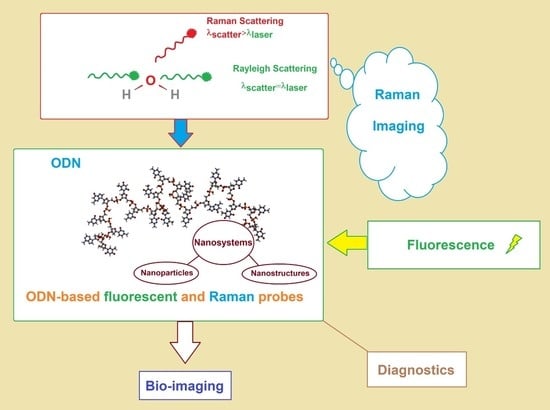
Nucleic Acid Probes in Bio-Imaging and Diagnostics: Recent Advances in ODN-Based Fluorescent and Surface-Enhanced Raman Scattering Nanoparticle and Nanostructured Systems
Abstract
1. Introduction
2. Nanoparticle-Based ODN Conjugates for Fluorescent Bio-Imaging
2.1. Gold ODN-Conjugated Nanoparticles for Fluorescent Bio-Imaging
2.2. Hybrid Gold-Inorganic ODN-Conjugated Nanoparticles for Fluorescent Bio-Imaging
2.3. Gold-Free ODN-Conjugated Nanoparticles
3. Self-Assembled ODN-Nanostructure Conjugates for Fluorescent Bio-Imaging
4. ODN-Nanostructure Conjugates for Raman Bio-Imaging
5. ODN-Nanostructure Conjugates for Dual Fluorescence–Raman Bio-Imaging and Detection
6. Conclusions and Perspectives
Author Contributions
Funding
 .
.Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, Y.; Du, Y.; Zhuo, Y.; Qiu, L. Functional Nucleic Acid-Based Live-Cell Fluorescence Imaging. Front. Chem. 2020, 8, 598013. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, A. Next-Generation Fluorescent Nucleic Acids Probes for Microscopic Analysis of Intracellular Nucleic Acids. Appl. Microsc. 2019, 49, 14. [Google Scholar] [CrossRef]
- Cruz Da Silva, E.; Foppolo, S.; Lhermitte, B.; Ingremeau, M.; Justiniano, H.; Klein, L.; Chenard, M.-P.; Vauchelles, R.; Abdallah, B.; Lehmann, M.; et al. Bioimaging Nucleic-Acid Aptamers with Different Specificities in Human Glioblastoma Tissues Highlights Tumoral Heterogeneity. Pharmaceutics 2022, 14, 1980. [Google Scholar] [CrossRef]
- Ebrahimi, S.B.; Samanta, D.; Mirkin, C.A. DNA-Based Nanostructures for Live-Cell Analysis. J. Am. Chem. Soc. 2020, 142, 11343–11356. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Mei, J.; Pan, S.; Xu, J.; Gu, T.; Li, Q.; Fan, X.; Li, Z. Chapter 7—Raman Spectroscopy to Study Biomolecules, Their Structure, and Dynamics. In Advanced Spectroscopic Methods to Study Biomolecular Structure and Dynamics; Saudagar, P., Tripathi, T., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 173–210. ISBN 978-0-323-99127-8. [Google Scholar]
- Samuel, A.Z.; Sugiyama, K.; Takeyama, H. Direct Intracellular Detection of Biomolecule Specific Bound-Water with Raman Spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 285, 121870. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Avni, A.; Joshi, A.; Walimbe, A.; Pattanashetty, S.G. A Deep Dive into Biomolecular Condensates Using Single-Droplet Surface-Enhanced Raman Spectroscopy. Biophys. J. 2023, 122, 60a. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; He, Y.; Liu, L.; Wang, X.; Jiang, S.; Yang, N.; Shi, N.; Li, Y. A Versatile Technique for Indiscriminate Detection of Unlabeled Biomolecules via Double-Enhanced Raman Scattering. Int. J. Biol. Macromol. 2023, 228, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Pan, R.; He, W.; Li, L.; Yang, Y.; Du, Z.; Luan, Z.; Zhang, X. In Situ Surface-Enhanced Raman Scattering Detection of Biomolecules in the Deep Ocean. Appl. Surf. Sci. 2023, 620, 156854. [Google Scholar] [CrossRef]
- Qiu, C.; Zhang, W.; Zhou, Y.; Cui, H.; Xing, Y.; Yu, F.; Wang, R. Highly Sensitive Surface-Enhanced Raman Scattering (SERS) Imaging for Phenotypic Diagnosis and Therapeutic Evaluation of Breast Cancer. Chem. Eng. J. 2023, 459, 141502. [Google Scholar] [CrossRef]
