Past, Present, and Future Perspectives on Computer-Aided Drug Design Methodologies
Abstract
1. Introduction: The Benefits of Computational Methods for Drug Discovery
1.1. The Drug Discovery Pipeline and the Problem of Candidate Selection
1.2. The Application of Computational Methods in Drug Discovery
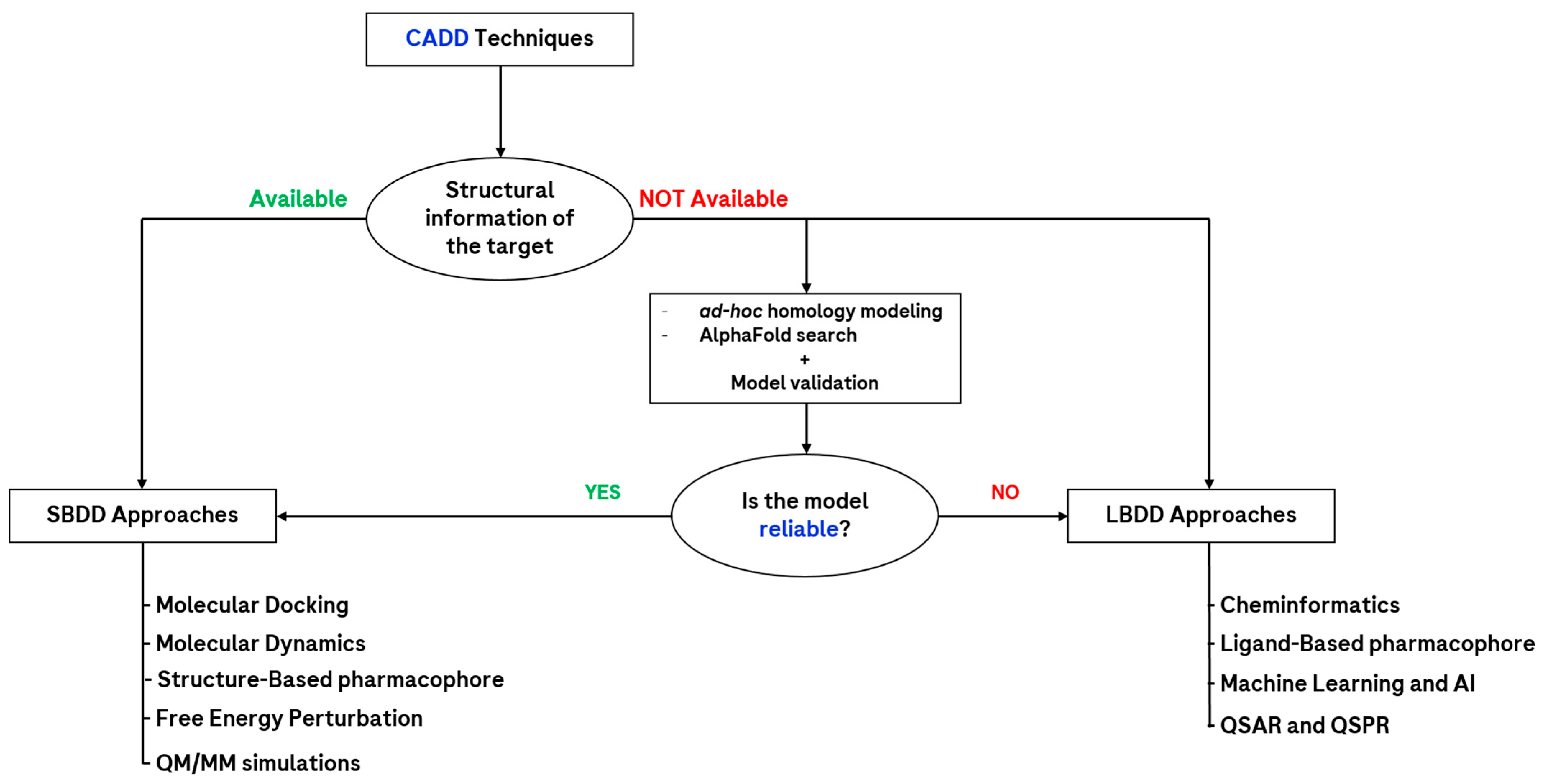
1.3. The Main Methodology Branches in CADD
2. Discussion
2.1. Ligand-Based Drug Design (LBDD)
Quantitative Structure–Activity Relationship (QSAR) Modeling and Cheminformatics
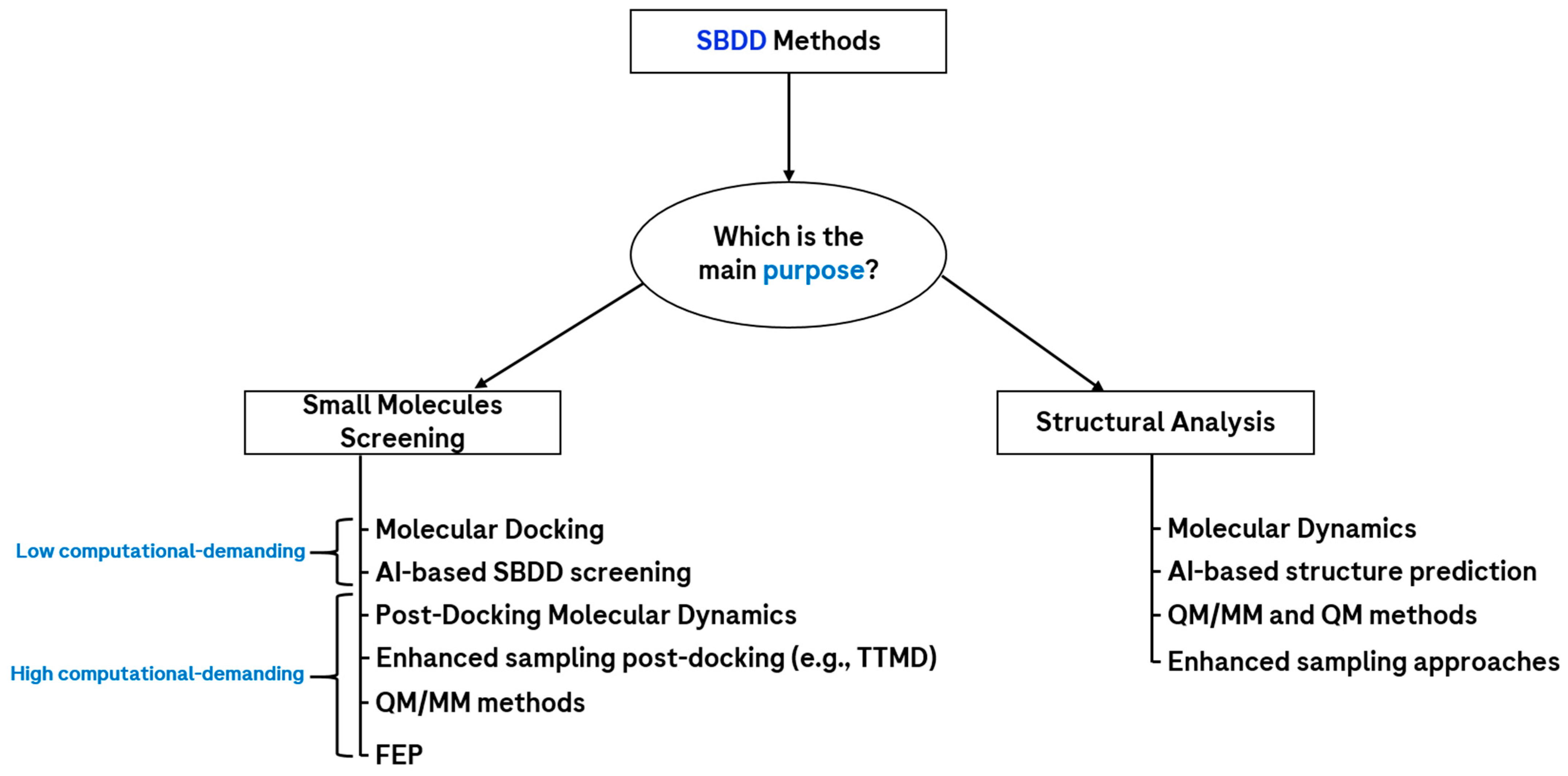
2.2. Structure-Based Drug Design (SBDD)
2.2.1. Molecular Docking
2.2.2. Molecular Dynamics
Enhanced Sampling Methods in Molecular Dynamics
Molecular Dynamics as a Post-Docking Approach
