Recognition of 8-Oxo-2′-deoxyguanosine in DNA Using the Triphosphate of 2′-Deoxycytidine Connecting the 1,3-Diazaphenoxazine Unit, dCdapTP
Abstract
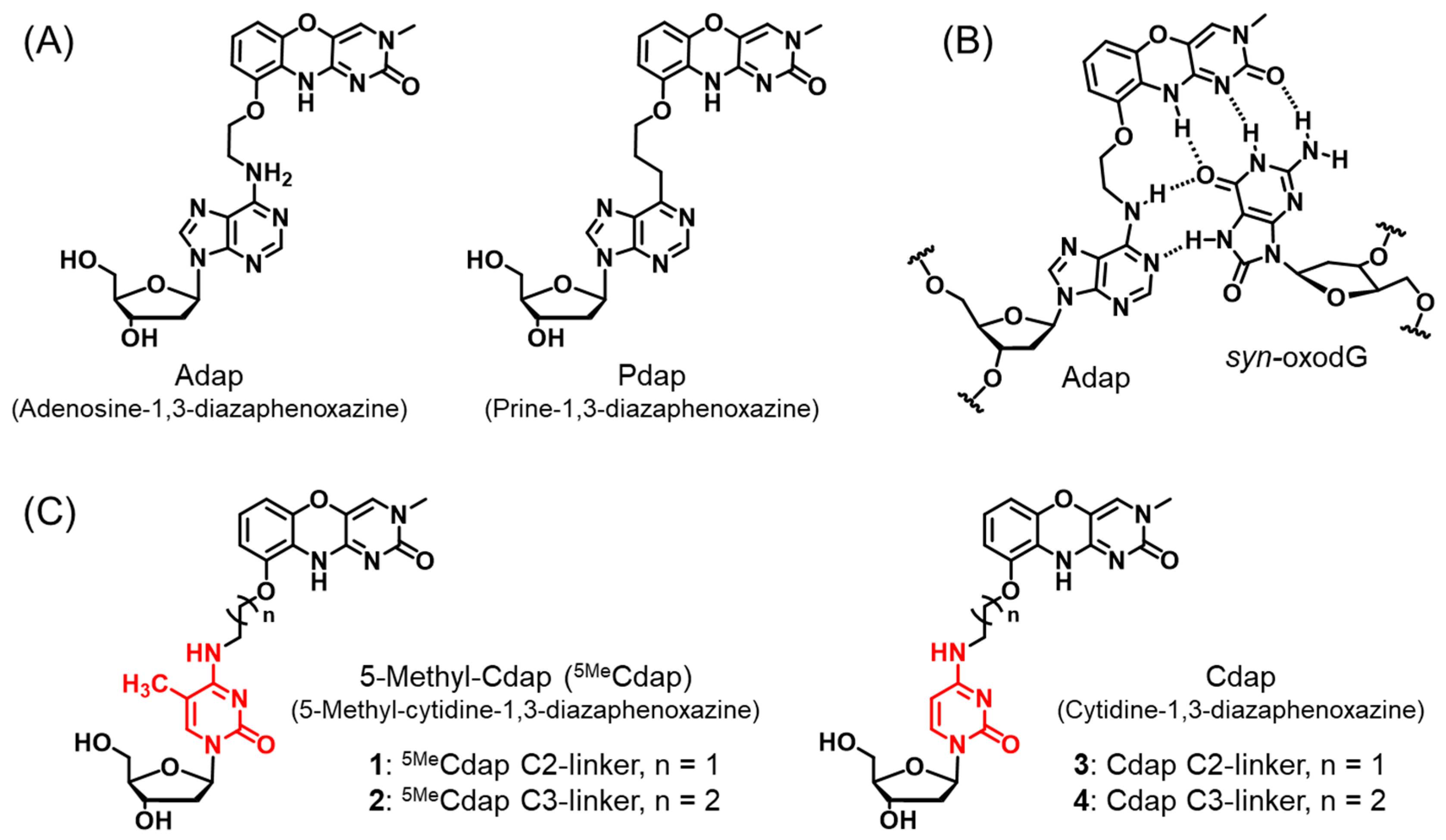
1. Introduction
2. Results and Discussion
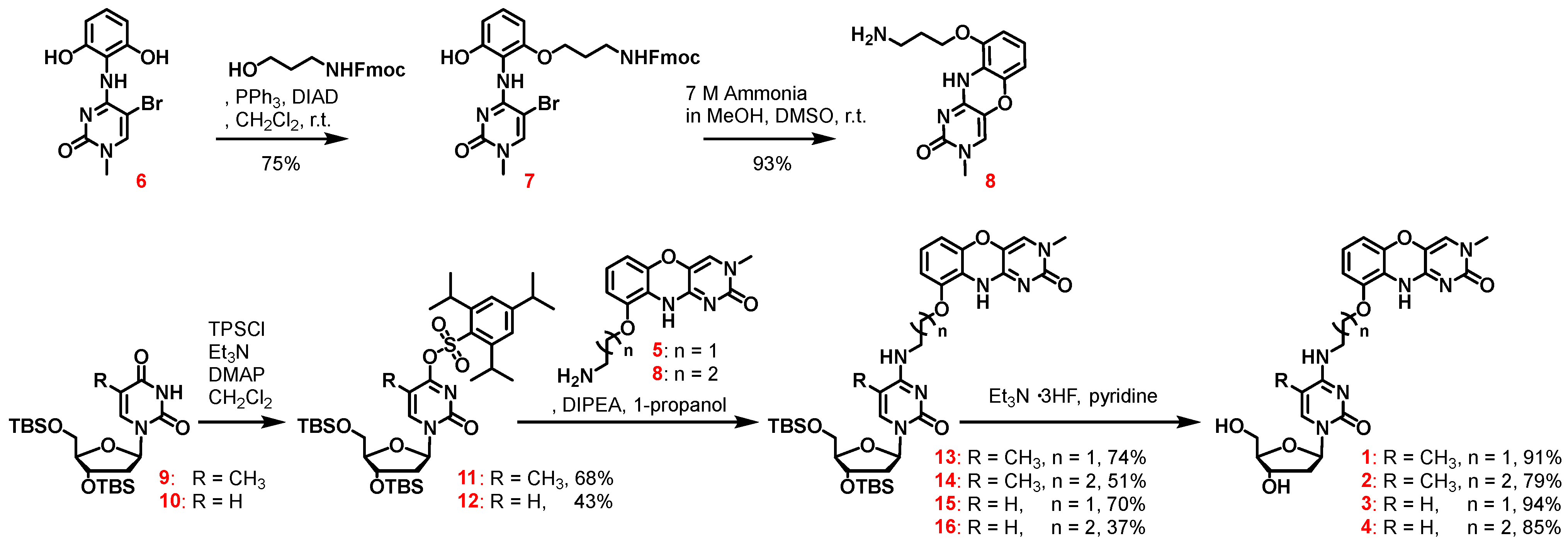
2.1. Synthesis of 3′-,5′-Diol Compounds
2.2. Synthesis of Triphosphate Compounds
2.3. Evaluation of oxodG Recognition Ability by Single Nucleotide Elongation Reaction
2.4. Steady-State Kinetic Study of dCapTP Using Klenow Fragment
3. Materials and Methods
3.1. General Methods of the Synthesis of Desired Compounds
3.2. Synthesis of the Phenoxazine Unit with a C3-Linker (8)
3.3. Synthesis of Compound 11
3.4. Synthesis of Compound 12
3.5. Synthesis of the TBS-Protected 5-Methyl-Cdap C2-Linker (13)
3.6. Synthesis of the TBS-Protected 5-Methyl-Cdap C3-Linker (14)
3.7. Synthesis of the TBS-Protected Cdap C2-Linker (15)
3.8. Synthesis of the TBS-Protected Cdap C3-Linker (16)
3.9. General Procedure of the Deprotection Reaction in TBS Groups
3.10. General Procedure of the Synthesis of the 5′-Triphosphate Compound
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Betteridge, D.J. What is oxidative stress? Metabolism 2000, 49, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Nakabeppu, Y.; Behmanesh, M.; Yamaguchi, H.; Yoshimura, D.; Sakumi, K. Oxidative Damage to Nucleic Acids; Evans, M.D., Cooke, M.S., Eds.; Springer: New York, NY, USA, 2007; pp. 40–53. ISBN 978-0-387-72973-2. [Google Scholar]
- Seidel, C.A.; Schulz, A.; Sauer, M.H. Nucleobase-Specific Quenching of Fluorescent Dyes. 1. Nucleobase One-Electron Redox Potentials and Their Correlation with Static and Dynamic Quenching Efficiencies. J. Phys. Chem. 1996, 100, 5541–5553. [Google Scholar] [CrossRef]
- Steenken, S.; Jovanovic, S.V. How Easily Oxidizable Is DNA? One-Electron Reduction Potentials of Adenosine and Guanosine Radicals in Aqueous Solution. J. Am. Chem. Soc. 1997, 119, 617–618. [Google Scholar] [CrossRef]