- Chen, M.; Solarska, R.; Li, M. Additional Important Considerations in Surface-Enhanced Raman Scattering Enhancement Factor Measurements. J. Phys. Chem. C 2023, 127, 2728–2734. [Google Scholar] [CrossRef]
- Nicholson, T.A.; Sagmeister, M.; Wijesinghe, S.N.; Farah, H.; Hardy, R.S.; Jones, S.W. Oligonucleotide Therapeutics for Age-Related Musculoskeletal Disorders: Successes and Challenges. Pharmaceutics 2023, 15, 237. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Zhou, X.; Yang, Z. Progress of Oligonucleotide Therapeutics Target to Rna. In Nucleic Acids in Medicinal Chemistry and Chemical Biology; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2023; pp. 373–427. ISBN 978-1-119-69279-9. [Google Scholar]
- Hall, J.; Hill, A. The MOE Modification of RNA: Origins and Widescale Impact on the Oligonucleotide Therapeutics Field. Helv. Chim. Acta 2023, 106, e202200169. [Google Scholar] [CrossRef]
- Amulya, E.; Sikder, A.; Vambhurkar, G.; Shah, S.; Khatri, D.K.; Raghuvanshi, R.S.; Singh, S.B.; Srivastava, S. Nanomedicine Based Strategies for Oligonucleotide Traversion across the Blood–Brain Barrier. J. Control. Release 2023, 354, 554–571. [Google Scholar] [CrossRef] [PubMed]
- Fàbrega, C.; Aviñó, A.; Navarro, N.; Jorge, A.F.; Grijalvo, S.; Eritja, R. Lipid and Peptide-Oligonucleotide Conjugates for Therapeutic Purposes: From Simple Hybrids to Complex Multifunctional Assemblies. Pharmaceutics 2023, 15, 320. [Google Scholar] [CrossRef]
- Paul, D.; Miller, M.H.; Born, J.; Samaddar, S.; Ni, H.; Avila, H.; Krishnamurthy, V.R.; Thirunavukkarasu, K. The Promising Therapeutic Potential of Oligonucleotides for Pulmonary Fibrotic Diseases. Expert Opin. Drug. Discov. 2023, 18, 193–206. [Google Scholar] [CrossRef]
- Goyenvalle, A.; Jimenez-Mallebrera, C.; van Roon, W.; Sewing, S.; Krieg, A.M.; Arechavala-Gomeza, V.; Andersson, P. Considerations in the Preclinical Assessment of the Safety of Antisense Oligonucleotides. Nucleic Acid Ther. 2023, 33, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ha, J.; Kim, M.; Kang, S.; Kang, M.; Lee, M. Antisense-Oligonucleotide Co-Micelles with Tumor Targeting Peptides Elicit Therapeutic Effects by Inhibiting MicroRNA-21 in the Glioblastoma Animal Models. J. Adv. Res. 2023; in press. [Google Scholar] [CrossRef]
- Xiang, Y.; Lu, Y. Using Personal Glucose Meters and Functional DNA Sensors to Quantify a Variety of Analytical Targets. Nat. Chem. 2011, 3, 697–703. [Google Scholar] [CrossRef]
- Huang, N.-H.; Li, R.-T.; Fan, C.; Wu, K.-Y.; Zhang, Z.; Chen, J.-X. Rapid Sequential Detection of Hg2+ and Biothiols by a Probe DNA-MOF Hybrid Sensory System. J. Inorg. Biochem. 2019, 197, 110690. [Google Scholar] [CrossRef]
- Bai, Y.; Shu, T.; Su, L.; Zhang, X. Functional Nucleic Acid-Based Fluorescence Polarization/Anisotropy Biosensors for Detection of Biomarkers. Anal. Bioanal. Chem. 2020, 412, 6655–6665. [Google Scholar] [CrossRef]
- Huang, G.; Su, C.; Wang, L.; Fei, Y.; Yang, J. The Application of Nucleic Acid Probe-Based Fluorescent Sensing and Imaging in Cancer Diagnosis and Therapy. Front. Chem. 2021, 9, 705458. [Google Scholar] [CrossRef] [PubMed]
- Saha, K.; Agasti, S.S.; Kim, C.; Li, X.; Rotello, V.M. Gold Nanoparticles in Chemical and Biological Sensing. Chem. Rev. 2012, 112, 2739–2779. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Gao, X.; Liu, D.; Chen, X. Gold Nanoparticles for In Vitro Diagnostics. Chem. Rev. 2015, 115, 10575–10636. [Google Scholar] [CrossRef] [PubMed]