Free-Energy Perturbation (FEP) and Thermodynamic Integration (TI)
Thermal Titration Molecular Dynamics (TTMD)
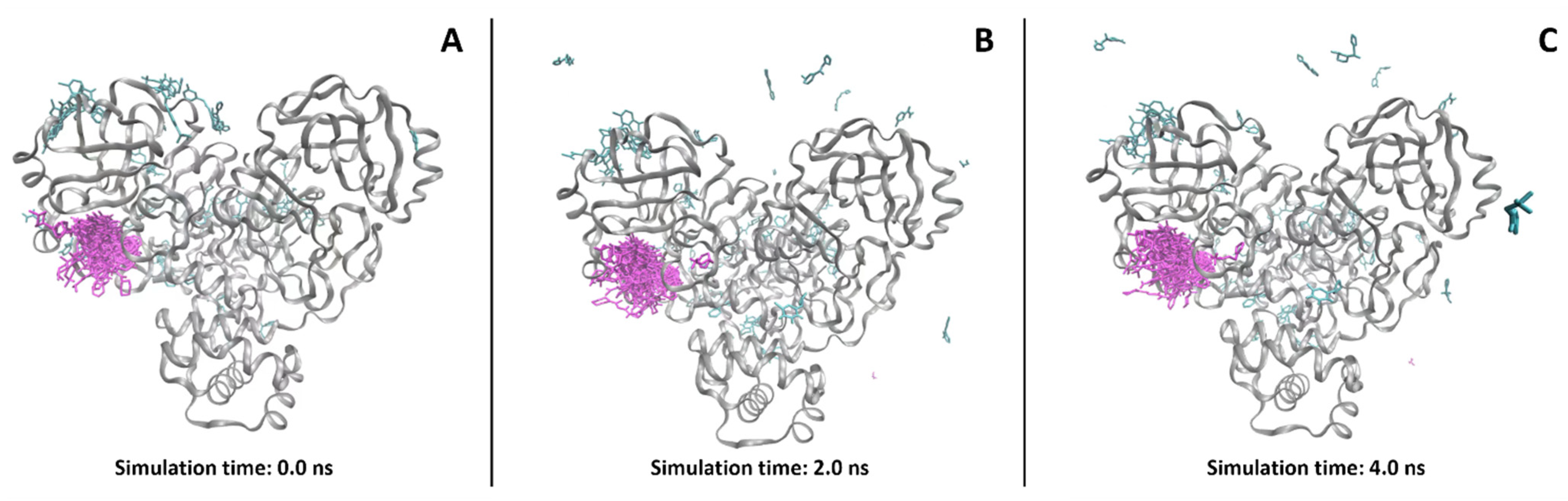
2.2.3. Supervised Molecular Dynamics
3. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martin, L.; Hutchens, M.; Hawkins, C. Clinical trial cycle times continue to increase despite industry efforts. Nat. Rev. Drug Discov. 2017, 16, 157. [Google Scholar] [CrossRef] [PubMed]
- Simoens, S.; Huys, I. R&D Costs of New Medicines: A Landscape Analysis. Front. Med. 2021, 8, 760762. [Google Scholar] [CrossRef]
- The Pharmaceutical Industry in Figures. Available online: https://www.efpia.eu/media/602709/the-pharmaceutical-industry-in-figures-2021.pdf (accessed on 2 March 2023).