- Burrows, C.J.; Muller, J.G. Oxidative Nucleobase Modifications Leading to Strand Scission. Chem. Rev. 1998, 98, 1109–1152. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.M. Radical and Radical Ion Reactivity in Nucleic Acid Chemistry; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2009. [Google Scholar]
- Kino, K.; Kawada, T.; Hirao-Suzuki, M.; Morikawa, M.; Miyazawa, H. Products of oxidative guanine damage form base pairs with guanine. Int. J. Mol. Sci. 2020, 21, 7645. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef] [PubMed]
- Santivasi, W.L.; Xia, F. Ionizing radiation-induced DNA damage, response, and repair. Antioxid. Redox Signal. 2014, 21, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Ames, B.N.; Gold, L.S. Endogenous mutagens and the causes of aging and cancer. Mutat. Res. 1991, 250, 3–16. [Google Scholar] [CrossRef]
- Steenken, S. Purine bases, nucleosides, and nucleotides: Aqueous solution redox chemistry and transformation reactions of their radical cations and e- and OH adducts. Chem. Rev. 1989, 89, 503–520. [Google Scholar] [CrossRef]
- Mingard, C.; Wu, J.; McKeague, M.; Sturla, S.J. Next-generation DNA damage sequencing. Chem. Soc. Rev. 2020, 49, 7354–7377. [Google Scholar] [CrossRef]
- Simoeki, A.; Rytarowska, A.; Poplawska, A.S.; Gackowski, D.; Rozalski, R.; Dziaman, T.; Szaflarska, M.C.; Olinski, R. Helicobacter pylori infection is associated with oxidatively damaged DNA in human leukocytes and decreased level of urinary 8-oxo-7,8-dihydroguanine. Carcinogenesis 2006, 27, 405–408. [Google Scholar] [CrossRef] [PubMed]
- Hah, S.S.; Mundt, J.M.; Kim, H.M.; Sumbad, R.A.; Turteltaub, K.W.; Henderson, P.T. Measurement of 7,8-dihydro-8-oxo-2-deoxyguanosine metabolism in MCF-7 cells at low concentrations using accelerator mass spectrometry. Proc. Natl. Acad. Sci. USA 2007, 104, 11203–11208. [Google Scholar] [CrossRef] [PubMed]
