’s Disease. PLoS ONE 2014, 9, e87923. [Google Scholar] [CrossRef]
- Liu, X.; Chan, C.B.; Qi, Q.; Xiao, G.; Luo, H.R.; He, X.L.; Ye, K.Q. Optimization of a Small Tropomyosin-Related Kinase B (TrkB) Agonist 7,8-Dihydroxyflavone Active in Mouse Models of Depression. J. Med. Chem. 2012, 55, 8524–8537. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Obianyo, O.; Chan, C.B.; Huang, J.; Xue, S.; Yang, J.J.; Zeng, F.; Goodman, M.; Ye, K. Biochemical and biophysical investigation of the brain-derived neurotrophic factor mimetic 7,8-dihydroxyflavone in the binding and activation of the TrkB receptor. J. Biol. Chem. 2014, 289, 27571–27584. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Gao, X.; Zhao, S.; Hu, W.; Chen, J. The Small-Molecule TrkB Agonist 7, 8-Dihydroxyflavone Decreases Hippocampal Newborn Neuron Death After Traumatic Brain Injury. J. Neuropathol. Exp. Neurol. 2015, 74, 557–567. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, X.; Schroeder, J.P.; Chan, C.B.; Song, M.; Yu, S.P.; Weinshenker, D.; Ye, K. 7,8-dihydroxyflavone prevents synaptic loss and memory deficits in a mouse model of Alzheime’s disease. Neuropsychopharmacology 2014, 39, 638–650. [Google Scholar] [CrossRef]
- Massa, S.M.; Yang, T.; Xie, Y.M.; Shi, J.; Bilgen, M.; Joyce, J.N.; Nehama, D.; Rajadas, J.; Longo, F.M. Small molecule BDNF mimetics activate TrkB signaling and prevent neuronal degeneration in rodents. J. Clin. Investig. 2010, 120, 1774–1785. [Google Scholar] [CrossRef] [PubMed]
- Edelbrock, A.N.; Alvarez, Z.; Simkin, D.; Fyrner, T.; Chin, S.M.; Sato, K.; Kiskinis, E.; Stupp, S.I. Supramolecular Nanostructure Activates TrkB Receptor Signaling of Neuronal Cells by Mimicking Brain-Derived Neurotrophic Factor. Nano Lett. 2018, 18, 6237–6247. [Google Scholar] [CrossRef]
- Casarotto, P.C.; Girych, M.; Fred, S.M.; Kovaleva, V.; Moliner, R.; Enkavi, G.; Biojone, C.; Cannarozzo, C.; Sahu, M.P.; Kaurinkoski, K.; et al. Antidepressant drugs act by directly binding to TRKB neurotrophin receptors. Cell 2021, 184, 1299–1313. [Google Scholar] [CrossRef]
- Jetsonen, E.; Didio, G.; Winkel, F.; Llach Pou, M.; Boj, C.; Kuczynski-Noyau, L.; Võikar, V.; Guirado, R.; Taira, T.; Lauri, S.E.; et al. Activation of TrkB in Parvalbumin interneurons is required for the promotion of reversal learning in spatial and fear memory by antidepressants. Neuropsychopharmacology 2023, 48, 1021–1030. [Google Scholar] [CrossRef]
- Sherwood, J.; Sowell, J.; Beyer, N.; Irvin, J.; Stephen, C.; Antone, A.J.; Bao, Y.P.; Ciesla, L.M. Cell-membrane coated iron oxide nanoparticles for isolation and specific identification of drug leads from complex matrices. Nanoscale 2019, 11, 6352–6359. [Google Scholar] [CrossRef] [PubMed]
- Arituluk, Z.C.; Horne, J.; Adhikari, B.; Steltzner, J.; Mansur, S.; Ahirwar, P.; Velu, S.E.; Gray, N.E.; Ciesla, L.M.; Bao, Y. Identification of TrkB binders from complex matrices using a magnetic drug screening nanoplatform. ACS Appl. Bio Mater. 2021, 4, 6244–6255. [Google Scholar] [CrossRef]