- Bohacek, R.S.; McMartin, C.; Guida, W.C. The art and practice of structure-based drug design: A molecular modeling perspective. Med. Res. Rev. 1996, 16, 3–50. [Google Scholar] [CrossRef]
- Szymański, P.; Markowicz, M.; Mikiciuk-Olasik, E. Adaptation of High-Throughput Screening in Drug Discovery—Toxicological Screening Tests. Int. J. Mol. Sci. 2011, 13, 427–452. [Google Scholar] [CrossRef]
- Moro, S.; Bacilieri, M.; Deflorian, F. Combining ligand-based and structure-based drug design in the virtual screening arena. Expert Opin. Drug Discov. 2007, 2, 37–49. [Google Scholar] [CrossRef]
- Petrović, D.; Scott, J.S.; Bodnarchuk, M.S.; Lorthioir, O.; Boyd, S.; Hughes, G.M.; Lane, J.; Wu, A.; Hargreaves, D.; Robinson, J.; et al. Virtual Screening in the Cloud Identifies Potent and Selective ROS1 Kinase Inhibitors. J. Chem. Inf. Model. 2022, 62, 3832–3843. [Google Scholar] [CrossRef]
- Luttens, A.; Gullberg, H.; Abdurakhmanov, E.; Vo, D.D.; Akaberi, D.; Talibov, V.O.; Nekhotiaeva, N.; Vangeel, L.; De Jonghe, S.; Jochmans, D.; et al. Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses. J. Am. Chem. Soc. 2022, 144, 2905–2920. [Google Scholar] [CrossRef]
- Gorgulla, C.; Boeszoermenyi, A.; Wang, Z.-F.; Fischer, P.D.; Coote, P.W.; Das, K.M.P.; Malets, Y.S.; Radchenko, D.S.; Moroz, Y.S.; Scott, D.A.; et al. An open-source drug discovery platform enables ultra-large virtual screens. Nature 2020, 580, 663–668. [Google Scholar] [CrossRef]
- Gupta, G. Racing the Clock, COVID Killer Sought among a Billion Molecules. Available online: https://blogs.nvidia.com/blog/2020/05/26/covid-autodock-summit-ornl/ (accessed on 2 March 2023).
- LeGrand, S.; Scheinberg, A.; Tillack, A.F.; Thavappiragasam, M.; Vermaas, J.V.; Agarwal, R.; Larkin, J.; Poole, D.; Santos-Martins, D.; Solis-Vasquez, L.; et al. GPU-Accelerated Drug Discovery with Docking on the Summit Supercomputer. In Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, Virtual Event, 21–24 September 2020; pp. 1–10. [Google Scholar] [CrossRef]
- FDA. The Drug Development Process. Available online: https://www.fda.gov/patients/learn-about-drug-and-device-approvals/drug-development-process (accessed on 2 March 2023).
- Zhu, T.; Cao, S.; Su, P.-C.; Patel, R.; Shah, D.; Chokshi, H.B.; Szukala, R.; Johnson, M.E.; Hevener, K.E. Hit Identification and Optimization in Virtual Screening: Practical Recommendations Based on a Critical Literature Analysis. J. Med. Chem. 2013, 56, 6560–6572. [Google Scholar] [CrossRef]
- Hughes, J.P.; Rees, S.; Kalindjian, S.B.; Philpott, K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef]
- Keseru, G.M.; Makara, G.M. Hit discovery and hit-to-lead approaches. Drug Discov. Today 2006, 11, 741–748. [Google Scholar] [CrossRef]
- Leelananda, S.P.; Lindert, S. Computational methods in drug discovery. Beilstein J. Org. Chem. 2016, 12, 2694–2718. [Google Scholar] [CrossRef]
- Dauter, Z.; Wlodawer, A. Progress in protein crystallography. Protein Pept. Lett. 2016, 23, 201–210. [Google Scholar] [CrossRef]
- Benjin, X.; Ling, L. Developments, applications, and prospects of cryo-electron microscopy. Protein Sci. 2020, 29, 872–882. [Google Scholar] [CrossRef]
- Cavasotto, C.N.; Phatak, S.S. Homology modeling in drug discovery: Current trends and applications. Drug Discov. Today 2009, 14, 676–683. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Hekkelman, M.L.; de Vries, I.; Joosten, R.P.; Perrakis, A. AlphaFill: Enriching AlphaFold models with ligands and cofactors. Nat. Methods 2023, 20, 205–213. [Google Scholar] [CrossRef]
- David, A.; Islam, S.; Tankhilevich, E.; Sternberg, M.J. The AlphaFold Database of Protein Structures: A Biologist’s Guide. J. Mol. Biol. 2021, 434, 167336. [Google Scholar] [CrossRef]
- Cherkasov, A.; Muratov, E.N.; Fourches, D.; Varnek, A.; Baskin, I.; Cronin, M.; Dearden, J.; Gramatica, P.; Martin, Y.C.; Todeschini, R.; et al. QSAR Modeling: Where Have You Been? Where Are You Going to? J. Med. Chem. 2014, 57, 4977–5010. [Google Scholar] [CrossRef]
- Yang, S.-Y. Pharmacophore modeling and applications in drug discovery: Challenges and recent advances. Drug Discov. 2010, 15, 444–450. [Google Scholar] [CrossRef]
- Kramer, C.; Fuchs, J.E.; Whitebread, S.; Gedeck, P.; Liedl, K.R. Matched Molecular Pair Analysis: Significance and the Impact of Experimental Uncertainty. J. Med. Chem. 2014, 57, 3786–3802. [Google Scholar] [CrossRef] [PubMed]
- Zhong, F.; Xing, J.; Li, X.; Liu, X.; Fu, Z.; Xiong, Z.; Lu, D.; Wu, X.; Zhao, J.; Tan, X.; et al. Artificial intelligence in drug design. Sci. China Life Sci. 2018, 61, 1191–1204. [Google Scholar] [CrossRef] [PubMed]
- Mouchlis, V.D.; Afantitis, A.; Serra, A.; Fratello, M.; Papadiamantis, A.G.; Aidinis, V.; Lynch, I.; Greco, D.; Melagraki, G. Advances in De Novo Drug Design: From Conventional to Machine Learning Methods. Int. J. Mol. Sci. 2021, 22, 1676. [Google Scholar] [CrossRef]
- Begam, B.; Kumar, J.S. A Study on Cheminformatics and its Applications on Modern Drug Discovery. Procedia Eng. 2012, 38, 1264–1275. [Google Scholar] [CrossRef]
- Kubinyi, H. Free Wilson Analysis. Theory, Applications and its Relationship to Hansch Analysis. Quant. Struct. Relatsh. 1988, 7, 121–133. [Google Scholar] [CrossRef]
- Silakari, O.; Singh, P.K. QSAR: Descriptor calculations, model generation, validation and their application. In Concepts and Experimental Protocols of Modelling and Informatics in Drug Design; Elsevier: Amsterdam, The Netherlands, 2021; pp. 29–63. [Google Scholar] [CrossRef]
- Ragno, R.; Esposito, V.; Di Mario, M.; Masiello, S.; Viscovo, M.; Cramer, R.D. Teaching and Learning Computational Drug Design: Student Investigations of 3D Quantitative Structure–Activity Relationships through Web Applications. J. Chem. Educ. 2020, 97, 1922–1930. [Google Scholar] [CrossRef]
- Dearden, J.C. The History and Development of Quantitative Structure-Activity Relationships (QSARs). Int. J. Quant. Struct. Relatsh. 2016, 1, 1–44. [Google Scholar] [CrossRef]
- TSoares, T.A.; Nunes-Alves, A.; Mazzolari, A.; Ruggiu, F.; Wei, G.-W.; Merz, K. The (Re)-Evolution of Quantitative Structure–Activity Relationship (QSAR) Studies Propelled by the Surge of Machine Learning Methods. J. Chem. Inf. Model. 2022, 62, 5317–5320. [Google Scholar] [CrossRef]
- Golbraikh, A.; Wang, X.S.; Zhu, H.; Tropsha, A. Predictive QSAR Modeling: Methods and Applications in Drug Discovery and Chemical Risk Assessment. In Handbook of Computational Chemistry; Springer: Dordrecht, The Netherlands, 2012; pp. 1309–1342. [Google Scholar] [CrossRef]
- Verma, J.; Khedkar, V.M.; Coutinho, E.C. 3D-QSAR in Drug Design-A Review. Curr. Top. Med. Chem. 2010, 10, 95–115. [Google Scholar] [CrossRef]
- Gasteiger, J.; Engel, T. Chemoinformatics: A Textbook; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2003. [Google Scholar]
- Muchmore, S.W.; Edmunds, J.J.; Stewart, K.D.; Hajduk, P.J. Cheminformatic Tools for Medicinal Chemists. J. Med. Chem. 2010, 53, 4830–4841. [Google Scholar] [CrossRef]
- Landrum, G. RDKit: Open-Source Cheminformatics. 2010. Available online: https://www.rdkit.org/ (accessed on 2 March 2023).