- Nakae, Y.; Stoward, P.J.; Bespalov, I.A.; Melamede, R.J.; Wallace, S.S. A new technique for the quantitative assessment of 8-oxoguanine in nuclear DNA as a marker of oxidative stress. Application to dystrophin-deficient DMD skeletal muscles. Histochem. Cell Biol. 2005, 124, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.Y.; Matteucci, M.D. A cytosine analogue capable of clamp-like binding to a guanine in helical nucleic acids. J. Am. Chem. Soc. 1998, 120, 8531–8532. [Google Scholar] [CrossRef]
- Bajacan, J.E.V.; Hong, I.S.; Penning, T.W.; Greenberg, M.M. Quantitative detection of 8-oxo-7,8-dihydro-2′-deoxyguanosine using chemical tagging and qPCR. Chem. Res. Toxicol. 2004, 27, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Guo, L.H.; Greenberg, M.M. Quantification of 8-oxodguo lesions in double-stranded DNA using a photoelectrochemical DNA sensor. Anal. Chem. 2012, 84, 6048–6053. [Google Scholar] [CrossRef] [PubMed]
- Schibel, A.E.P.; An, N.; Jin, Q.; Fleming, A.M.; Burrows, C.J.; White, H.S. Nanopore detection of 8-oxo-7,8-dihydro-2′-deoxyguanosine in immobilized single-stranded DNA via adduct formation to the DNA damage site. J. Am. Chem. Soc. 2010, 132, 17992–17995. [Google Scholar] [CrossRef] [PubMed]
- Riedl, J.; Fleming, A.M.; Burrows, C.J. Sequencing of DNA lesions facilitated by site-specific excision via base excision repair DNA glycosylases yielding ligatable gaps. J. Am. Chem. Soc. 2016, 138, 491–494. [Google Scholar] [CrossRef] [PubMed]
- Fromme, J.C.; Bruner, S.D.; Yang, W.; Karplus, M.; Verdine, G.L. Product-assisted catalysis in base-excision DNA repair. Nat. Struct. Mol. Biol. 2003, 10, 204–211. [Google Scholar] [CrossRef]
