Discovery of N,4-Di(1H-pyrazol-4-yl)pyrimidin-2-amine-Derived CDK2 Inhibitors as Potential Anticancer Agents: Design, Synthesis, and Evaluation
Abstract
1. Introduction
2. Results and Discussion
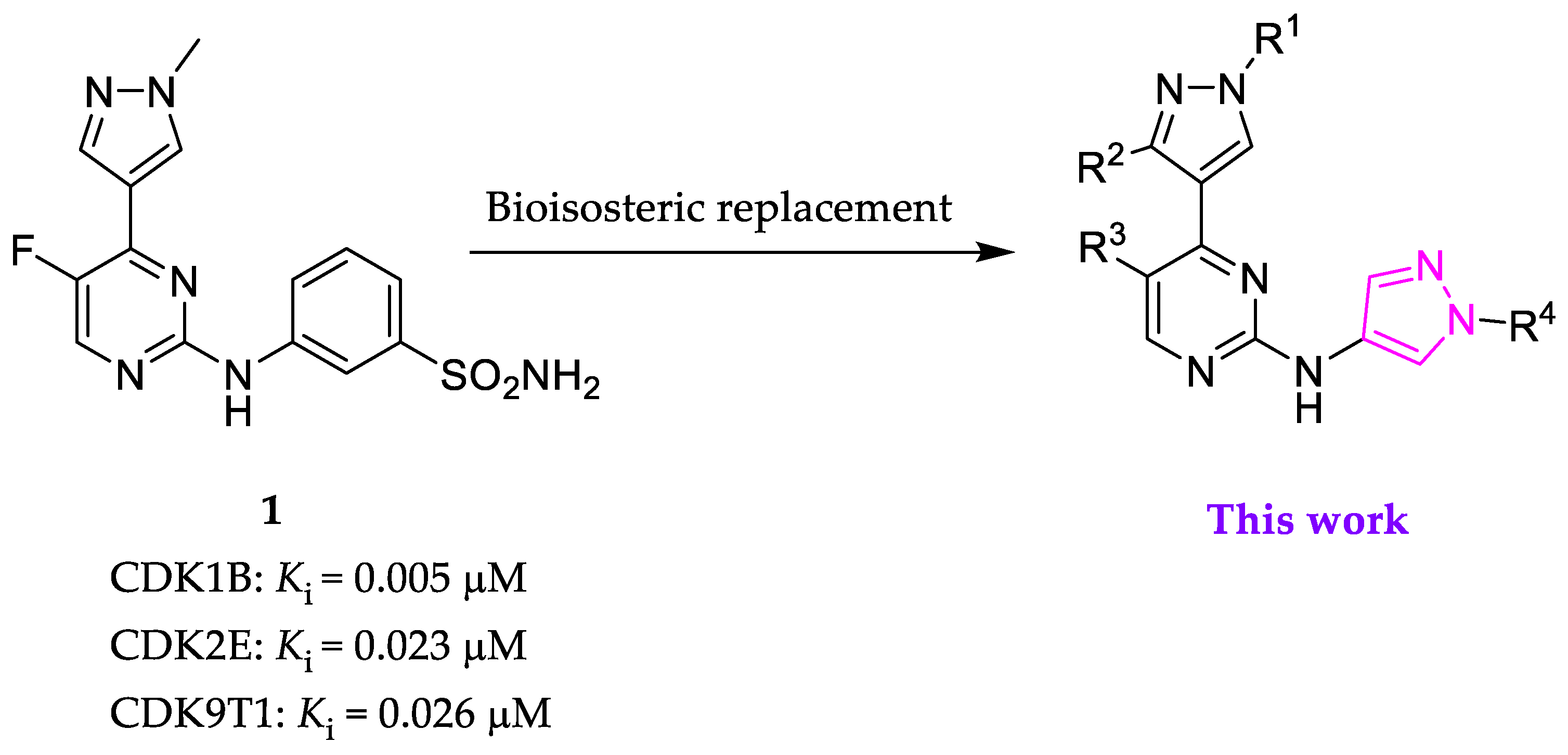
2.1. Inhibitor Design
2.2. Synthesis
2.3. Structure-Activity Relationships
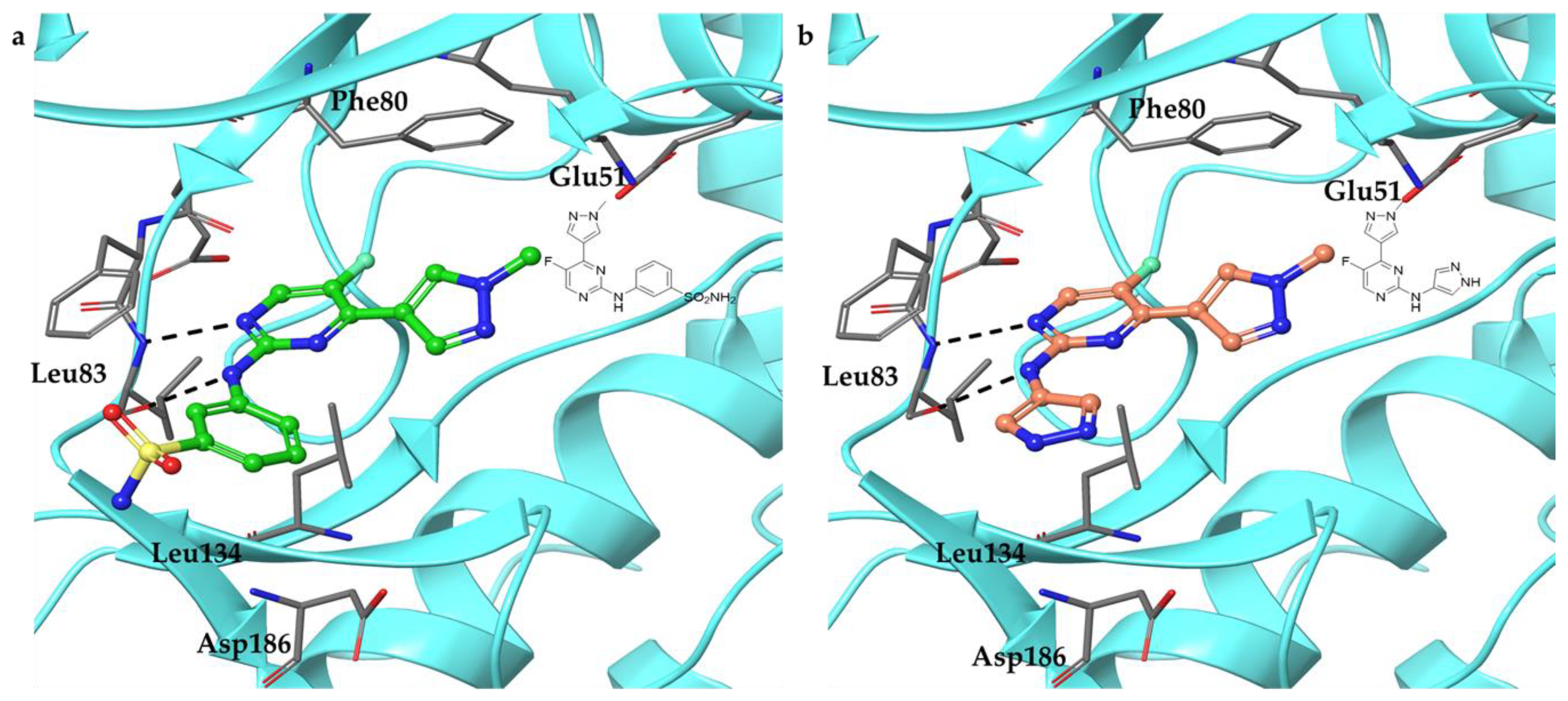
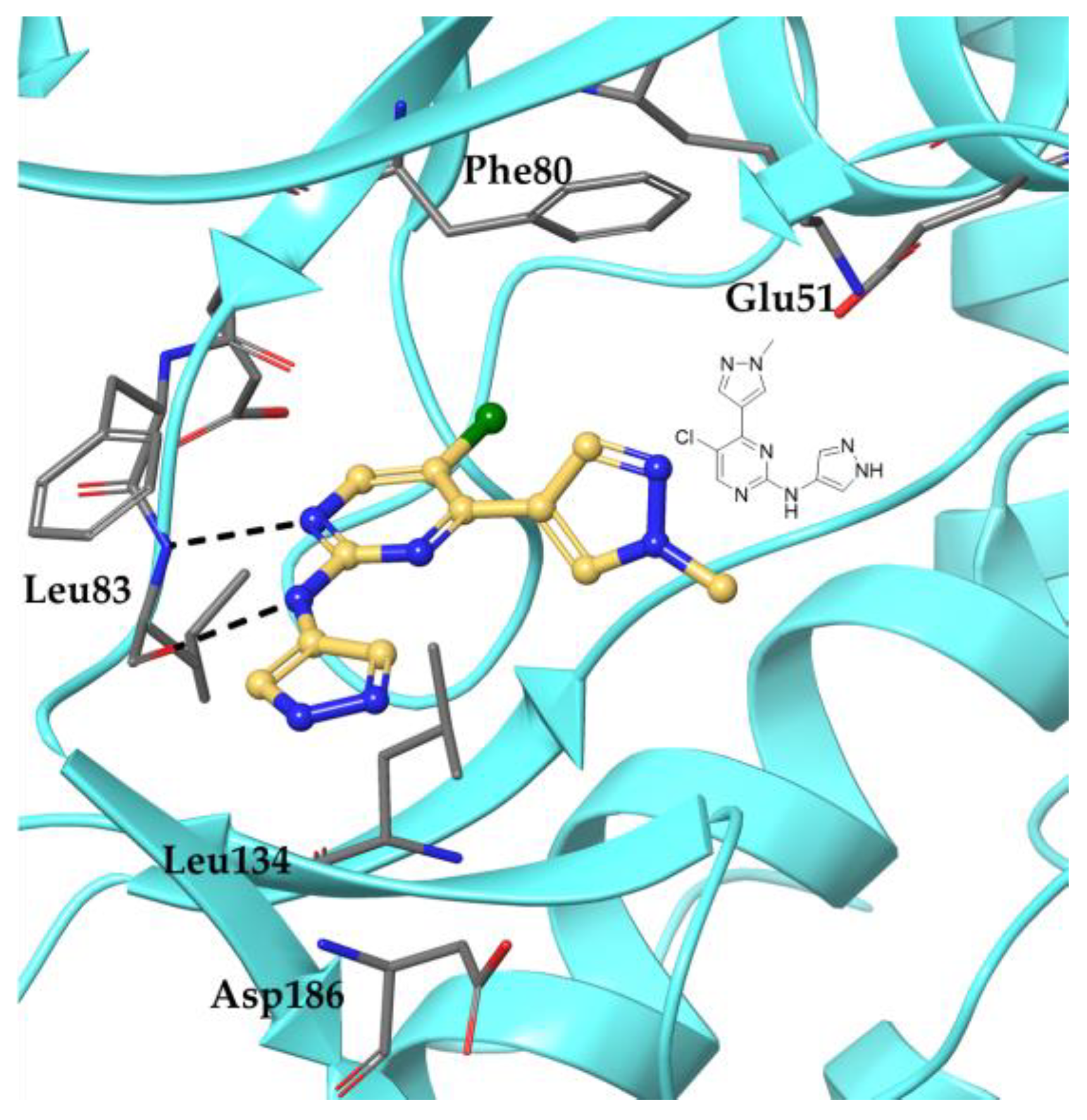
2.4. Docking Study on 15
2.5. Antiproliferative Activities of 15
2.6. Cellular Mechanistic Studies of 15
2.6.1. Effect of 15 on the Cell Cycle Progression
2.6.2. Effect of 15 on Apoptosis in Cancer Cells
2.6.3. Effect of 15 on Colony Formation
2.6.4. Western Blot Analysis
3. Conclusions
4. Experimental Section
4.1. Method for Computational Modeling
4.2. Chemistry
4.2.1. General Synthetic Procedure A: Suzuki Coupling [33]
4.2.2. General Synthetic Procedure B: Buchwald–Hartwig Amination [33]
4.3. Biology
4.3.1. Kinase Assays
4.3.2. Cell Lines
4.3.3. Cell Viability Assays
4.3.4. Cell Cycle Analysis
4.3.5. Apoptosis Assay
4.3.6. Colony Formation
4.3.7. Western Blot Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tadesse, S.; Caldon, E.C.; Tilley, W.; Wang, S. Cyclin-dependent kinase 2 inhibitors in cancer therapy: An update. J. Med. Chem. 2019, 62, 4233–4251. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Kaldis, P. CDKs, cyclins and CKIs: Roles beyond cell cycle regulation. Development 2013, 140, 3079–3093. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M. Cyclin-dependent kinases. Genome Biol. 2014, 15, 122. [Google Scholar] [CrossRef]
- Yang, L.; Fang, D.; Chen, H.; Lu, Y.; Dong, Z.; Ding, H.F.; Jing, Q.; Su, S.B.; Huang, S. Cyclin-dependent kinase 2 is an ideal target for ovary tumors with elevated cyclin E1 expression. Oncotarget 2015, 6, 20801–20812. [Google Scholar] [CrossRef]