- Obergrussberger, A.; Friis, S.; Bruggemann, A.; Fertig, N. Automated patch clamp in drug discovery: Major breakthroughs and innovation in the last decade. Expert Opin. Drug Discov. 2021, 16, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Ciesla, L.; Moaddel, R. Comparison of analytical techniques for the identification of bioactive compounds from natural products. Nat. Prod. Rep. 2016, 33, 1131–1145. [Google Scholar] [CrossRef] [PubMed]
- Ciesla, L.; Okine, M.; Rosenberg, A.; Dossou, K.S.S.; Toll, L.; Wainer, I.W.; Moaddel, R. Development and characterization of the alpha3beta4alpha5 nicotinic receptor cellular membrane affinity chromatography column and its application for on line screening of plant extracts. J. Chromatogr. A 2016, 1431, 138–144. [Google Scholar] [CrossRef]
- Thornburg, C.C.; Britt, J.R.; Evans, J.R.; Akee, R.K.; Whitt, J.A.; Trinh, S.K.; Harris, M.J.; Thompson, J.R.; Ewing, T.L.; Shipley, S.M.; et al. NCI Program for Natural Product Discovery: A Publicly-Accessible Library of Natural Product Fractions for High-Throughput Screening. ACS Chem. Biol. 2018, 13, 2484–2497. [Google Scholar] [CrossRef]
- McGann, M. FRED pose prediction and virtual screening accuracy. J. Chem. Inf. Model. 2011, 51, 578–596. [Google Scholar] [CrossRef] [PubMed]
- Zagrebelsky, M.; Korte, M. Are TrkB receptor agonists the right tool to fulfill the promises for a therapeutic value of the brain-derived neurotrophic factor? Neural Regen. Res. 2024, 19, 29–34. [Google Scholar] [CrossRef]
- Ademuwagun, I.A.; Oduselu, G.O.; Rotimi, S.O.; Adebiyi, E. Pharmacophore-Aided Virtual Screening and Molecular Dynamics Simulation Identifies TrkB Agonists for Treatment of CDKL5-Deficiency Disorders. Bioinform. Biol. Insights 2023, 17, 11779322231158254. [Google Scholar] [CrossRef]
- Enkavi, G.; Girych, M.; Moliner, R.; Vattulainen, I.; Castrén, E. TrkB transmembrane domain: Bridging structural understanding with therapeutic strategy. Trends Biochem. Sci. 2024, 49, 445–446. [Google Scholar] [CrossRef]
- Pattarawarapan, M.; Burgess, K. Molecular Basis of Neurotrophin−Receptor Interactions. J. Med. Chem. 2003, 46, 5277–5291. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, C.F. Emerging themes in structural biology of neurotrophic factors. Trends Neurosci. 1998, 21, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Banfield, M.J.; Naylor, R.L.; Robertson, A.G.; Allen, S.J.; Dawbarn, D.; Brady, R.L. Specificity in Trk receptor: Neurotrophin interactions: The crystal structure of TrkB-d5 in complex with neurotrophin-4/5. Structure 2001, 9, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the worldwide protein data bank. Nat. Struct. Mol. Biol. 2003, 10, 980. [Google Scholar] [CrossRef] [PubMed]
- Beaman, E.E.; Bonde, A.N.; Larsen, S.M.U.; Ozenne, B.; Lohela, T.J.; Nedergaard, M.; Gíslason, G.H.; Knudsen, G.M.; Holst, S.C. Blood–brain barrier permeable β-blockers linked to lower risk of Alzheimer’s disease in hypertension. Brain 2023, 146, 1141–1151. [Google Scholar] [CrossRef]
- CuraSen Therapeutics, I.s.P. A Study of CST-2032 and CST-107 in Subjects with Mild Cognitive Impairment or Mild Dementia Due to Parkinson's or Alzheimer's Disease; United States National Library of Medicine (NLM): Bethesda, MD, USA, 2023. Available online: https://clinicaltrials.gov/study/NCT05104463 (accessed on 16 August 2024).