- Lane, L.A.; Qian, X.; Nie, S. SERS Nanoparticles in Medicine: From Label-Free Detection to Spectroscopic Tagging. Chem. Rev. 2015, 115, 10489–10529. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.; Ho, W.K.H.; Xia, X.; Chan, C.K.W.; Zhang, Q.; Ng, Y.M.; Lam, C.Y.K.; Cheung, J.C.W.; Wang, J.; Yang, M.; et al. A Multilayered Mesoporous Gold Nanoarchitecture for Ultraeffective Near-Infrared Light-Controlled Chemo/Photothermal Therapy for Cancer Guided by SERS Imaging. Small 2023, 19, 2206762. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Das, N.; Rayavarapu, R.G. Role of Tunable Gold Nanostructures in Cancer Nanotheranostics: Implications on Synthesis, Toxicity, Clinical Applications and Their Associated Opportunities and Challenges. J. Nanotheranostics 2023, 4, 1–34. [Google Scholar] [CrossRef]
- Li, W.; Zhou, T.; Sun, W.; Liu, M.; Wang, X.; Wang, F.; Zhang, G.; Zhang, Z. A Conjugated Aptamer and Oligonucleotides-Stabilized Gold Nanoclusters Nanoplatform for Targeted Fluorescent Imaging and Efficient Drug Delivery. Colloids. Surf. A Physicochem. Eng. Asp. 2023, 657, 130521. [Google Scholar] [CrossRef]
- Sondhi, P.; Lingden, D.; Bhattarai, J.K.; Demchenko, A.V.; Stine, K.J. Applications of Nanoporous Gold in Therapy, Drug Delivery, and Diagnostics. Metals 2023, 13, 78. [Google Scholar] [CrossRef]
- Cândido, M.; Vieira, P.; Campos, A.; Soares, C.; Raniero, L. Gold-Coated Superparamagnetic Iron Oxide Nanoparticles Functionalized to EGF and Ce6 Complexes for Breast Cancer Diagnoses and Therapy. Pharmaceutics 2022, 15, 100. [Google Scholar] [CrossRef]
- Fujita, H.; Ohta, S.; Nakamura, N.; Somiya, M.; Horie, M. Progress of Endogenous and Exogenous Nanoparticles for Cancer Therapy and Diagnostics. Genes 2023, 14, 259. [Google Scholar] [CrossRef]
- Mirkin, C.A.; Letsinger, R.L.; Mucic, R.C.; Storhoff, J.J. A DNA-Based Method for Rationally Assembling Nanoparticles into Macroscopic Materials. Nature 1996, 382, 607–609. [Google Scholar] [CrossRef] [PubMed]
- Cutler, J.I.; Auyeung, E.; Mirkin, C.A. Spherical Nucleic Acids. J. Am. Chem. Soc. 2012, 134, 1376–1391. [Google Scholar] [CrossRef]
- Liu, Z.; Zhao, J.; Zhang, R.; Han, G.; Zhang, C.; Liu, B.; Zhang, Z.; Han, M.-Y.; Gao, X. Cross-Platform Cancer Cell Identification Using Telomerase-Specific Spherical Nucleic Acids. ACS Nano 2018, 12, 3629–3637. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. Identification of a Specific Telomere Terminal Transferase Activity in Tetrahymena Extracts. Cell 1985, 43, 405–413. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. A Telomeric Sequence in the RNA of Tetrahymena Telomerase Required for Telomere Repeat Synthesis. Nature 1989, 337, 331–337. [Google Scholar] [CrossRef]