- Akli, S.; Van Pelt, C.S.; Bui, T.; Meijer, L.; Keyomarsi, K. CDK2 is required for breast cancer mediated by the low-molecular-weight isoform of cyclin E. Cancer Res. 2011, 71, 3377–3386. [Google Scholar] [CrossRef]
- Molenaar, J.J.; Ebus, M.E.; Geerts, D.; Koster, J.; Lamers, F.; Valentijn, L.J.; Westerhout, E.M.; Versteeg, R.; Caron, H.N. Inactivation of CDK2 is synthetically lethal to MYCN over-expressing cancer cells. Proc. Natl. Acad. Sci. USA 2009, 106, 12968–12973. [Google Scholar] [CrossRef]
- Guarducci, C.; Bonechi, M.; Benelli, M.; Biagioni, C.; Boccalini, G.; Romagnoli, D.; Verardo, R.; Schiff, R.; Osborne, C.K.; De Angelis, C.; et al. Cyclin E1 and Rb modulation as common events at time of resistance to palbociclib in hormone receptor-positive breast cancer. NPJ Breast Cancer 2018, 4, 38. [Google Scholar] [CrossRef]
- Turner, N.C.; Liu, Y.; Zhu, Z.; Loi, S.; Colleoni, M.; Loibl, S.; DeMichele, A.; Harbeck, N.; Andre, F.; Bayar, M.A.; et al. Cyclin E1 expression and palbociclib efficacy in previously treated hormone receptor-positive metastatic breast cancer. J. Clin. Oncol. 2019, 37, 1169–1178. [Google Scholar] [CrossRef]
- Tadesse, S.; Anshabo, A.T.; Portman, N.; Lim, E.; Tilley, W.; Caldon, C.E.; Wang, S. Targeting CDK2 in cancer: Challenges and opportunities for therapy. Drug Discov. Today 2020, 25, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Pandey, K.; Park, N.; Park, K.S.; Hur, J.; Cho, Y.B.; Kang, M.; An, H.J.; Kim, S.; Hwang, S.; Moon, Y.W. Combined CDK2 and CDK4/6 inhibition overcomes palbociclib resistance in breast cancer by enhancing senescence. Cancers 2020, 12, 3566. [Google Scholar] [CrossRef]