- Dalke, A.; Hert, J.; Kramer, C. mmpdb: An Open-Source Matched Molecular Pair Platform for Large Multiproperty Data Sets. J. Chem. Inf. Model. 2018, 58, 902–910. [Google Scholar] [CrossRef] [PubMed]
- Bolcato, G.; Heid, E.; Boström, J. On the Value of Using 3D Shape and Electrostatic Similarities in Deep Generative Methods. J. Chem. Inf. Model. 2022, 62, 1388–1398. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, J.O.; Durrant, J.D. AutoGrow4: An open-source genetic algorithm for de novo drug design and lead optimization. J. Cheminform. 2020, 12, 25. [Google Scholar] [CrossRef]
- Riniker, S.; Landrum, G.A. Similarity maps-a visualization strategy for molecular fingerprints and machine-learning methods. J. Cheminform. 2013, 5, 43. [Google Scholar] [CrossRef]
- Daré, J.K.; Freitas, M.P. Is conformation relevant for QSAR purposes? 2D Chemical representation in a 3D-QSAR perspective. J. Comput. Chem. 2022, 43, 917–922. [Google Scholar] [CrossRef]
- Nikonenko, A.; Zankov, D.; Baskin, I.; Madzhidov, T.; Polishchuk, P. Multiple Conformer Descriptors for QSAR Modeling. Mol. Inform. 2021, 40, 2060030. [Google Scholar] [CrossRef]
- Günther, S.; Senger, C.; Michalsky, E.; Goede, A.; Preissner, R. Representation of target-bound drugs by computed conformers: Implications for conformational libraries. BMC Bioinform. 2006, 7, 293. [Google Scholar] [CrossRef]
- Bosc, N.; Atkinson, F.; Felix, E.; Gaulton, A.; Hersey, A.; Leach, A.R. Large scale comparison of QSAR and conformal prediction methods and their applications in drug discovery. J. Cheminform. 2019, 11, 4. [Google Scholar] [CrossRef]
- Neves, B.J.; Braga, R.C.; Melo-Filho, C.C.; Moreira-Filho, J.T.; Muratov, E.N.; Andrade, C.H. QSAR-Based Virtual Screening: Advances and Applications in Drug Discovery. Front. Pharmacol. 2018, 9, 1275. [Google Scholar] [CrossRef]
- Kwon, S.; Bae, H.; Jo, J.; Yoon, S. Comprehensive ensemble in QSAR prediction for drug discovery. BMC Bioinform. 2019, 20, 521. [Google Scholar] [CrossRef]
- Anderson, A.C. The Process of Structure-Based Drug Design. Chem. Biol. 2003, 10, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Pavan, M.; Bassani, D.; Sturlese, M.; Moro, S. From the Wuhan-Hu-1 strain to the XD and XE variants: Is targeting the SARS-CoV-2 spike protein still a pharmaceutically relevant option against COVID-19? J. Enzyme Inhib. Med. Chem. 2022, 37, 1704–1714. [Google Scholar] [CrossRef]
- Kuntz, I.D.; Blaney, J.M.; Oatley, S.J.; Langridge, R.; Ferrin, T.E. A geometric approach to macromolecule-ligand interactions. J. Mol. Biol. 1982, 161, 269–288. [Google Scholar] [CrossRef]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Curr. Comput. Aided-Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef]
- Pavan, M.; Menin, S.; Bassani, D.; Sturlese, M.; Moro, S. Implementing a Scoring Function Based on Interaction Fingerprint for Autogrow4: Protein Kinase CK1δ as a Case Study. Front. Mol. Biosci. 2022, 9, 909499. [Google Scholar] [CrossRef]
- Liu, J.; Wang, R. Classification of Current Scoring Functions. J. Chem. Inf. Model. 2015, 55, 475–482. [Google Scholar] [CrossRef]
- Guedes, I.A.; Pereira, F.S.S.; Dardenne, L.E. Empirical Scoring Functions for Structure-Based Virtual Screening: Applications, Critical Aspects, and Challenges. Front. Pharmacol. 2018, 9, 1089. [Google Scholar] [CrossRef]
- Shen, Q.; Xiong, B.; Zheng, M.; Luo, X.; Luo, C.; Liu, X.; Du, Y.; Li, J.; Zhu, W.; Shen, J.; et al. Knowledge-Based Scoring Functions in Drug Design: 2. Can the Knowledge Base Be Enriched? J. Chem. Inf. Model. 2010, 51, 386–397. [Google Scholar] [CrossRef]
- Li, J.; Fu, A.; Zhang, L. An Overview of Scoring Functions Used for Protein–Ligand Interactions in Molecular Docking. Interdiscip. Sci. Comput. Life Sci. 2019, 11, 320–328. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking 1 1Edited by F. E. Cohen. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Korb, O.; Stützle, T.; Exner, T.E. PLANTS: Application of Ant Colony Optimization to Structure-Based Drug Design. In Ant Colony Optimization and Swarm Intelligence; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4150, pp. 247–258. [Google Scholar] [CrossRef]
- Pecoraro, C.; De Franco, M.; Carbone, D.; Bassani, D.; Pavan, M.; Cascioferro, S.; Parrino, B.; Cirrincione, G.; Dall’acqua, S.; Moro, S.; et al. 1,2,4-Amino-triazine derivatives as pyruvate dehydrogenase kinase inhibitors: Synthesis and pharmacological evaluation. Eur. J. Med. Chem. 2023, 249, 115134. [Google Scholar] [CrossRef] [PubMed]
- Torres, P.H.M.; Sodero, A.C.R.; Jofily, P.; Silva, F.P., Jr. Key Topics in Molecular Docking for Drug Design. Int. J. Mol. Sci. 2019, 20, 4574. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Cosconati, S.; Forli, S.; Perryman, A.L.; Harris, R.; Goodsell, D.S.; Olson, A.J. Virtual screening with AutoDock: Theory and practice. Expert Opin. Drug Discov. 2010, 5, 597–607. [Google Scholar] [CrossRef]