- Pan, W.; Zhang, T.; Yang, H.; Diao, W.; Li, N.; Tang, B. Multiplexed Detection and Imaging of Intracellular MRNAs Using a Four-Color Nanoprobe. Anal. Chem. 2013, 85, 10581–10588. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-J.; Cui, M.-R.; Li, X.-L.; Chen, H.-Y.; Xu, J.-J. A Self-Powered 3D DNA Walker with Programmability and Signal-Amplification for Illuminating MicroRNA in Living Cells. Chem. Commun. 2020, 56, 2135–2138. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.-P.; Ma, P.-Q.; Liu, H.; Guo, X.; Yin, B.-C.; Ye, B.-C. Rational Engineering of a Dynamic, Entropy-Driven DNA Nanomachine for Intracellular MicroRNA Imaging. Angew. Chem. Int. Ed. Engl. 2017, 56, 9077–9081. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Liu, B.; Pan, W.; Li, N.; Tang, B. A Spherical Nucleic Acid Probe Based on the Au-Se Bond. Anal. Chem. 2020, 92, 8459–8463. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jiang, Q.; Liu, Y.; Wang, W.; Yu, W.; Wang, F.; Liu, X. Precision Spherical Nucleic Acids Enable Sensitive FEN1 Imaging and Controllable Drug Delivery for Cancer-Specific Therapy. Anal. Chem. 2021, 93, 11275–11283. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Zhao, D.; Huang, J.; Zhu, Y.; Chao, J.; Su, S.; Li, J.; Wang, L.; Shi, J.; Zuo, X.; et al. Poly-Adenine-Mediated Fluorescent Spherical Nucleic Acid Probes for Live-Cell Imaging of Endogenous Tumor-Related MRNA. Nanomedicine 2018, 14, 1797–1807. [Google Scholar] [CrossRef]
- Cui, M.-R.; Li, X.-L.; Xu, J.-J.; Chen, H.-Y. Acid-Switchable DNAzyme Nanodevice for Imaging Multiple Metal Ions in Living Cells. ACS Appl. Mater. Interfaces. 2020, 12, 13005–13012. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Hao, C.; Chen, C.; Ma, W.; Sun, M.; Xu, L.; Xu, C.; Kuang, H. Ratiometric FRET Encoded Hierarchical ZrMOF @ Au Cluster for Ultrasensitive Quantifying MicroRNA In Vivo. Adv. Mater. 2022, 34, 2107449. [Google Scholar] [CrossRef]
- Qu, A.; Sun, M.; Kim, J.-Y.; Xu, L.; Hao, C.; Ma, W.; Wu, X.; Liu, X.; Kuang, H.; Kotov, N.A.; et al. Stimulation of Neural Stem Cell Differentiation by Circularly Polarized Light Transduced by Chiral Nanoassemblies. Nat. Biomed. Eng. 2021, 5, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, W.; Zhang, Y.; Zhang, X.; Liu, Y.; Ju, H. A Near-Infrared Photo-Switched MicroRNA Amplifier for Precise Photodynamic Therapy of Early-Stage Cancers. Angew. Chem. Int. Ed. Engl. 2020, 59, 21454–21459. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, P.; Ma, X.; Lin, X.; Xu, S.; Niazi, S.; Wang, Z. Real-Time in Situ Observation of P53-Mediated Cascade Activation of Apoptotic Pathways with Nucleic Acid Multicolor Fluorescent Probes Based on Symmetrical Gold Nanostars. Nano. Res. 2022, 1–10. [Google Scholar] [CrossRef]
- Ma, Y.; Mao, G.; Wu, G.; Fan, J.; He, Z.; Huang, W. A Novel Nano-Beacon Based on DNA Functionalized QDs for Intracellular Telomerase Activity Monitoring. Sens. Actuators B Chem. 2020, 304, 127385. [Google Scholar] [CrossRef]
- Zheng, Y.; He, S.; Jin, P.; Gao, Y.; Di, Y.; Gao, L.; Wang, J. Construction of Multifunctional Carboxymethyl Cellulose Nanohydrogel Carriers Based on Near-Infrared DNA-Templated Quantum Dots for Tumor Theranostics. RSC Adv. 2022, 12, 31869–31877. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Liu, Y.; Zhou, Q.; Du, H.; Zhao, X.; Ye, F.; Zhao, H. Catalytic Hairpin Assembly Indirectly Covalent on Fe3O4@C Nanoparticles with Signal Amplification for Intracellular Detection of MiRNA. Talanta 2021, 223, 121675. [Google Scholar] [CrossRef]