- Nash, H.M.; Lu, R.; Lane, W.S.; Verdine, G.L. The critical active-site amine of the human 8-oxoguanine DNA glycosylase, hOgg1: Direct identification, ablation and chemical reconstitution. Chem Biol. 1997, 4, 693–702. [Google Scholar] [CrossRef]
- Lu, R.; Nash, H.M.; Verdine, G.L. A mammalian DNA repair enzyme that excises oxidatively damaged guanines maps to a locus frequently lost in lung cancer. Curr. Biol. 1997, 7, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, Y.; Kawaguchi, R.; Sasaki, S. Adenosine-1,3-diazaphenoxazine Derivative for Selective Base Pair Formation with 8-Oxo-2′-deoxyguanosine in DNA. J. Am. Chem. Soc. 2011, 133, 7272–7275. [Google Scholar] [CrossRef]
- Taniguchi, Y.; Kikukawa, Y.; Sasaki, S. Discrimination between 8-oxo-2′-deoxyguanosine and 2′-deoxyguanosine in DNA by the single nucleotide primer extension reaction with Adap triphosphate. Angew. Chem. Int. Ed. 2015, 54, 5147–5151. [Google Scholar] [CrossRef] [PubMed]
- Kikukawa, Y.; Kawazoe, R.; Miyahara, R.; Sakurada, T.; Nagata, Y.; Sasaki, S.; Taniguchi, Y. Multiple-turnover single nucleotide primer extension reactions to detect of 8-oxo-2′-deoxyguanosine. ChemComm 2022, 58, 5399–5402. [Google Scholar] [CrossRef]
- Ohkubo, A.; Yamada, K.; Ito, Y.; Yoshimura, K.; Miyauchi, K.; Kanamori, T.; Masaki, Y.; Seio, K.; Yuasa, H.; Sekine, M. Synthesis and triplex-forming properties of oligonucleotides capable of recognizing corresponding DNA duplexes containing four base pairs. Nucleic Acids Res. 2015, 43, 5675–5686. [Google Scholar] [CrossRef] [PubMed]
- Steinert, H.S.; Schäfer, F.; Jonker, H.R.A.; Heckel, A.; Schwalbe, H. Influence of the absolute configuration of NPE-caged cytosine on DNA single base pair stability. Angew. Chem. Int. Ed. 2014, 53, 1072–1075. [Google Scholar] [CrossRef]
- Caton-Williams, J.; Hoxhaj, R.; Fiaz, B.; Huang, Z. Use of a novel 5′-regioselective phosphitylating reagent for one-pot synthesis of nucleoside 5′-triphosphates from unprotected nucleosides. Curr. Protoc. Nucleic Acid Chem. 2013, 52, 1.30.1–1.30.21. [Google Scholar] [CrossRef]