- Beale, G.; Haagensen, E.J.; Thomas, H.D.; Wang, L.Z.; Revill, C.H.; Payne, S.L.; Golding, B.T.; Hardcastle, I.R.; Newell, D.R.; Griffin, R.J.; et al. Combined PI3K and CDK2 inhibition induces cell death and enhances in vivo antitumour activity in colorectal cancer. Br. J. Cancer 2016, 115, 682–690. [Google Scholar] [CrossRef]
- Rao, S.S.; Stoehr, J.; Dokic, D.; Wan, L.; Decker, J.T.; Konopka, K.; Thomas, A.L.; Wu, J.; Kaklamani, V.G.; Shea, L.D.; et al. Synergistic effect of eribulin and CDK inhibition for the treatment of triple negative breast cancer. Oncotarget 2017, 8, 83925–83939. [Google Scholar] [CrossRef] [PubMed]
- Chohan, T.A.; Qian, H.; Pan, Y.; Chen, J.Z. Cyclin-dependent kinase-2 as a target for cancer therapy: Progress in the development of CDK2 inhibitors as anti-cancer agents. Curr. Med. Chem. 2015, 22, 237–263. [Google Scholar] [CrossRef]
- Wang, S.D.; Wood, G.; Meades, C.; Griffiths, G.; Midgley, C.; McNae, I.; McInnes, C.; Anderson, S.; Jackson, W.; Mezna, M.; et al. Synthesis and biological activity of 2-anilino-4-(1H-pyrrol-3-yl) pyrimidine CDK inhibitors. Bioorg. Med. Chem. Lett. 2004, 14, 4237–4240. [Google Scholar] [CrossRef]
- Wang, S.D.; Meades, C.; Wood, G.; Osnowski, A.; Anderson, S.; Yuill, R.; Thomas, M.; Mezna, M.; Jackson, W.; Midgley, C.; et al. 2-anilino-4-(thiazol-5-yl)pyrimidine CDK inhibitors: Synthesis, SAR analysis, X-ray crystallography, and biological activity. J. Med. Chem. 2004, 47, 1662–1675. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.D.; Griffiths, G.; Midgley, C.A.; Barnett, A.L.; Cooper, M.; Grabarek, J.; Ingram, L.; Jackson, W.; Kontopidis, G.; McClue, S.J.; et al. Discovery and characterization of 2-Anilino-4-(Thiazol-5-yl)pyrimidine transcriptional CDK inhibitors as anticancer agents. Chem. Biol. 2010, 17, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Shi, S.; Huang, S.; Hole, A.J.; Abbas, A.Y.; Baumli, S.; Liu, X.; Lam, F.; Foley, D.W.; Fischer, P.M.; et al. Substituted 4-(thiazol-5-yl)-2-(phenylamino)pyrimidines are highly active CDK9 inhibitors: Synthesis, X-ray crystal structures, structure-activity relationship, and anticancer activities. J. Med. Chem. 2013, 56, 640–659. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, S.; Bantie, L.; Tomusange, K.; Yu, M.F.; Islam, S.; Bykovska, N.; Noll, B.; Zhu, G.; Li, P.; Lam, F.; et al. Discovery and pharmacological characterization of a novel series of highly selective inhibitors of cyclin-dependent kinases 4 and 6 as anticancer agents. Br. J. Pharmacol. 2018, 175, 2399–2413. [Google Scholar] [CrossRef]
- Yu, M.F.; Teo, T.; Yang, Y.C.; Li, M.J.; Long, Y.; Philip, S.; Noll, B.; Heinemann, G.K.; Diab, S.; Eldi, P.; et al. Potent and orally bioavailable CDK8 inhibitors: Design, synthesis, structure-activity relationship analysis and biological evaluation. Eur. J. Med. Chem. 2021, 214, 113248. [Google Scholar] [CrossRef]
- Fanta, B.S.; Mekonnen, L.; Basnet, S.K.C.; Teo, T.; Lenjisa, J.; Khair, N.Z.; Kou, L.; Tadesse, S.; Sykes, M.J.; Yu, M.; et al. 2-Anilino-4-(1-methyl-1H-pyrazol-4-yl)pyrimidine-derived CDK2 inhibitors as anticancer agents: Design, synthesis & evaluation. Bioorg. Med. Chem. 2023, 80, 117158. [Google Scholar]
- Roskoski, R. Properties of FDA-approved small molecule protein kinase inhibitors: A 2020 update. Pharmacol. Res. 2020, 152, 104609. [Google Scholar] [CrossRef]
- Bhutani, P.; Joshi, G.; Raja, N.; Bachhav, N.; Rajanna, P.K.; Bhutani, H.; Paul, A.T.; Kumar, R. U.S. FDA approved drugs from 2015-June 2020: A perspective. J. Med. Chem. 2021, 64, 2339–2381. [Google Scholar] [CrossRef] [PubMed]