- Huo, M.; Li, S.; Zhang, P.; Feng, Y.; Liu, Y.; Wu, N.; Ju, H.; Ding, L. Nanoamplicon Comparator for Live-Cell MicroRNA Imaging. Anal. Chem. 2019, 91, 3374–3381. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Hu, L.; He, H.; Zhang, F.; Wang, M.; Wei, W.; Xia, Z. Y-Shaped DNA-Mediated Hybrid Nanoflowers as Efficient Gene Carriers for Fluorescence Imaging of Tumor-Related MRNA in Living Cells. Anal. Chim. Acta. 2019, 1057, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Lin, L.; Xu, C.; Tian, H.; Chen, X. A GSH-Gated DNA Nanodevice for Tumor-Specific Signal Amplification of MicroRNA and MR Imaging–Guided Theranostics. Small 2019, 15, 1903016. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.-J.; Huang, Z.-M.; Xiao, H.-Y.; Wu, Z.-K.; Tang, L.-J.; Jiang, J.-H. Protein Scaffolded DNA Tetrads Enable Efficient Delivery and Ultrasensitive Imaging of MiRNA through Crosslinking Hybridization Chain Reaction. Chem. Sci. 2018, 9, 4892–4897. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.-M.; Lin, M.-Y.; Zhang, C.-H.; Wu, Z.; Yu, R.-Q.; Jiang, J.-H. Recombinant Fusion Streptavidin as a Scaffold for DNA Nanotetrads for Nucleic Acid Delivery and Telomerase Activity Imaging in Living Cells. Anal. Chem. 2019, 91, 9361–9365. [Google Scholar] [CrossRef]
- Andersen, V.L.; Vinther, M.; Kumar, R.; Ries, A.; Wengel, J.; Nielsen, J.S.; Kjems, J. A Self-Assembled, Modular Nucleic Acid-Based Nanoscaffold for Multivalent Theranostic Medicine. Theranostics 2019, 9, 2662–2677. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Liao, R.; Guo, J. Highly Multiplexed Single-Cell In Situ RNA and DNA Analysis by Consecutive Hybridization. Molecules 2020, 25, 4900. [Google Scholar] [CrossRef]
- Song, J.; Mou, H.-Z.; Li, X.-Q.; Liu, Y.; Yang, X.-J.; Chen, H.-Y.; Xu, J.-J. Self-Assembled DNA/RNA Nanospheres with Cascade Signal Amplification for Intracellular MicroRNA Imaging. Sens. Actuators B Chem. 2022, 360, 131644. [Google Scholar] [CrossRef]
- Xiao, F.; Lin, L.; Chao, Z.; Shao, C.; Chen, Z.; Wei, Z.; Lu, J.; Huang, Y.; Li, L.; Liu, Q.; et al. Organic Spherical Nucleic Acids for the Transport of a NIR-II-Emitting Dye Across the Blood-Brain Barrier. Angew. Chem. Int. Ed. Engl. 2020, 59, 9702–9710. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Fang, X.; Li, H.; Xue, H.; Wei, Z.; Zhang, W.; Zhu, Y.; Lin, L.; Zhao, Y.; Wu, C.; et al. Light-Harvesting Fluorescent Spherical Nucleic Acids Self-Assembled from a DNA-Grafted Conjugated Polymer for Amplified Detection of Nucleic Acids. Angew. Chem. Int. Ed. 2022, 61, e202115812. [Google Scholar] [CrossRef]
- Abramczyk, H.; Brozek-Pluska, B. Raman Imaging in Biochemical and Biomedical Applications. Diagnosis and Treatment of Breast Cancer. Chem. Rev. 2013, 113, 5766–5781. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.R.; Gambhir, S.S. Nanomaterials for In Vivo Imaging. Chem. Rev. 2017, 117, 901–986. [Google Scholar] [CrossRef]
- Sheikhzadeh, E.; Beni, V.; Zourob, M. Nanomaterial Application in Bio/Sensors for the Detection of Infectious Diseases. Talanta 2021, 230, 122026. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Li, X.; Li, K.; Nie, Z.; Tan, W. DNA-Modulated Plasmon Resonance: Methods and Optical Applications. ACS Appl. Mater. Interfaces. 2020, 12, 14741–14760. [Google Scholar] [CrossRef]