- Huang, S.-Y. Comprehensive assessment of flexible-ligand docking algorithms: Current effectiveness and challenges. Brief. Bioinform. 2018, 19, 982–994. [Google Scholar] [CrossRef]
- Sotriffer, C.A. Accounting for Induced-Fit Effects in Docking: What is Possible and What is Not? Curr. Top. Med. Chem. 2011, 11, 179–191. [Google Scholar] [CrossRef]
- Amaro, R.E.; Baudry, J.; Chodera, J.; Demir, Ö.; McCammon, J.A.; Miao, Y.; Smith, J.C. Ensemble Docking in Drug Discovery. Biophys. J. 2018, 114, 2271–2278. [Google Scholar] [CrossRef]
- Spinaci, A.; Buccioni, M.; Catarzi, D.; Cui, C.; Colotta, V.; Ben, D.D.; Cescon, E.; Francucci, B.; Grieco, I.; Lambertucci, C.; et al. Dual Anta-Inhibitors’ of the A2A Adenosine Receptor and Casein Kinase CK1delta: Synthesis, Biological Evaluation, and Molecular Modeling Studies. Pharmaceuticals 2023, 16, 167. [Google Scholar] [CrossRef]
- Sartore, G.; Bassani, D.; Ragazzi, E.; Traldi, P.; Lapolla, A.; Moro, S. In silico evaluation of the interaction between ACE2 and SARS-CoV-2 Spike protein in a hyperglycemic environment. Sci. Rep. 2021, 11, 22860. [Google Scholar] [CrossRef]
- Roberts, B.C.; Mancera, R.L. Ligand−Protein Docking with Water Molecules. J. Chem. Inf. Model. 2008, 48, 397–408. [Google Scholar] [CrossRef]
- Deng, N.; Forli, S.; He, P.; Perryman, A.; Wickstrom, L.; Vijayan, R.S.K.; Tiefenbrunn, T.; Stout, D.; Gallicchio, E.; Olson, A.J.; et al. Distinguishing Binders from False Positives by Free Energy Calculations: Fragment Screening Against the Flap Site of HIV Protease. J. Phys. Chem. B 2015, 119, 976–988. [Google Scholar] [CrossRef]
- Poli, G.; Tuccinardi, T. Consensus Docking in Drug Discovery. Curr. Bioact. Compd. 2020, 16, 182–190. [Google Scholar] [CrossRef]
- Houston, D.R.; Walkinshaw, M.D. Consensus Docking: Improving the Reliability of Docking in a Virtual Screening Context. J. Chem. Inf. Model. 2013, 53, 384–390. [Google Scholar] [CrossRef]
- Bolcato, G.; Cescon, E.; Pavan, M.; Bissaro, M.; Bassani, D.; Federico, S.; Spalluto, G.; Sturlese, M.; Moro, S. A Computational Workflow for the Identification of Novel Fragments Acting as Inhibitors of the Activity of Protein Kinase CK1δ. Int. J. Mol. Sci. 2021, 22, 9741. [Google Scholar] [CrossRef]
- Rastelli, G.; Pinzi, L. Refinement and Rescoring of Virtual Screening Results. Front. Chem. 2019, 7, 498. [Google Scholar] [CrossRef]
- Peach, M.L.; Nicklaus, M.C. Combining docking with pharmacophore filtering for improved virtual screening. J. Cheminform. 2009, 1, 6. [Google Scholar] [CrossRef]
- Shivanika, C.; Kumar, D.; Ragunathan, V.; Tiwari, P.; Sumitha, A. Molecular docking, validation, dynamics simulations, and pharmacokinetic prediction of natural compounds against the SARS-CoV-2 main-protease. J. Biomol. Struct. Dyn. 2022, 40, 585–611. [Google Scholar] [CrossRef]
- Pavan, M.; Menin, S.; Bassani, D.; Sturlese, M.; Moro, S. Qualitative Estimation of Protein–Ligand Complex Stability through Thermal Titration Molecular Dynamics Simulations. J. Chem. Inf. Model. 2022, 62, 5715–5728. [Google Scholar] [CrossRef]
- Menin, S.; Pavan, M.; Salmaso, V.; Sturlese, M.; Moro, S. Thermal Titration Molecular Dynamics (TTMD): Not Your Usual Post-Docking Refinement. Int. J. Mol. Sci. 2023, 24, 3596. [Google Scholar] [CrossRef] [PubMed]
- Pavan, M.; Bassani, D.; Bolcato, G.; Bissaro, M.; Sturlese, M.; Moro, S. Computational Strategies to Identify New Drug Candidates against Neuroinflammation. Curr. Med. Chem. 2022, 29, 4756–4775. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, S.A.; Dror, R.O. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef] [PubMed]
- Pavan, M.; Bassani, D.; Sturlese, M.; Moro, S. Investigating RNA–protein recognition mechanisms through supervised molecular dynamics (SuMD) simulations. NAR Genom. Bioinform. 2022, 4, lqac088. [Google Scholar] [CrossRef] [PubMed]
- De Vivo, M.; Masetti, M.; Bottegoni, G.; Cavalli, A. Role of Molecular Dynamics and Related Methods in Drug Discovery. J. Med. Chem. 2016, 59, 4035–4061. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Durrant, J.D.; McCammon, J.A. Molecular dynamics simulations and drug discovery. BMC Biol. 2011, 9, 71. [Google Scholar] [CrossRef]
- Tzeliou, C.E.; Mermigki, M.A.; Tzeli, D. Review on the QM/MM Methodologies and Their Application to Metalloproteins. Molecules 2022, 27, 2660. [Google Scholar] [CrossRef]
- Gorgulla, C.; Jayaraj, A.; Fackeldey, K.; Arthanari, H. Emerging frontiers in virtual drug discovery: From quantum mechanical methods to deep learning approaches. Curr. Opin. Chem. Biol. 2022, 69, 102156. [Google Scholar] [CrossRef]