- El-Gamal, M.I.; Zaraei, S.O.; Madkour, M.M.; Anbar, H.S. Evaluation of substituted pyrazole-based kinase inhibitors in one decade (2011–2020): Current status and future prospects. Molecules 2022, 27, 330. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Zhou, M.Z.; Ye, T.; Wang, P.P.; Lu, R.; Wang, Y.L.; Liu, C.X.; Xiao, W.; Li, J.Y.; Meng, Z.B.; et al. Discovery of a series of 5-amide-1H-pyrazole-3-carboxyl derivatives as potent P2Y(14)R antagonists with anti-inflammatory characters. J. Med. Chem. 2022, 65, 15967–15990. [Google Scholar] [CrossRef]
- Cuozzo, A.; Daina, A.; Perez, M.A.S.; Michielin, O.; Zoete, V. SwissBioisostere 2021: Updated structural, bioactivity and physicochemical data delivered by a reshaped web interface. Nucleic Acids Res. 2021, 50, D1382–D1390. [Google Scholar] [CrossRef]
- Wirth, M.; Zoete, V.; Michielin, O.; Sauer, W.H.B. SwissBioisostere: A database of molecular replacements for ligand design. Nucleic Acids Res. 2012, 41, D1137–D1143. [Google Scholar] [CrossRef]
- Wyatt, P.G.; Woodhead, A.J.; Berdini, V.; Boulstridge, J.A.; Carr, M.G.; Cross, D.M.; Davis, D.J.; Devine, L.A.; Early, T.R.; Feltell, R.E.; et al. Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H-pyrazole-3-carboxamide (AT7519), a novel cyclin dependent kinase inhibitor using fragment-based X-ray crystallography and structure based drug design. J. Med. Chem. 2008, 51, 4986–4999. [Google Scholar] [CrossRef]
- Cho, Y.S.; Borland, M.; Brain, C.; Chen, C.H.T.; Cheng, H.; Chopra, R.; Chung, K.; Groarke, J.; He, G.; Hou, Y.; et al. 4-(Pyrazol-4-yl)-pyrimidines as selective inhibitors of cyclin-dependent kinase 4/6. J. Med. Chem. 2010, 53, 7938–7957. [Google Scholar] [CrossRef] [PubMed]