- Si, Y.; Bai, Y.; Qin, X.; Li, J.; Zhong, W.; Xiao, Z.; Li, J.; Yin, Y. Alkyne-DNA-Functionalized Alloyed Au/Ag Nanospheres for Ratiometric Surface-Enhanced Raman Scattering Imaging Assay of Endonuclease Activity in Live Cells. Anal. Chem. 2018, 90, 3898–3905. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, Y.; Lu, W.; Liu, C.; Ge, S.; Zhou, X.; Bi, C.; Cao, X. A Novel Surface-Enhanced Raman Scattering Probe Based on Au Nanoboxes for Dynamic Monitoring of Caspase-3 during Cervical Cancer Cell Apoptosis. J. Mater. Chem. B 2021, 9, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Wang, Z.; Bi, L.; Bi, C.; Du, Q. Gold Nanocage-Based Surface-Enhanced Raman Scattering Probes for Long-Term Monitoring of Intracellular MicroRNA during Bone Marrow Stem Cell Differentiation. Nanoscale 2020, 12, 1513–1527. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wu, X.; Sun, M.; Xu, L.; Kuang, H.; Xu, C. Tetrahedron Probes for Ultrasensitive In Situ Detection of Telomerase and Surface Glycoprotein Activity in Living Cells. Anal. Chem. 2020, 92, 2310–2315. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Qiao, Z.; Wang, X.; Bi, S. Enzyme-Free Catalyzed Self-Assembly of Three-Dimensional Hyperbranched DNA Structures for in Situ SERS Imaging and Molecular Logic Operations. Chem. Eng. J. 2022, 446, 136838. [Google Scholar] [CrossRef]
- Liu, C.; Chen, C.; Li, S.; Dong, H.; Dai, W.; Xu, T.; Liu, Y.; Yang, F.; Zhang, X. Target-Triggered Catalytic Hairpin Assembly-Induced Core-Satellite Nanostructures for High-Sensitive “Off-to-On” SERS Detection of Intracellular MicroRNA. Anal. Chem. 2018, 90, 10591–10599. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Lee, J.U.; Jeon, M.; Park, K.; Sim, S. Three-Dimensional Hierarchical Plasmonic Nano-Architecture Based Label-Free Surface-Enhanced Raman Spectroscopy Detection of Urinary Exosomal MiRNA for Clinical Diagnosis of Prostate Cancer. Biosens. Bioelectron. 2022, 205, 114116. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Ren, J.-Q.; Shen, A.-G.; Hu, J.-M. Splicing Nanoparticles-Based “Click” SERS Could Aid Multiplex Liquid Biopsy and Accurate Cellular Imaging. J. Am. Chem. Soc. 2018, 140, 10649–10652. [Google Scholar] [CrossRef]
- Zhu, W.; Wang, C.-Y.; Hu, J.-M.; Shen, A.-G. Promoted “Click” SERS Detection for Precise Intracellular Imaging of Caspase-3. Anal. Chem. 2021, 93, 4876–4883. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Jin, Q.; Yan, N.; Feng, J.; Wang, J.; Tang, X. SERS Nanoprobes in Biologically Raman Silent Region for Tumor Cell Imaging and In Vivo Tumor Spectral Detection in Mice. Adv. Biosyst. 2018, 2, 1800100. [Google Scholar] [CrossRef]
- Wolf, J.M.; Wolf, L.M.; Bello, G.L.; Maccari, J.G.; Nasi, L.A. Molecular Evolution of SARS-CoV-2 from December 2019 to August 2022. J. Med. Virol. 2023, 95, e28366. [Google Scholar] [CrossRef] [PubMed]
- Roviello, V.; Gilhen-Baker, M.; Vicidomini, C.; Roviello, G.N. Forest-Bathing and Physical Activity as Weapons against COVID-19: A Review. Env. Chem. Lett. 2022, 20, 131–140. [Google Scholar] [CrossRef]
- Borbone, N.; Piccialli, I.; Falanga, A.P.; Piccialli, V.; Roviello, G.N.; Oliviero, G. Nucleic Acids as Biotools at the Interface between Chemistry and Nanomedicine in the COVID-19 Era. Int. J. Mol. Sci. 2022, 23, 4359. [Google Scholar] [CrossRef] [PubMed]
- Roviello, V.; Roviello, G.N. Less COVID-19 Deaths in Southern and Insular Italy Explained by Forest Bathing, Mediterranean Environment, and Antiviral Plant Volatile Organic Compounds. Env. Chem. Lett. 2022, 20, 7–17. [Google Scholar] [CrossRef]