- Bassani, D.; Pavan, M.; Bolcato, G.; Sturlese, M.; Moro, S. Re-Exploring the Ability of Common Docking Programs to Correctly Reproduce the Binding Modes of Non-Covalent Inhibitors of SARS-CoV-2 Protease Mpro. Pharmaceuticals 2022, 15, 180. [Google Scholar] [CrossRef]
- Lotz, S.D.; Dickson, A. Unbiased Molecular Dynamics of 11 min Timescale Drug Unbinding Reveals Transition State Stabilizing Interactions. J. Am. Chem. Soc. 2018, 140, 618–628. [Google Scholar] [CrossRef]
- Shaw, D.E.; Adams, P.J.; Azaria, A.; Bank, J.A.; Batson, B.; Bell, A.; Bergdorf, M.; Bhatt, J.; Butts, J.A.; Correia, T.; et al. Anton 3. In Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis, St. Louis, MO, USA, 14–19 November 2021; pp. 1–11. [Google Scholar] [CrossRef]
- Hartmann, C.; Banisch, R.; Sarich, M.; Badowski, T.; Schütte, C. Characterization of Rare Events in Molecular Dynamics. Entropy 2013, 16, 350–376. [Google Scholar] [CrossRef]
- Lazim, R.; Suh, D.; Choi, S. Advances in Molecular Dynamics Simulations and Enhanced Sampling Methods for the Study of Protein Systems. Int. J. Mol. Sci. 2020, 21, 6339. [Google Scholar] [CrossRef]
- Patel, J.S.; Berteotti, A.; Ronsisvalle, S.; Rocchia, W.; Cavalli, A. Steered Molecular Dynamics Simulations for Studying Protein–Ligand Interaction in Cyclin-Dependent Kinase 5. J. Chem. Inf. Model. 2014, 54, 470–480. [Google Scholar] [CrossRef]
- Sinko, W.; Miao, Y.; de Oliveira, C.A.F.; McCammon, J.A. Population Based Reweighting of Scaled Molecular Dynamics. J. Phys. Chem. B 2013, 117, 12759–12768. [Google Scholar] [CrossRef]
- Sugita, Y.; Okamoto, Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999, 314, 141–151. [Google Scholar] [CrossRef]
- Bussi, G.; Laio, A. Using metadynamics to explore complex free-energy landscapes. Nat. Rev. Phys. 2020, 2, 200–212. [Google Scholar] [CrossRef]
- Miao, Y.; Feher, V.A.; McCammon, J.A. Gaussian Accelerated Molecular Dynamics: Unconstrained Enhanced Sampling and Free Energy Calculation. J. Chem. Theory Comput. 2015, 11, 3584–3595. [Google Scholar] [CrossRef]
- Yu, Z.; Su, H.; Chen, J.; Hu, G. Deciphering Conformational Changes of the GDP-Bound NRAS Induced by Mutations G13D, Q61R, and C118S through Gaussian Accelerated Molecular Dynamic Simulations. Molecules 2022, 27, 5596. [Google Scholar] [CrossRef]
- Chen, J.; Zeng, Q.; Wang, W.; Sun, H.; Hu, G. Decoding the Identification Mechanism of an SAM-III Riboswitch on Ligands through Multiple Independent Gaussian-Accelerated Molecular Dynamics Simulations. J. Chem. Inf. Model. 2022, 62, 6118–6132. [Google Scholar] [CrossRef]
- Sabbadin, D.; Moro, S. Supervised Molecular Dynamics (SuMD) as a Helpful Tool To Depict GPCR–Ligand Recognition Pathway in a Nanosecond Time Scale. J. Chem. Inf. Model. 2014, 54, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Alonso, H.; Bliznyuk, A.A.; Gready, J.E. Combining docking and molecular dynamic simulations in drug design. Med. Res. Rev. 2006, 26, 531–568. [Google Scholar] [CrossRef] [PubMed]
- Fratev, F.; Sirimulla, S. An Improved Free Energy Perturbation FEP+ Sampling Protocol for Flexible Ligand-Binding Domains. Sci. Rep. 2019, 9, 16829. [Google Scholar] [CrossRef] [PubMed]
- Cournia, Z.; Allen, B.; Sherman, W. Relative Binding Free Energy Calculations in Drug Discovery: Recent Advances and Practical Considerations. J. Chem. Inf. Model. 2017, 57, 2911–2937. [Google Scholar] [CrossRef] [PubMed]
- Lovering, F.; Aevazelis, C.; Chang, J.; Dehnhardt, C.; Fitz, L.; Han, S.; Janz, K.; Lee, J.; Kaila, N.; McDonald, J.; et al. Imidazotriazines: Spleen Tyrosine Kinase (Syk) Inhibitors Identified by Free-Energy Perturbation (FEP). ChemMedChem 2016, 11, 217–233. [Google Scholar] [CrossRef]
- Ngo, S.T.; Nguyen, H.M.; Huong, L.T.T.; Quan, P.M.; Truong, V.K.; Tung, N.T.; Vu, V.V. Assessing potential inhibitors of SARS-CoV-2 main protease from available drugs using free energy perturbation simulations. RSC Adv. 2020, 10, 40284–40290. [Google Scholar] [CrossRef]
- Deflorian, F.; Perez-Benito, L.; Lenselink, E.B.; Congreve, M.; Van Vlijmen, H.W.T.; Mason, J.S.; De Graaf, C.; Tresadern, G. Accurate Prediction of GPCR Ligand Binding Affinity with Free Energy Perturbation. J. Chem. Inf. Model. 2020, 60, 5563–5579. [Google Scholar] [CrossRef]
- Gapsys, V.; Yildirim, A.; Aldeghi, M.; Khalak, Y.; van der Spoel, D.; de Groot, B.L. Accurate absolute free energies for ligand–protein binding based on non-equilibrium approaches. Commun. Chem. 2021, 4, 61. [Google Scholar] [CrossRef]
- Azimi, S.; Khuttan, S.; Wu, J.Z.; Pal, R.K.; Gallicchio, E. Relative Binding Free Energy Calculations for Ligands with Diverse Scaffolds with the Alchemical Transfer Method. J. Chem. Inf. Model. 2022, 62, 309–323. [Google Scholar] [CrossRef]
- Wang, L.; Wu, Y.; Deng, Y.; Kim, B.; Pierce, L.; Krilov, G.; Lupyan, D.; Robinson, S.; Dahlgren, M.K.; Greenwood, J.; et al. Accurate and Reliable Prediction of Relative Ligand Binding Potency in Prospective Drug Discovery by Way of a Modern Free-Energy Calculation Protocol and Force Field. J. Am. Chem. Soc. 2015, 137, 2695–2703. [Google Scholar] [CrossRef]