- Barvian, M.; Boschelli, D.H.; Cossrow, J.; Dobrusin, E.; Fattaey, A.; Fritsch, A.; Fry, D.; Harvey, P.; Keller, P.; Garrett, M.; et al. Pyrido[2,3-d]pyrimidin-7-one inhibitors of cyclin-dependent kinases. J. Med. Chem. 2000, 43, 4606–4616. [Google Scholar] [CrossRef]
- Frame, S.; Saladino, C.; MacKay, C.; Atrash, B.; Sheldrake, P.; McDonald, E.; Clarke, P.A.; Workman, P.; Blake, D.; Zheleva, D. Fadraciclib (CYC065), a novel CDK inhibitor, targets key pro-survival and oncogenic pathways in cancer. PLoS ONE 2020, 15, e0234103. [Google Scholar] [CrossRef]
- Hawkins, P.C.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer generation with OMEGA: Algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 2010, 50, 572–584. [Google Scholar] [CrossRef]
- McGann, M. FRED pose prediction and virtual screening accuracy. J. Chem. Inf. Model. 2011, 51, 578–596. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Hu, M.S.; Qi, W.Y.; Yang, Z.; Tang, M.H.; He, J.; Chen, Y.; Bai, P.; Yuan, X.; Zhang, C.F.; et al. Discovery of potent and orally effective dual Janus Kinase 2/FLT3 inhibitors for the treatment of acute myelogenous leukemia and myeloproliferative neoplasms. J. Med. Chem. 2019, 62, 10305–10320. [Google Scholar] [CrossRef] [PubMed]
- Bantie, L.; Tadesse, S.; Likisa, J.; Yu, M.F.; Noll, B.; Heinemann, G.; Lokman, N.A.; Ricciardelli, C.; Oehler, M.K.; Beck, A.; et al. A first-in-class CDK4 inhibitor demonstrates in vitro, ex-vivo and in vivo efficacy against ovarian cancer. Gynecol. Oncol. 2020, 159, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Teo, T.; Yu, M.F.; Yang, Y.C.; Gillam, T.; Lam, F.; Sykes, M.J.; Wang, S.D. Pharmacologic co-inhibition of Mnks and mTORC1 synergistically suppresses proliferation and perturbs cell cycle progression in blast crisis-chronic myeloid leukemia cells. Cancer Lett. 2015, 357, 612–623. [Google Scholar] [CrossRef] [PubMed]