- Vicidomini, C.; Roviello, V.; Roviello, G.N. In Silico Investigation on the Interaction of Chiral Phytochemicals from Opuntia Ficus-Indica with SARS-CoV-2 Mpro. Symmetry 2021, 13, 1041. [Google Scholar] [CrossRef]
- Moitra, P.; Chaichi, A.; Abid Hasan, S.M.; Dighe, K.; Alafeef, M.; Prasad, A.; Gartia, M.R.; Pan, D. Probing the Mutation Independent Interaction of DNA Probes with SARS-CoV-2 Variants through a Combination of Surface-Enhanced Raman Scattering and Machine Learning. Biosens. Bioelectron. 2022, 208, 114200. [Google Scholar] [CrossRef]
- Liu, J.; Zheng, T.; Tian, Y. Functionalized H-BN Nanosheets as a Theranostic Platform for SERS Real-Time Monitoring of MicroRNA and Photodynamic Therapy. Angew. Chem. Int. Ed. 2019, 58, 7757–7761. [Google Scholar] [CrossRef]
- Jin, Q.; Fan, X.; Chen, C.; Huang, L.; Wang, J.; Tang, X. Multicolor Raman Beads for Multiplexed Tumor Cell and Tissue Imaging and in Vivo Tumor Spectral Detection. Anal. Chem. 2019, 91, 3784–3789. [Google Scholar] [CrossRef]
- Zhang, N.; Ye, S.; Wang, Z.; Li, R.; Wang, M. A Dual-Signal Twinkling Probe for Fluorescence-SERS Dual Spectrum Imaging and Detection of MiRNA in Single Living Cell via Absolute Value Coupling of Reciprocal Signals. ACS Sens. 2019, 4, 924–930. [Google Scholar] [CrossRef]
- Liu, X.; Wang, X.; Ye, S.; Li, R.; Li, H. A One-Two-Three Multifunctional System for Enhanced Imaging and Detection of Intracellular MicroRNA and Chemogene Therapy. ACS Appl. Mater. Interfaces. 2021, 13, 27825–27835. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, H.; Ye, S.; Wang, X.; Ma, L. Fluorescent-Raman Binary Star Ratio Probe for MicroRNA Detection and Imaging in Living Cells. Anal. Chem. 2021, 93, 1466–1471. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Han, W.; Bi, C.; Song, W.; Niu, S.; Zhou, H.; Zhang, X. Many Birds, One Stone: A Smart Nanodevice for Ratiometric Dual-Spectrum Assay of Intracellular MicroRNA and Multimodal Synergetic Cancer Therapy. ACS Nano 2021, 15, 6961–6976. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Ma, L.; Tian, F.; Gu, Y.; Li, J.; Zhang, P.; Yang, G.; Li, H.; Qu, L.-L. Fluorescence and Surface-Enhanced Raman Scattering Dual-Mode Nanoprobe for Monitoring Telomerase Activity in Living Cells. Microchem. J. 2022, 175, 107171. [Google Scholar] [CrossRef]
- Li, C.; Chen, P.; Wang, Z.; Ma, X. A DNAzyme-Gold Nanostar Probe for SERS-Fluorescence Dual-Mode Detection and Imaging of Calcium Ions in Living Cells. Sens. Actuators B Chem. 2021, 347, 130596. [Google Scholar] [CrossRef]
- Roviello, G.N.; Musumeci, D.; Pedone, C.; Bucci, E.M. Synthesis, characterization and hybridization studies of an alternate nucleo-ε/γ-peptide: Complexes formation with natural nucleic acids. Amino. Acids 2010, 38, 103–111. [Google Scholar] [CrossRef]
- Roviello, G.N.; Musumeci, D.; Moccia, M.; Castiglione, M.; Sapio, R.; Valente, M.; Bucci, E.M.; Perretta, G.; Pedone, C. dab Pna: Design, Synthesis, And Dna Binding Studies. Nucleosides Nucleotides Nucleic Acids 2007, 26, 1307–1310. [Google Scholar] [CrossRef]
- Scognamiglio, P.L.; Riccardi, C.; Palumbo, R.; Gale, T.F.; Musumeci, D.; Roviello, G.N. Self-assembly of thyminyl l-tryptophanamide (TrpT) building blocks for the potential development of drug delivery nanosystems. J. Nanostruct. Chem. 2023, 1–19. [Google Scholar] [CrossRef]