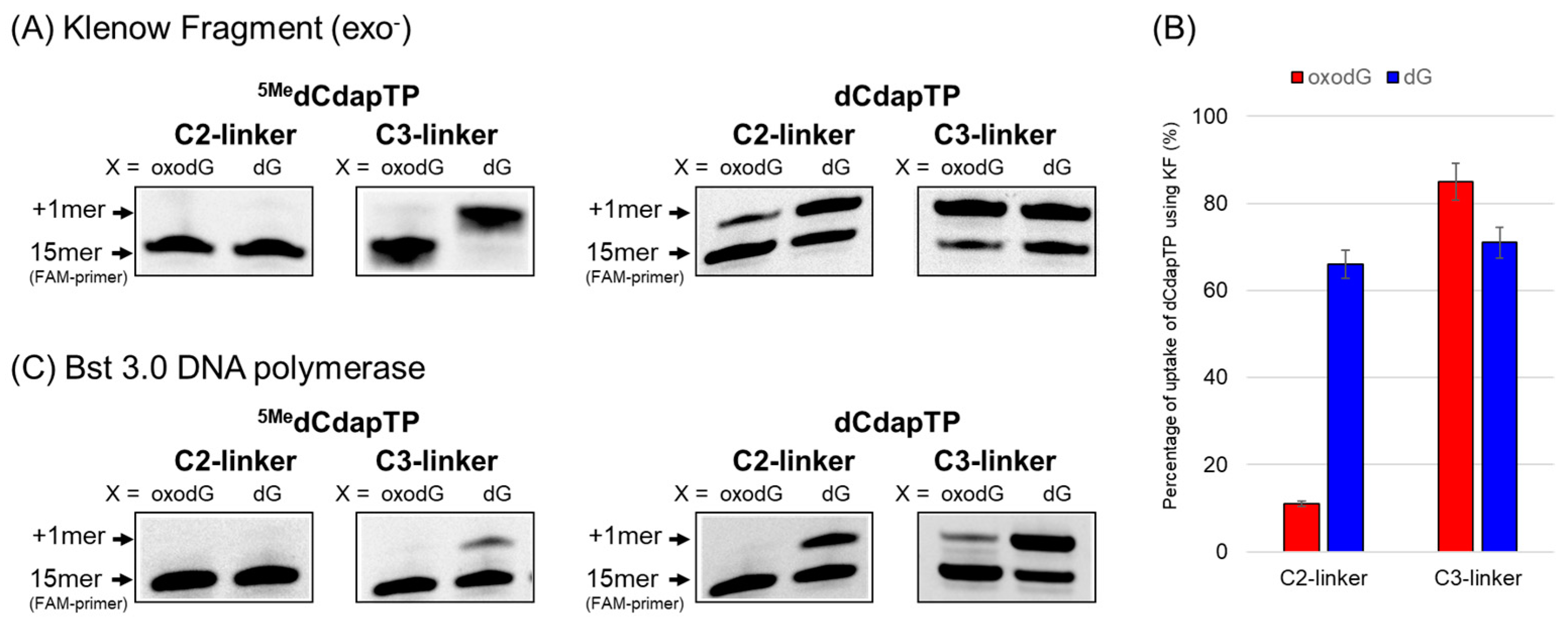

| Entry | dNTP | X= | Vmax (%min−1) | Km (µM) | Efficiency (%min−1M−1) |
|---|---|---|---|---|---|
| 1 | dCdapTP C2-linker | 8-oxodG | 0.85 | 0.29 | 2.91 × 106 |
| 2 | dG | 1.95 | 0.27 | 7.22 × 106 | |
| 3 | dA | 0.09 | 0.13 | 0.69 × 106 | |
| 4 | dC | 0.08 | 0.07 | 1.13 × 106 | |
| 5 | T | 0.11 | 0.13 | 0.88 × 106 | |
| 6 | dCdapTP C3-linker | 8-oxodG | 2.86 | 0.36 | 8.00 × 106 |
| 7 | dG | 2.20 | 0.28 | 7.64 × 106 | |
| 8 | dA | 0.06 | 0.23 | 0.28 × 106 | |
| 9 | dC | 0.07 | 0.15 | 0.50 × 106 | |
| 10 | T | 0.09 | 0.14 | 0.64 × 106 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakurada, T.; Chikada, Y.; Miyahara, R.; Taniguchi, Y. Recognition of 8-Oxo-2′-deoxyguanosine in DNA Using the Triphosphate of 2′-Deoxycytidine Connecting the 1,3-Diazaphenoxazine Unit, dCdapTP. Molecules 2024, 29, 2270. https://doi.org/10.3390/molecules29102270
Sakurada T, Chikada Y, Miyahara R, Taniguchi Y. Recognition of 8-Oxo-2′-deoxyguanosine in DNA Using the Triphosphate of 2′-Deoxycytidine Connecting the 1,3-Diazaphenoxazine Unit, dCdapTP. Molecules. 2024; 29(10):2270. https://doi.org/10.3390/molecules29102270
Chicago/Turabian StyleSakurada, Takato, Yuta Chikada, Ryo Miyahara, and Yosuke Taniguchi. 2024. "Recognition of 8-Oxo-2′-deoxyguanosine in DNA Using the Triphosphate of 2′-Deoxycytidine Connecting the 1,3-Diazaphenoxazine Unit, dCdapTP" Molecules 29, no. 10: 2270. https://doi.org/10.3390/molecules29102270
APA StyleSakurada, T., Chikada, Y., Miyahara, R., & Taniguchi, Y. (2024). Recognition of 8-Oxo-2′-deoxyguanosine in DNA Using the Triphosphate of 2′-Deoxycytidine Connecting the 1,3-Diazaphenoxazine Unit, dCdapTP. Molecules, 29(10), 2270. https://doi.org/10.3390/molecules29102270