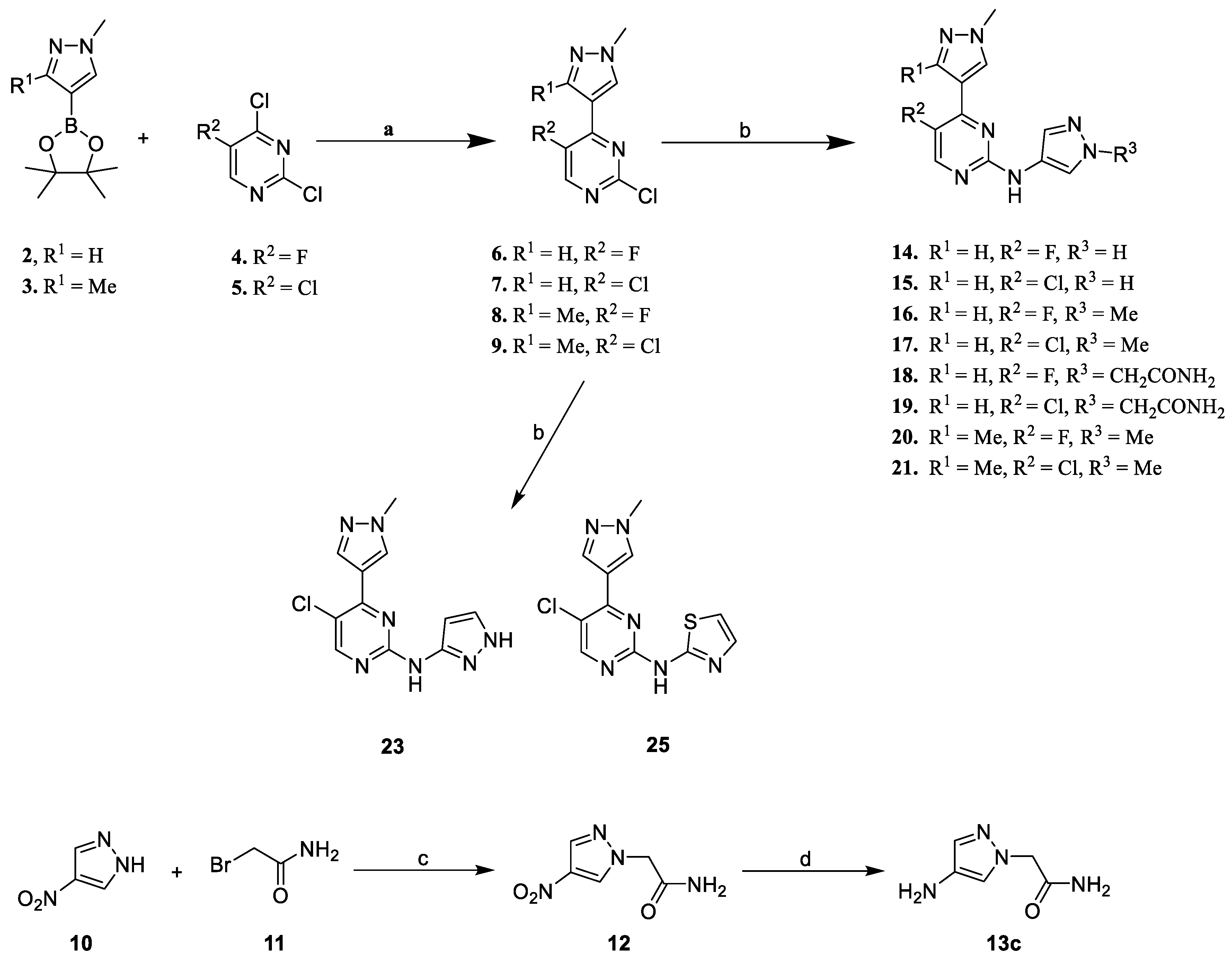

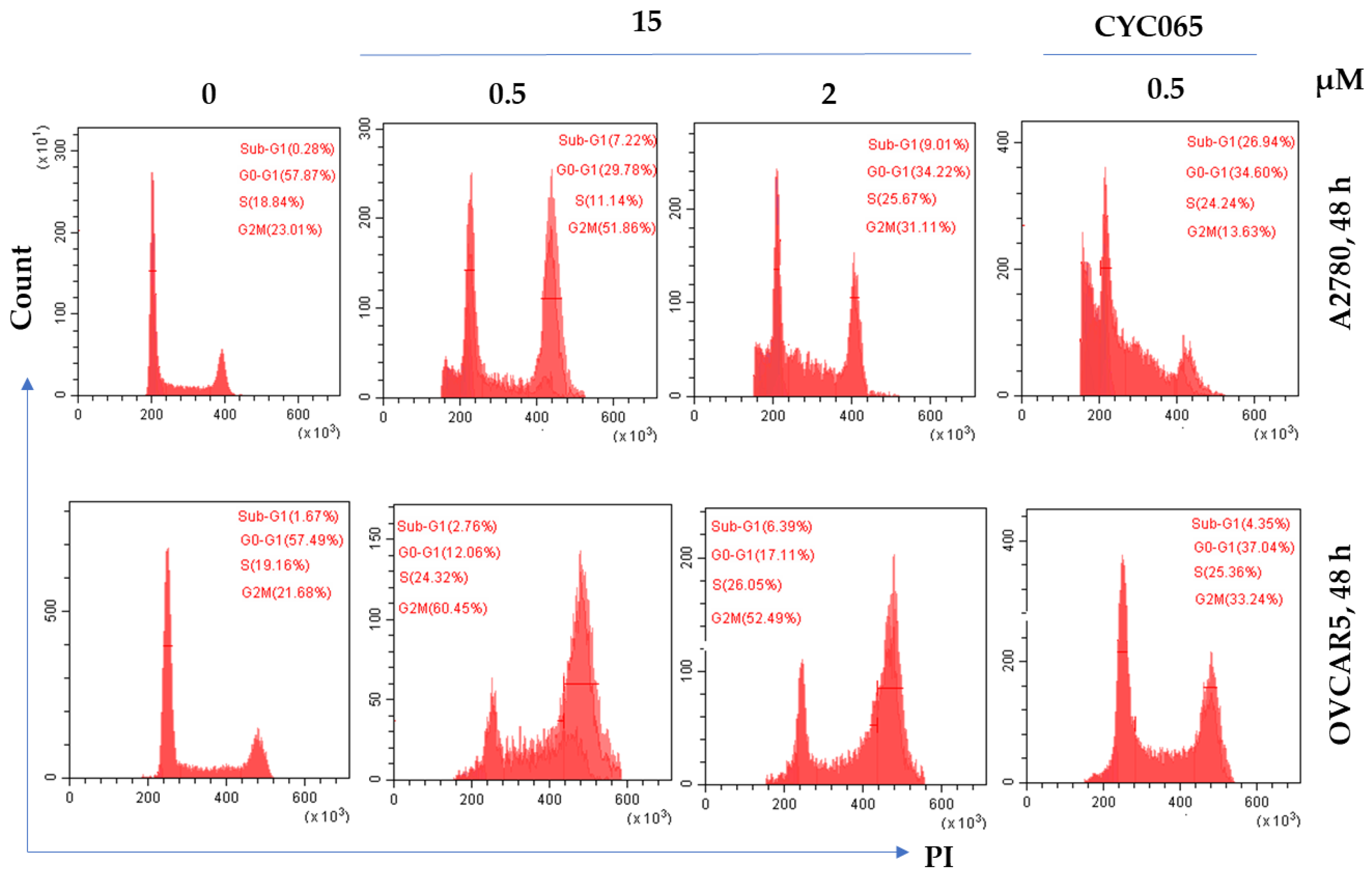
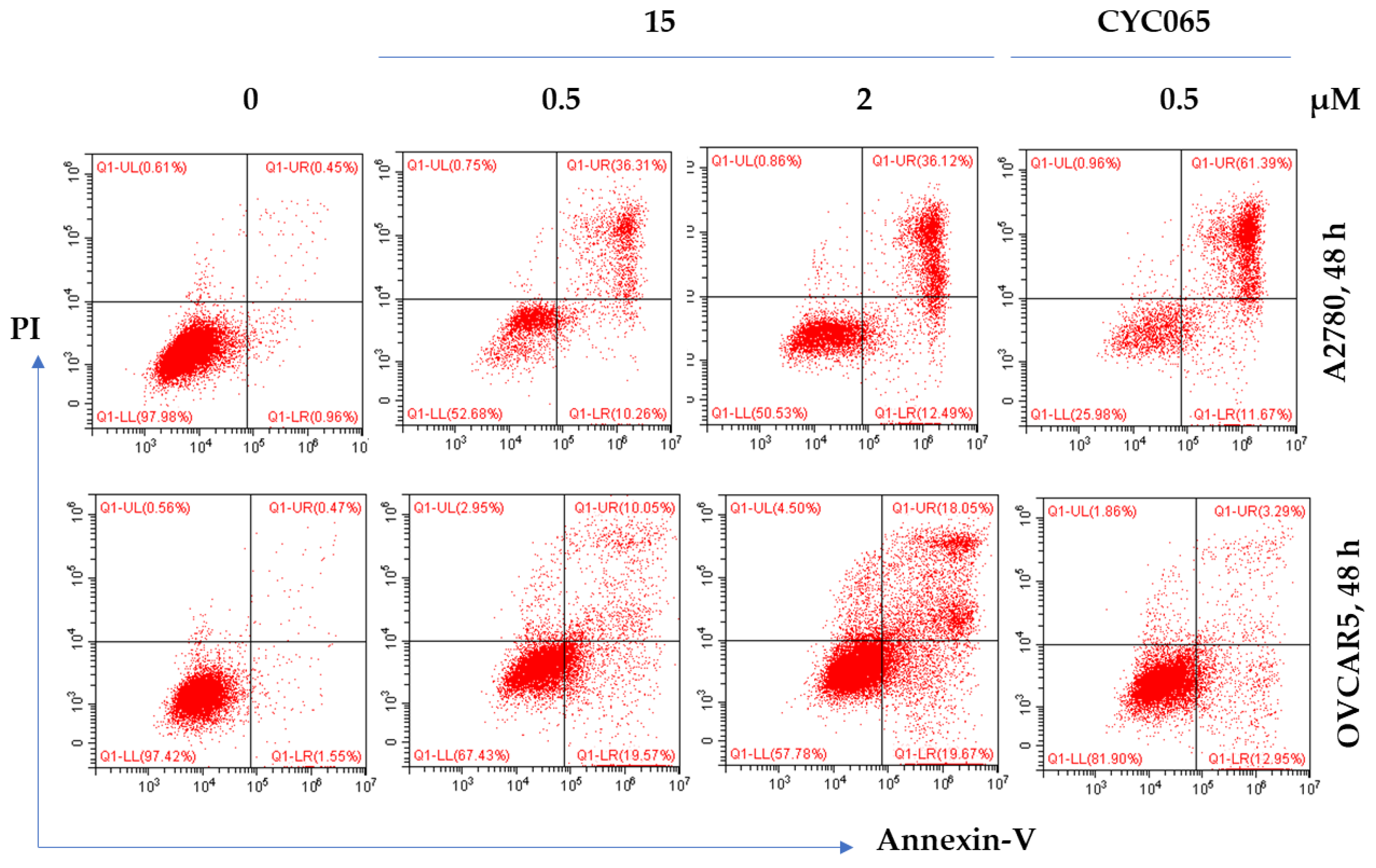
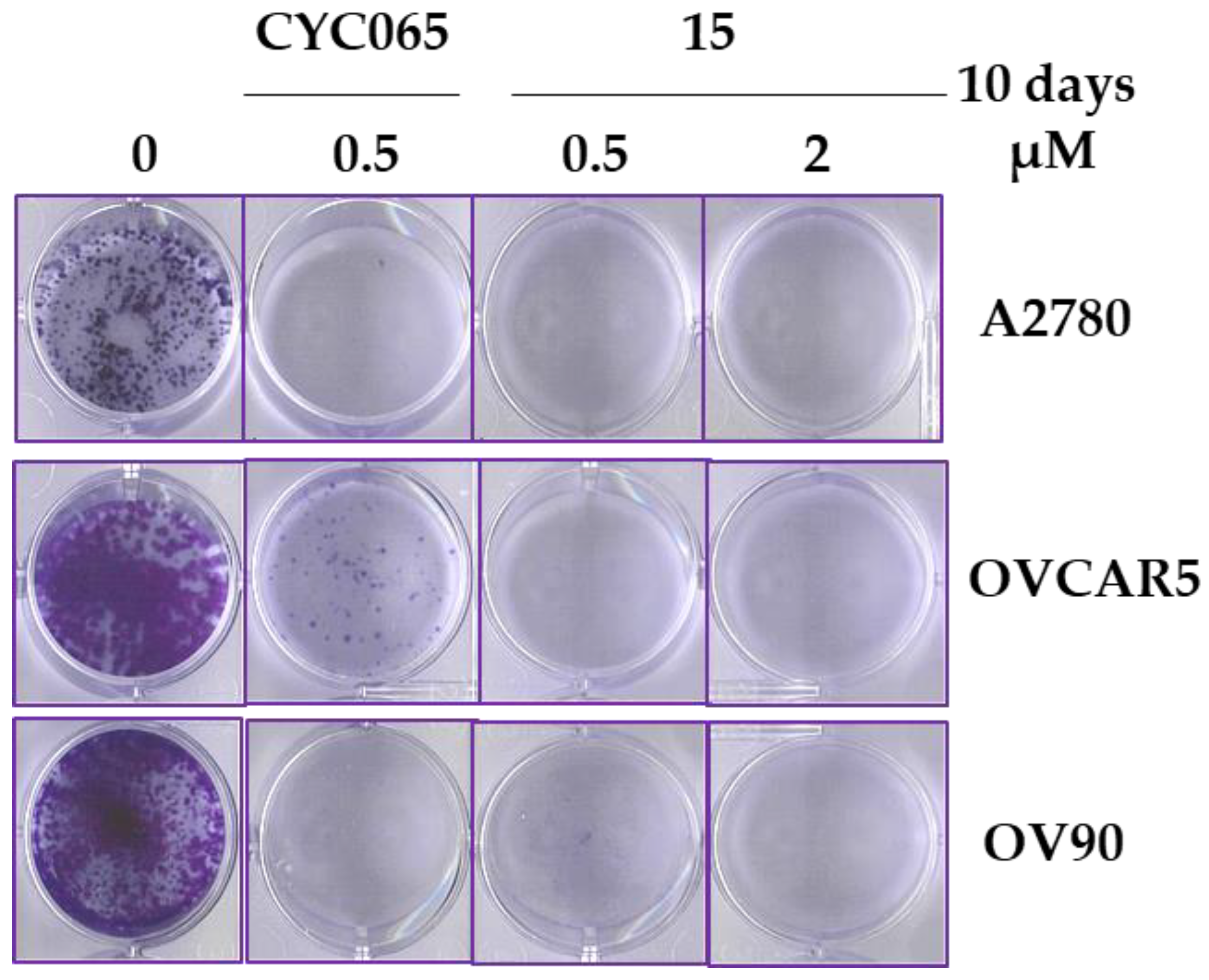
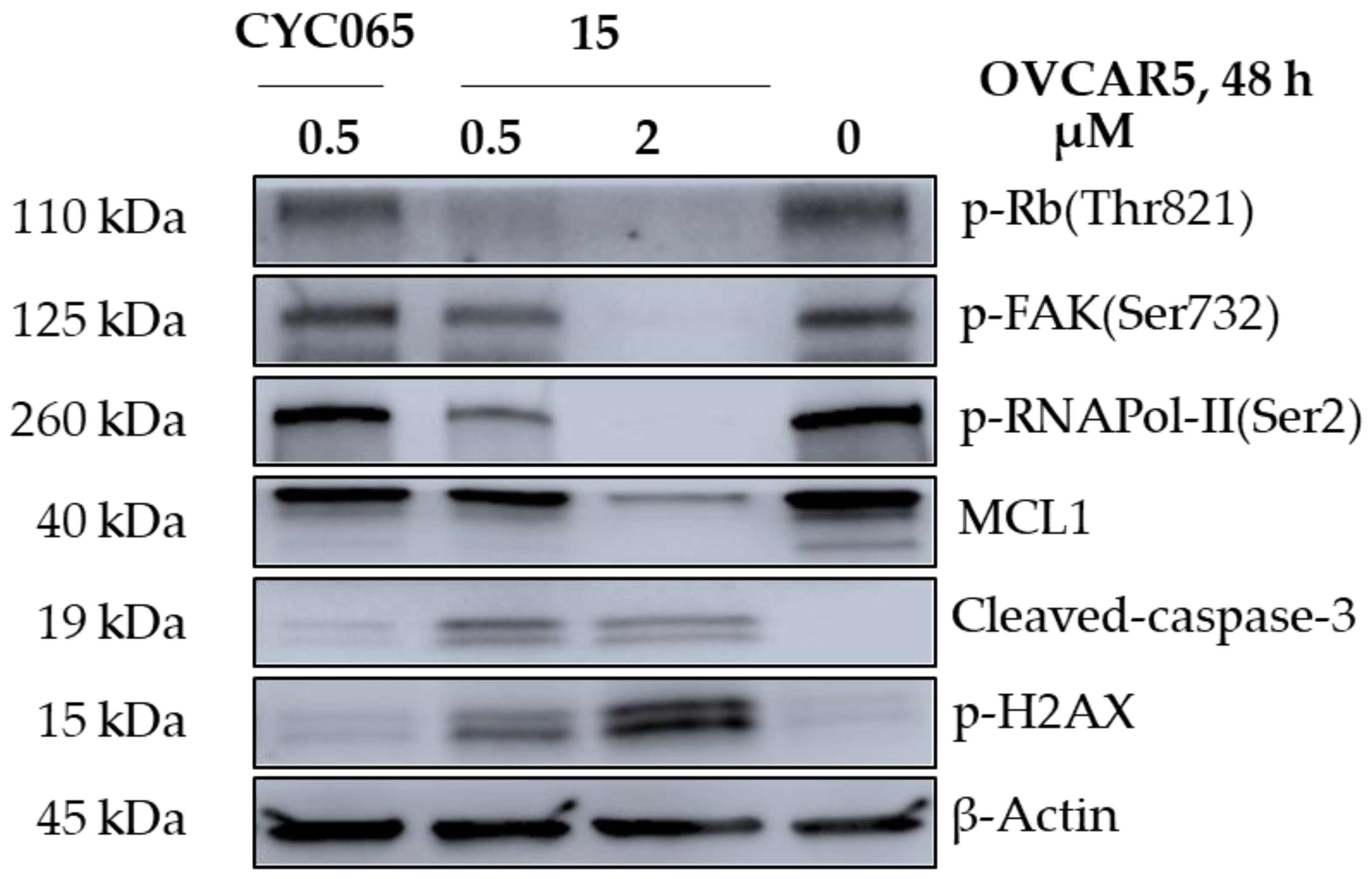
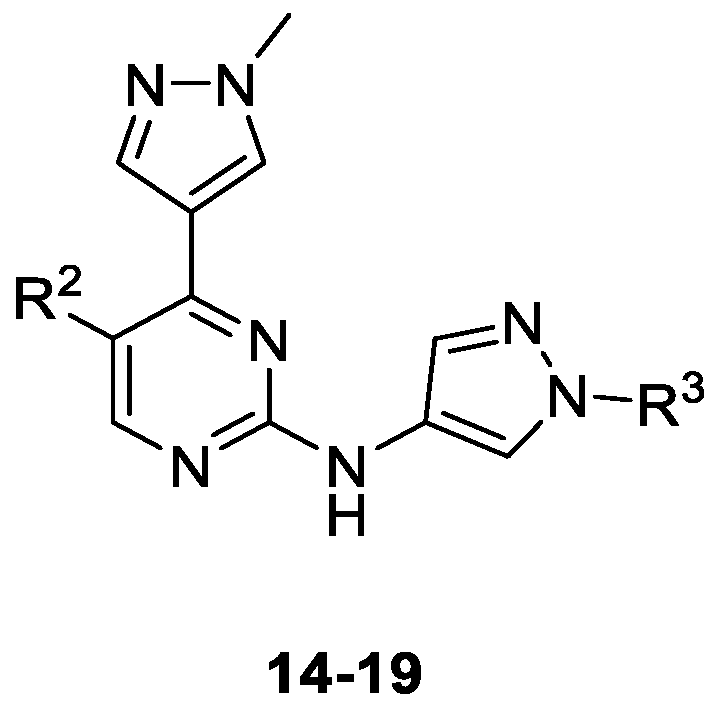
| Compound | Substituent | Ki (µM) or % Enzymatic Inhibition at 1 µM * | GI50 (µM) or % Growth Inhibition (GI) at 1 µM * | ||||
|---|---|---|---|---|---|---|---|
| R2 | R3 | CDK1B | CDK2E | CDK5p25 | CDK9T1 | A2780 | |
| 14 | F | H | 0.018 | 0.007 | 0.003 | 0.016 | 0.498 |
| 15 | Cl | H | 0.025 | 0.005 | 0.003 | 0.020 | 0.158 |
| 16 | F | Me | 0.092 | 0.023 | 0.018 | 0.098 | 9 * |
| 17 | Cl | Me | 0.034 | 0.011 | 0.006 | 0.056 | 1.023 |
| 18 | F | CH2CONH2 | 0.112 | 0.085 | 0.193 | 68* | 0 * |
| 19 | Cl | CH2CONH2 | 0.018 | 0.025 | 0.052 | 0.101 | 44 * |
| 1 | - | 0.005 | 0.023 | 0.077 | 0.026 | 0.018 | |
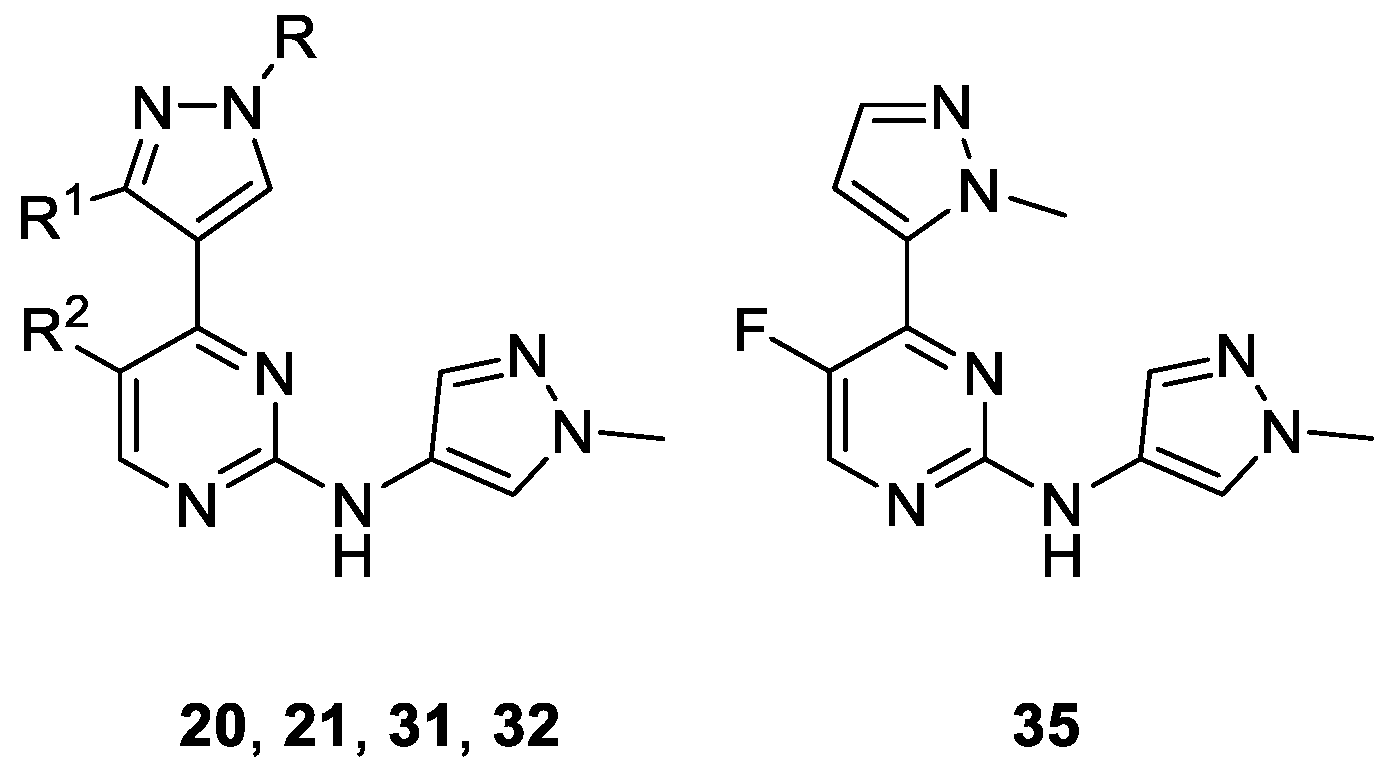
| Compound | Substituent | Ki (µM) or % Enzymatic Inhibition at 1 µM * | %GI at 1 µM * | |||||
|---|---|---|---|---|---|---|---|---|
| R | R1 | R2 | CDK1B | CDK2E | CDK5p25 | CDK9T1 | A2780 | |
| 20 | Me | Me | F | 0.164 | 0.026 | 0.018 | 0.343 | 7 * |
| 21 | Me | Me | Cl | 0.225 | 0.023 | 0.035 | 0.062 | 8 * |
| 31 | 2-morpholinoethyl | H | F | 26 * | 48 * | 43 * | 24 * | 0 * |
| 32 | 2-morpholinoethyl | H | Cl | 36 * | 0.252 | 0.126 | 21 * | 3 * |
| 35 | - | 49 * | 64 * | 58 * | 45 * | 19 * | ||
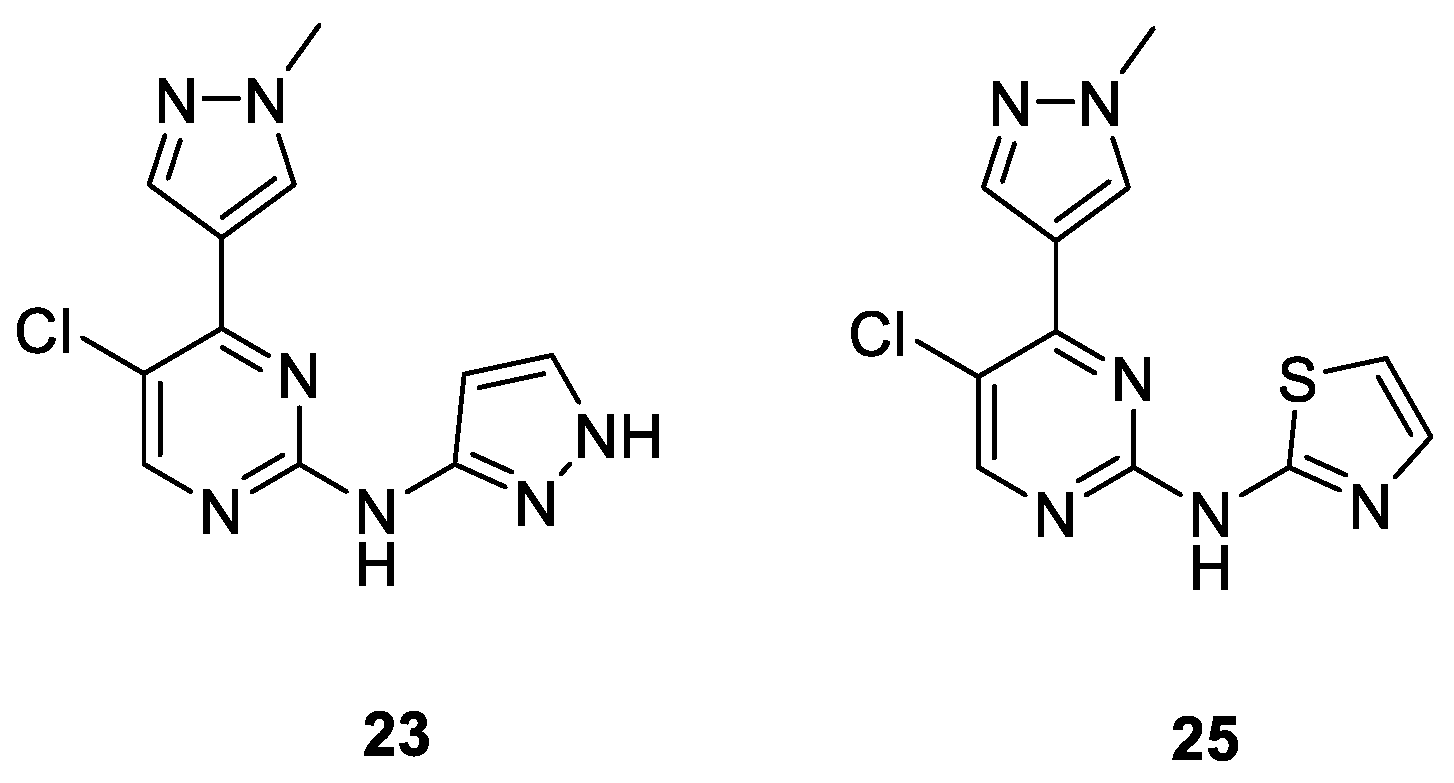
| Compound | Ki (µM) or % Enzymatic Inhibition at 1 µM * | GI50 (µM) or %GI at 1 µM * | |||
|---|---|---|---|---|---|
| CDK1B | CDK2E | CDK5p25 | CDK9T1 | A2780 | |
| 23 | 9 * | 0.090 | 0.040 | 40 * | 7.350 |
| 25 | 31 * | 46 * | 0.177 | 31 * | 9 * |
| Cancer Type | Cancer Cell Line | Antiproliferative Activity, GI50 (µM) |
|---|---|---|