- Abel, R.; Wang, L.; Harder, E.D.; Berne, B.J.; Friesner, R.A. Advancing Drug Discovery through Enhanced Free Energy Calculations. Acc. Chem. Res. 2017, 50, 1625–1632. [Google Scholar] [CrossRef]
- LMartins, L.C.; Cino, E.A.; Ferreira, R.S. PyAutoFEP: An Automated Free Energy Perturbation Workflow for GROMACS Integrating Enhanced Sampling Methods. J. Chem. Theory Comput. 2021, 17, 4262–4273. [Google Scholar] [CrossRef]
- Mey, A.S.J.S.; Allen, B.K.; Macdonald, H.E.B.; Chodera, J.D.; Hahn, D.F.; Kuhn, M.; Michel, J.; Mobley, D.L.; Naden, L.N.; Prasad, S.; et al. Best Practices for Alchemical Free Energy Calculations [Article v1.0]. Living J. Comput. Mol. Sci. 2020, 2, 18378. [Google Scholar] [CrossRef]
- Wu, D.; Zheng, X.; Liu, R.; Li, Z.; Jiang, Z.; Zhou, Q.; Huang, Y.; Wu, X.-N.; Zhang, C.; Huang, Y.-Y.; et al. Free energy perturbation (FEP)-guided scaffold hopping. Acta Pharm. Sin. B 2021, 12, 1351–1362. [Google Scholar] [CrossRef]
- Steinbrecher, T.B.; Dahlgren, M.; Cappel, D.; Lin, T.; Wang, L.; Krilov, G.; Abel, R.; Friesner, R.; Sherman, W. Accurate Binding Free Energy Predictions in Fragment Optimization. J. Chem. Inf. Model. 2015, 55, 2411–2420. [Google Scholar] [CrossRef]
- Resat, H.; Mezei, M. Studies on free energy calculations. I. Thermodynamic integration using a polynomial path. J. Chem. Phys. 1993, 99, 6052–6061. [Google Scholar] [CrossRef]
- Bruckner, S.; Boresch, S. Efficiency of alchemical free energy simulations. II. Improvements for thermodynamic integration. J. Comput. Chem. 2010, 32, 1320–1333. [Google Scholar] [CrossRef]
- Zhang, Q.; Yang, Y.; Gong, X.; Zhao, N.; Zhang, Y.; Liu, H. Thermodynamic integration combined with molecular dynamic simulations to explore the cross-resistance mechanism of isoniazid and ethionamide. Proteins Struct. Funct. Bioinform. 2022, 90, 1142–1151. [Google Scholar] [CrossRef]
- Mishra, S.K.; Calabró, G.; Loeffler, H.H.; Michel, J.; Koča, J. Evaluation of Selected Classical Force Fields for Alchemical Binding Free Energy Calculations of Protein-Carbohydrate Complexes. J. Chem. Theory Comput. 2015, 11, 3333–3345. [Google Scholar] [CrossRef]
- Huai, Z.; Shen, Z.; Sun, Z. Binding Thermodynamics and Interaction Patterns of Inhibitor-Major Urinary Protein-I Binding from Extensive Free-Energy Calculations: Benchmarking AMBER Force Fields. J. Chem. Inf. Model. 2020, 61, 284–297. [Google Scholar] [CrossRef]
- Christ, C.D.; Fox, T. Accuracy Assessment and Automation of Free Energy Calculations for Drug Design. J. Chem. Inf. Model. 2014, 54, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Garbett, N.C.; Chaires, J.B. Thermodynamic studies for drug design and screening. Expert Opin. Drug Discov. 2012, 7, 299–314. [Google Scholar] [CrossRef] [PubMed]
- Decherchi, S.; Cavalli, A. Thermodynamics and Kinetics of Drug-Target Binding by Molecular Simulation. Chem. Rev. 2020, 120, 12788–12833. [Google Scholar] [CrossRef] [PubMed]
- Endter, L.J.; Smirnova, Y.; Risselada, H.J. Density Field Thermodynamic Integration (DFTI): A ‘Soft’ Approach to Calculate the Free Energy of Surfactant Self-Assemblies. J. Phys. Chem. B 2020, 124, 6775–6785. [Google Scholar] [CrossRef]
- Kaästner, J.; Thiel, W. Bridging the gap between thermodynamic integration and umbrella sampling provides a novel analysis method: “Umbrella integration”. J. Chem. Phys. 2005, 123, 144104. [Google Scholar] [CrossRef]
- Pan, A.C.; Xu, H.; Palpant, T.; Shaw, D.E. Quantitative Characterization of the Binding and Unbinding of Millimolar Drug Fragments with Molecular Dynamics Simulations. J. Chem. Theory Comput. 2017, 13, 3372–3377. [Google Scholar] [CrossRef]
- ALong, A.; Zhao, H.; Huang, X. Structural Basis for the Interaction between Casein Kinase 1 Delta and a Potent and Selective Inhibitor. J. Med. Chem. 2012, 55, 956–960. [Google Scholar] [CrossRef]
- Ursu, A.; Illich, D.J.; Takemoto, Y.; Porfetye, A.T.; Zhang, M.; Brockmeyer, A.; Janning, P.; Watanabe, N.; Osada, H.; Vetter, I.R.; et al. Epiblastin A Induces Reprogramming of Epiblast Stem Cells Into Embryonic Stem Cells by Inhibition of Casein Kinase 1. Cell Chem. Biol. 2016, 23, 494–507. [Google Scholar] [CrossRef]
- Husic, B.E.; Pande, V.S. Markov State Models: From an Art to a Science. J. Am. Chem. Soc. 2018, 140, 2386–2396. [Google Scholar] [CrossRef]
- Bernardi, R.C.; Melo, M.C.; Schulten, K. Enhanced sampling techniques in molecular dynamics simulations of biological systems. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2015, 1850, 872–877. [Google Scholar] [CrossRef]