- Dedoni, S.; Marras, L.; Olianas, M.C.; Ingianni, A.; Onali, P. Downregulation of TrkB Expression and Signaling by Valproic Acid and Other Histone Deacetylase Inhibitors. J. Pharmacol. Exp. Ther. 2019, 370, 490–503. [Google Scholar] [CrossRef]
- Mikitsh, J.L.; Chacko, A.M. Pathways for small molecule delivery to the central nervous system across the blood-brain barrier. Perspect. Med. Chem. 2014, 6, 11–24. [Google Scholar] [CrossRef]
- Laurens, C.; Abot, A.; Delarue, A.; Knauf, C. Central Effects of Beta-Blockers May Be Due to Nitric Oxide and Hydrogen Peroxide Release Independently of Their Ability to Cross the Blood-Brain Barrier. Front. Neurosci. 2019, 13, 33. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, Z.; Zhang, Z.; Liu, X.; Kang, S.S.; Zhang, Y.; Ye, K. The prodrug of 7,8-dihydroxyflavone development and therapeutic efficacy for treating Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2018, 115, 578–583. [Google Scholar] [CrossRef] [PubMed]
- de Pins, B.; Cifuentes-Díaz, C.; Farah, A.T.; López-Molina, L.; Montalban, E.; Sancho-Balsells, A.; López, A.; Ginés, S.; Delgado-García, J.M.; Alberch, J.; et al. Conditional BDNF Delivery from Astrocytes Rescues Memory Deficits, Spine Density, and Synaptic Properties in the 5xFAD Mouse Model of Alzheimer Disease. J. Neurosci. 2019, 39, 2441–2458. [Google Scholar] [CrossRef]
- Moliner, R.; Girych, M.; Brunello, C.A.; Kovaleva, V.; Biojone, C.; Enkavi, G.; Antenucci, L.; Kot, E.F.; Goncharuk, S.A.; Kaurinkoski, K.; et al. Psychedelics promote plasticity by directly binding to BDNF receptor TrkB. Nat. Neurosci. 2023, 26, 1032–1041. [Google Scholar] [CrossRef] [PubMed]
- Chitranshi, N.; Gupta, V.; Kumar, S.; Graham, S.L. Exploring the molecular interactions of 7, 8-dihydroxyflavone and its derivatives with TrkB and VEGFR2 proteins. Int. J. Mol. Sci. 2015, 16, 21087–21108. [Google Scholar] [CrossRef]
- Chiang, N.-N.; Lin, T.-H.; Teng, Y.-S.; Sun, Y.-C.; Chang, K.-H.; Lin, C.-Y.; Hsieh-Li, H.M.; Su, M.-T.; Chen, C.-M.; Lee-Chen, G.-J. Flavones 7, 8-DHF, quercetin, and apigenin against Tau toxicity via activation of TRKB signaling in ΔK280 TauRD-DsRed SH-SY5Y cells. Front. Aging Neurosci. 2021, 13, 758895. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.-J.; Lin, T.-H.; Chang, K.-H.; Lin, W.; Hsieh-Li, H.M.; Su, M.-T.; Chen, C.-M.; Sun, Y.-C.; Lee-Chen, G.-J. Novel TRKB agonists activate TRKB and downstream ERK and AKT signaling to protect Aβ-GFP SH-SY5Y cells against Aβ toxicity. Aging 2022, 14, 7568. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery; ACS Publications: Washington, DC, USA, 2011. [Google Scholar]
- ChemAxon. Marvin Was Used for Drawing, Displaying and Characterizing Chemical Structures, Substructures and Reactions; Marvin 24.1.2; ChemAxon: Budapest, Hungary, 2024. [Google Scholar]
- Cazorla, M.; Prémont, J.; Mann, A.; Girard, N.; Kellendonk, C.; Rognan, D. Identification of a low-molecular weight TrkB antagonist with anxiolytic and antidepressant activity in mice. J. Clin. Investig. 2011, 121, 1846–1857. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, P.C.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer generation with OMEGA: Algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 2010, 50, 572–584. [Google Scholar] [CrossRef]
- OpenEye. QUACPAC, Version 2.2.3.3; Cadence Molecular Sciences: Santa Fe, NM, USA, 2024. [Google Scholar]
- Xiang, J.Z.; Honig, B. Jackal: A Protein Structure Modeling Package; Columbia University and Howard Hughes Medical Institute: New York, NY, USA, 2002. [Google Scholar]
- Kaech, S.; Banker, G. Culturing hippocampal neurons. Nat. Protoc. 2006, 1, 2406–2415. [Google Scholar] [CrossRef]