| Leukemia | MV4-11 | 0.127 |
| U937 | 0.186 | |
| Breast | MCF7 | 0.342 |
| MDA-MB-231 | 0.458 | |
| Ovarian | OVCAR5 | 0.150 |
| OAW28 | 0.534 | |
| OV90 | 0.390 | |
| Cov318 | 0.488 | |
| Cov504 | 0.540 | |
| Glioblastoma | U87 | 0.560 |
| T98G | 0.380 | |
| U251 | 0.350 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fanta, B.S.; Lenjisa, J.; Teo, T.; Kou, L.; Mekonnen, L.; Yang, Y.; Basnet, S.K.C.; Hassankhani, R.; Sykes, M.J.; Yu, M.; et al. Discovery of N,4-Di(1H-pyrazol-4-yl)pyrimidin-2-amine-Derived CDK2 Inhibitors as Potential Anticancer Agents: Design, Synthesis, and Evaluation. Molecules 2023, 28, 2951. https://doi.org/10.3390/molecules28072951
Fanta BS, Lenjisa J, Teo T, Kou L, Mekonnen L, Yang Y, Basnet SKC, Hassankhani R, Sykes MJ, Yu M, et al. Discovery of N,4-Di(1H-pyrazol-4-yl)pyrimidin-2-amine-Derived CDK2 Inhibitors as Potential Anticancer Agents: Design, Synthesis, and Evaluation. Molecules. 2023; 28(7):2951. https://doi.org/10.3390/molecules28072951
Chicago/Turabian StyleFanta, Biruk Sintayehu, Jimma Lenjisa, Theodosia Teo, Lianmeng Kou, Laychiluh Mekonnen, Yuchao Yang, Sunita K. C. Basnet, Ramin Hassankhani, Matthew J. Sykes, Mingfeng Yu, and et al. 2023. "Discovery of N,4-Di(1H-pyrazol-4-yl)pyrimidin-2-amine-Derived CDK2 Inhibitors as Potential Anticancer Agents: Design, Synthesis, and Evaluation" Molecules 28, no. 7: 2951. https://doi.org/10.3390/molecules28072951
APA StyleFanta, B. S., Lenjisa, J., Teo, T., Kou, L., Mekonnen, L., Yang, Y., Basnet, S. K. C., Hassankhani, R., Sykes, M. J., Yu, M., & Wang, S. (2023). Discovery of N,4-Di(1H-pyrazol-4-yl)pyrimidin-2-amine-Derived CDK2 Inhibitors as Potential Anticancer Agents: Design, Synthesis, and Evaluation. Molecules, 28(7), 2951. https://doi.org/10.3390/molecules28072951