- Cuzzolin, A.; Sturlese, M.; Deganutti, G.; Salmaso, V.; Sabbadin, D.; Ciancetta, A.; Moro, S. Deciphering the Complexity of Ligand-Protein Recognition Pathways Using Supervised Molecular Dynamics (SuMD) Simulations. J. Chem. Inf. Model. 2016, 56, 687–705. [Google Scholar] [CrossRef]
- Sabbadin, D.; Ciancetta, A.; Deganutti, G.; Cuzzolin, A.; Moro, S. Exploring the recognition pathway at the human A 2A adenosine receptor of the endogenous agonist adenosine using supervised molecular dynamics simulations. Medchemcomm 2015, 6, 1081–1085. [Google Scholar] [CrossRef]
- Bolcato, G.; Pavan, M.; Bassani, D.; Sturlese, M.; Moro, S. Ribose and Non-Ribose A2A Adenosine Receptor Agonists: Do They Share the Same Receptor Recognition Mechanism? Biomedicines 2022, 10, 515. [Google Scholar] [CrossRef]
- Pavan, M.; Bolcato, G.; Bassani, D.; Sturlese, M.; Moro, S. Supervised Molecular Dynamics (SuMD) Insights into the mechanism of action of SARS-CoV-2 main protease inhibitor PF-07321332. J. Enzyme Inhib. Med. Chem. 2021, 36, 1645–1649. [Google Scholar] [CrossRef]
- Panday, S.K.; Sturlese, M.; Salmaso, V.; Ghosh, I.; Moro, S. Coupling Supervised Molecular Dynamics (SuMD) with Entropy Estimations To Shine Light on the Stability of Multiple Binding Sites. ACS Med. Chem. Lett. 2019, 10, 444–449. [Google Scholar] [CrossRef]
- Salmaso, V.; Sturlese, M.; Cuzzolin, A.; Moro, S. Exploring Protein-Peptide Recognition Pathways Using a Supervised Molecular Dynamics Approach. Structure 2017, 25, 655–662.e2. [Google Scholar] [CrossRef]
- Jayatunga, M.K.; Xie, W.; Ruder, L.; Schulze, U.; Meier, C. AI in small-molecule drug discovery: A coming wave? Nat. Rev. Drug Discov. 2022, 21, 175–176. [Google Scholar] [CrossRef]
- Boniolo, F.; Dorigatti, E.; Ohnmacht, A.J.; Saur, D.; Schubert, B.; Menden, M.P. Artificial intelligence in early drug discovery enabling precision medicine. Expert Opin. Drug Discov. 2021, 16, 991–1007. [Google Scholar] [CrossRef]
- Cavasotto, C.N.; Di Filippo, J.I. Artificial intelligence in the early stages of drug discovery. Arch. Biochem. Biophys. 2020, 698, 108730. [Google Scholar] [CrossRef]
- Füzi, B.; Mathai, N.; Kirchmair, J.; Ecker, G.F. Toxicity prediction using target, interactome, and pathway profiles as descriptors. Toxicol. Lett. 2023, 381, 20–26. [Google Scholar] [CrossRef]
- Lysenko, A.; Sharma, A.; Boroevich, K.A.; Tsunoda, T. An integrative machine learning approach for prediction of toxicity-related drug safety. Life Sci. Alliance 2018, 1, e201800098. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Moore, J.H. TargetTox: A Feature Selection Pipeline for Identifying Predictive Targets Associated with Drug Toxicity. J. Chem. Inf. Model. 2021, 61, 5386–5394. [Google Scholar] [CrossRef] [PubMed]
- Atz, K.; Isert, C.; Böcker, M.N.A.; Jiménez-Luna, J.; Schneider, G. Δ-Quantum machine-learning for medicinal chemistry. Phys. Chem. Chem. Phys. 2022, 24, 10775–10783. [Google Scholar] [CrossRef] [PubMed]
- Pu, L.; Govindaraj, R.G.; Lemoine, J.M.; Wu, H.-C.; Brylinski, M. DeepDrug3D: Classification of ligand-binding pockets in proteins with a convolutional neural network. PLoS Comput. Biol. 2019, 15, e1006718. [Google Scholar] [CrossRef]
- Ain, Q.U.; Aleksandrova, A.; Roessler, F.D.; Ballester, P.J. Machine-learning scoring functions to improve structure-based binding affinity prediction and virtual screening. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2015, 5, 405–424. [Google Scholar] [CrossRef]
- Graff, D.E.; Shakhnovich, E.I.; Coley, C.W. Accelerating high-throughput virtual screening through molecular pool-based active learning. Chem. Sci. 2021, 12, 7866–7881. [Google Scholar] [CrossRef]
- Pozzan, A. QM Calculations in ADMET Prediction. Quantum Mech. Drug Discov. 2020, 2114, 285–305. [Google Scholar] [CrossRef]
- Isert, C.; Atz, K.; Jiménez-Luna, J.; Schneider, G. QMugs, quantum mechanical properties of drug-like molecules. Sci. Data 2022, 9, 273. [Google Scholar] [CrossRef]
- Böselt, L.; Thürlemann, M.; Riniker, S. Machine Learning in QM/MM Molecular Dynamics Simulations of Condensed-Phase Systems. J. Chem. Theory Comput. 2021, 17, 2641–2658. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bassani, D.; Moro, S. Past, Present, and Future Perspectives on Computer-Aided Drug Design Methodologies. Molecules 2023, 28, 3906. https://doi.org/10.3390/molecules28093906
Bassani D, Moro S. Past, Present, and Future Perspectives on Computer-Aided Drug Design Methodologies. Molecules. 2023; 28(9):3906. https://doi.org/10.3390/molecules28093906
Chicago/Turabian StyleBassani, Davide, and Stefano Moro. 2023. "Past, Present, and Future Perspectives on Computer-Aided Drug Design Methodologies" Molecules 28, no. 9: 3906. https://doi.org/10.3390/molecules28093906
APA StyleBassani, D., & Moro, S. (2023). Past, Present, and Future Perspectives on Computer-Aided Drug Design Methodologies. Molecules, 28(9), 3906. https://doi.org/10.3390/molecules28093906