| Detected miRNA | Technique | Strategy | LOD | Ref. |
|---|---|---|---|---|
| miR-21 | SERS | Boron nitride nanosheets-conjugated DNA oligonucleotide-copper(II) phthalocyanine. | 0.7 fM | [82] |
| miR-21 | SERS | DNA tetrahedron-mediated branched catalytic hairpin assembly reaction on AuNPs. | 0.33 fM | [70] |
| miR-21 | SERS | Two types of hairpin DNA-three-way branch DNA on AuNP. | 3.26 fM | [85] |
| miR-21 | SERS | DNA hairpins on plasmonic copper-sulfide-polydopamine AuNP. | 4.95 aM | [87] |
| miR-21 | Fluorescence | DNA hairpins on plasmonic copper-sulfide-polydopamine AuNP. | 0.11 pM | [87] |
| miR-21 | Fluorescence | Protein scaffolded DNA tetrad. | 6 pM | [55] |
| miR-21 | Fluorescence | Spherical nucleic acids with fluorescent π-conjugated polymer. | 1.7 pM | [61] |
| miR-21 | Fluorescence | Fluorescent nucleic acid probe and polymer-modified MnO2 nanosheets. | 30 pM | [54] |
| miR-21 | Fluorescence | Zirconium metal–organic frameworks @ gold architecture functionalized with fluorophore-labeled DNA. | 4.51 zmol/ngRNA | [45] |
| miR-21 miR-10a | SERS | Gold nanopillar with self-assembling DNA probe-conjugated AuNP. | ~10 aM | [72] |
| miR-21-D | Fluorescence | Hairpin-conjugated core-shell NaYF4:Er/Gd/Yb@NaGdF4 nanoparticles. | 1.02 nM | [52] |
| miR-20a | Fluorescence | Hairpin-conjugated Fe3O4@C nanoparticles. | 0.491 pM | [51] |
| miR-155 | Fluorescence | Self-assembled nucleic acid nanosphere. | 0.031 fM | [59] |
| miR-144-3p | SERS | Gold nanocage functionalized with DNA hairpins. | 13.6 aM | [68] |
| miR-203 | Fluorescence-SERS | DNA hairpins on AuNP. | ≤0.13 nM * | [86] |
| miR-203 | SERS | Triple helix structures immobilized on AuNP’. | 0.63 pM | [84] |
| miR-1246 | SERS | Core-satellite nanostructure: DNA-modified gold nanodumbells and DNA-modified AuNPs. | 0.85 aM | [71] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sardaru, M.-C.; Marangoci, N.-L.; Palumbo, R.; Roviello, G.N.; Rotaru, A. Nucleic Acid Probes in Bio-Imaging and Diagnostics: Recent Advances in ODN-Based Fluorescent and Surface-Enhanced Raman Scattering Nanoparticle and Nanostructured Systems. Molecules 2023, 28, 3561. https://doi.org/10.3390/molecules28083561
Sardaru M-C, Marangoci N-L, Palumbo R, Roviello GN, Rotaru A. Nucleic Acid Probes in Bio-Imaging and Diagnostics: Recent Advances in ODN-Based Fluorescent and Surface-Enhanced Raman Scattering Nanoparticle and Nanostructured Systems. Molecules. 2023; 28(8):3561. https://doi.org/10.3390/molecules28083561
Chicago/Turabian StyleSardaru, Monica-Cornelia, Narcisa-Laura Marangoci, Rosanna Palumbo, Giovanni N. Roviello, and Alexandru Rotaru. 2023. "Nucleic Acid Probes in Bio-Imaging and Diagnostics: Recent Advances in ODN-Based Fluorescent and Surface-Enhanced Raman Scattering Nanoparticle and Nanostructured Systems" Molecules 28, no. 8: 3561. https://doi.org/10.3390/molecules28083561
APA StyleSardaru, M.-C., Marangoci, N.-L., Palumbo, R., Roviello, G. N., & Rotaru, A. (2023). Nucleic Acid Probes in Bio-Imaging and Diagnostics: Recent Advances in ODN-Based Fluorescent and Surface-Enhanced Raman Scattering Nanoparticle and Nanostructured Systems. Molecules, 28(8), 3561. https://doi.org/10.3390/molecules28083561