- Long, H.S.; Stander, M.A.; Van Wyk, B.E. Notes on the occurrence and significance of triterpenoids (asiaticoside and related compounds) and caffeoylquinic acids in Centella species. S. Afr. J. Bot. 2012, 82, 53–59. [Google Scholar] [CrossRef]

| Compound Names | BP 1 | BP 2 | BP3 | BP4 | BP5 |
|---|---|---|---|---|---|
| Dicaffeopllauinic Acid | −7.63 | 0.74 | −3.06 | 1.15 | 0.2 |
| Castillicetin | −7.32 | 2.59 | −3.71 | 0.02 | −2.10 |
| O-Caffeoylquinic Acid | −7.02 | −3.77 | −5.82 | −4.14 | −4.45 |
| Stigmasterol | −6.70 | - | 0.85 | - | 3.63 |
| 7,8-dihydroxyflavone | −6.27 | −2.26 | −5.12 | −3.91 | −3.26 |
| Quercetin | −5.86 | −3.35 | −5.09 | −3.25 | −3.20 |
| Pomolic Acid | −4.99 | 5.91 | −0.27 | 4.66 | 1.60 |
| Madecassic Acid | −4.89 | 5.94 | −1.30 | 8.57 | 1.90 |
| Naringin | −3.52 | - | - | - | - |
| Compound Names | BP1 | BP2 | BP3 | BP4 | BP5 |
|---|---|---|---|---|---|
| Nadolol | −8.13 | −3.0 | −4.75 | −2.96 | −4.95 |
| Benproperine Phosphate | −7.63 | −0.51 | −4.49 | −2.44 | −3.7 |
| Clofazimine | −7.36 | 6.46 | 2.77 | - | −4.79 |
| Brimonidine | −7.32 | −4.77 | −4.56 | −4.51 | −5.15 |
| Amiodarone Hydrochloride | −6.95 | - | - | - | - |
| 3,5,3′-Triiodothyronine | −6.67 | −0.95 | −4.69 | −3.00 | −3.13 |
| 7,8-dihydroxyflavone | −6.27 | −2.26 | −5.12 | −3.91 | −3.26 |
| Mestranol | −6.18 | −0.91 | −3.45 | −1.81 | −1.48 |
| Levonorgestrel | −5.86 | −0.08 | −3.76 | −2.57 | −1.22 |
| Ondansetron | −5.84 | −0.73 | −3.53 | −3.68 | −3.29 |
| Vecuronium Bromide | −5.16 | - | 3.64 | - | 9.9 |
| Raloxifene HCl | −5.54 | 9.62 | 2.24 | 7.45 | 5.01 |
| Oxaprozin | −5.00 | −1.2 | −3.53 | −3.07 | −3.89 |
| Valproic Acid | −4.83 | −3.06 | −3.80 | −2.96 | −3.28 |
| Deferiprone | −3.66 | −3.79 | - | - | - |
| Diazepam | −4.4 | −1.31 | −3.13 | −1.39 | −2.57 |
| Loperamide Hydrochloride | −4.50 | 8.24 | −0.32 | 7.97 | 3.98 |
| Hydrogen Bond | Distance (Å) | Angle (°) |
|---|---|---|
| 7,8-dihydroxyflavone | ||
| O3-H…N(Gly344) | 2.69 | 46.0 |
| O3-H…O(Phe305) | 2.85 | 110.6 |
| O4-H…ND1(His335) | 3.18 | 45.6 |
| Nadolol | ||
| O21-H…OD2(Asp298) | 2.14 | 96.9 |
| O21…H-OG1(Thr296) | 3.06 | 62.5 |
| O20-H…N(Gly344) | 3.15 | 82.2 |
| O19…H-ND1(His343) | 2.73 | 118.4 |
| O19…H-OG1(Thr306) | 2.8 | 61.3 |
| O19-H…O(Phe305) | 2.64 | 57.2 |
| Valproic Acid | ||
| O10…H-ND1(His335) | 2.84 | 149.3 |
| Dicaffeoylquinic Acid | ||
| O32-H…OD2(Asp298) | 2.37 | 45.4 |
| O27…H-ND1(His335) | 2.99 | 129.7 |
| O35-H…O(Gly344) | 2.76 | 134.7 |
| O35…H-N(Gly344) | 2.55 | 96.4 |
| O35-H…O(Phe305) | 3.27 | 65.2 |
| O34-H…OG1(Thr306) | 2.65 | 91.5 |
| O34…H-ND1(His343) | 2.86 | 122.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, D.D.; Mansur, S.; Ciesla, L.; Gray, N.E.; Zhao, S.; Bao, Y. A Combined Computational and Experimental Approach to Studying Tropomyosin Kinase Receptor B Binders for Potential Treatment of Neurodegenerative Diseases. Molecules 2024, 29, 3992. https://doi.org/10.3390/molecules29173992
Nguyen DD, Mansur S, Ciesla L, Gray NE, Zhao S, Bao Y. A Combined Computational and Experimental Approach to Studying Tropomyosin Kinase Receptor B Binders for Potential Treatment of Neurodegenerative Diseases. Molecules. 2024; 29(17):3992. https://doi.org/10.3390/molecules29173992
Chicago/Turabian StyleNguyen, Duc D., Shomit Mansur, Lukasz Ciesla, Nora E. Gray, Shan Zhao, and Yuping Bao. 2024. "A Combined Computational and Experimental Approach to Studying Tropomyosin Kinase Receptor B Binders for Potential Treatment of Neurodegenerative Diseases" Molecules 29, no. 17: 3992. https://doi.org/10.3390/molecules29173992
APA StyleNguyen, D. D., Mansur, S., Ciesla, L., Gray, N. E., Zhao, S., & Bao, Y. (2024). A Combined Computational and Experimental Approach to Studying Tropomyosin Kinase Receptor B Binders for Potential Treatment of Neurodegenerative Diseases. Molecules, 29(17), 3992. https://doi.org/10.3390/molecules29173992









