Abstract
Understanding the non-covalent interactions in host-guest complexes is crucial to their stability, design and applications. Here, we use density functional theory to compare the ability of β-cyclodextrin (β-CD) and heptakis(2,6-di-O-methyl)-β-cyclodextrin (DM-β-CD) to encapsulate the model guest phenol. For both macrocycles, we quantify the intramolecular interactions before and after the formation of the complex, as well as the intermolecular host-guest and host-host dimer interactions. These are individually classified as van der Waals interactions or hydrogen bonds, respectively. The results show a stronger intramolecular binding energy of β-CD, with the absolute difference being −5.53 kcal/mol relative to DM-β-CD. Consequently, the intermolecular interactions of both cyclodextrins with phenol are affected, such that the free binding energy calculated for the DM-β-CD/phenol complex (−5.23 kcal/mol) is ≈50% more negative than for the complex with β-CD (−2.62 kcal/mol). The latter is in excellent agreement with the experimental data (−2.69 kcal/mol), which validates the level of theory (B97-3c) used. Taken together, the methylation of β-CD increases the stability of the host-guest complex with the here studied guest phenol through stronger van der Waals interactions and hydrogen bonds. We attribute this to the disruption of the hydrogen bond network in the primary face of β-CD upon methylation, which influences the flexibility of the host toward the guest as well as the strength of the intermolecular interactions. Our work provides fundamental insights into the impact of different non-covalent interactions on host-guest stability, and we suggest that this theoretical framework can be adapted to other host-guest complexes to evaluate and quantify their non-covalent interactions.
1. Introduction
Cyclodextrins (CDs) consist of circularly connected glucose units, joined by α-1,4 glycosidic bonds (Figure 1a), and are well-known to encapsulate small molecules forming host-guest complexes [1,2,3,4]. The general structure of CDs is toroidal, with two openings (“faces”), and the primary and secondary alcohol groups are located at the smaller and larger opening, respectively (Figure 1). The inside of CDs is hydrophilic, but significantly less hydrophilic than the common aqueous environment, and hence both apolar and polar guests can be hosted in their cavity.
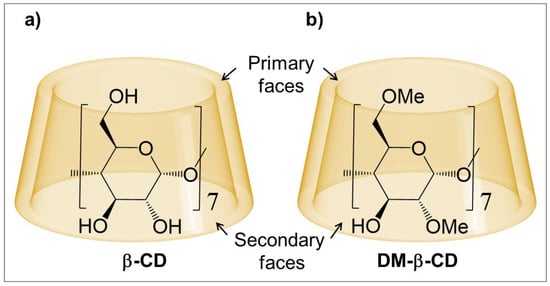
Figure 1.
Schematic of the macrocycles: (a) β-Cyclodextrin (β-CD) and (b) Heptakis(2,6-di-O-methyl)-β-cyclodextrin (DM-β-CD).
Host-guest complexes with CDs have attracted broad interest in the last decades, driven by myriad applications, including drug delivery [5,6,7,8], food science [9,10], sensing [11,12], petroleum chemistry [13] and pollutant removal [14,15]. These inclusion complexes have been studied and extensively characterized by a range of analytical techniques [16]; however, decoupling their non-covalent interactions on a fundamental level is often difficult with experimental solution phase techniques, where all non-covalent interactions contribute simultaneously [17]. Interactions in CD inclusion complexes have been extensively modelled with computational techniques [18], predominantly with molecular dynamics simulations [19] and density functional theory (DFT) [20]. The latter was shown to be ideally suited to model the non-covalent interactions of CD complexes with a high level of accuracy [20], and recently a method to estimate the energies and nature of non-covalent interactions from the density of critical bonding points has been reported [21]. We suggest that this methodology can also be applied to CD-based host-guest complexes.
In CDs, the number of glucose units controls the size of their cavity, and possibly the most prevalent cyclodextrin is β-cyclodextrin (β-CD), which involves seven glucopyranoside monomers (Figure 1a). One major drawback of β-CD is its low solubility in water [22]; however, a significant increase is observed when substituting the hydroxyl groups with neutral or ionic substituents [23], e.g., with methyl groups [24]. Methylation of CDs not only increases their solubility, but also enhances the stability of their host-guest complexes [25,26,27,28,29,30,31,32,33,34], for example up to ≈1.2 kcal/mol for the guests methyl orange and 8-anilino-1-naphthalenesulfonate [31,32].
Methylated (β)-CDs have been proposed for applications in polymer chemistry [35,36,37], soil decontamination [38,39], batteries [40], pharmaceutics [41] and as chiral selectors in electrophoresis [42,43], and for most of these the formation of highly stable host-guest complexes is essential. Hence, a precise understanding of their non-covalent interactions is warranted in order to tune their inclusion complex properties. Previously, Wenz and Höfler investigated the effect of methylation on the energetics of β-CD host-guest complexes using isothermal titration microcalorimetry, showing that binding free energies and enthalpies significantly depend on the degree and place of β-CD methylation [33,44]. More precisely, Wenz found that methylation in the primary face enhances the binding constant, whereas the opposite was observed for methylation in the secondary face [33].
Applying DFT, we extend the work by Wenz to reveal the impact of methylation on the hosting ability of β-CD, using the methylated cyclodextrin heptakis(2,6-di-O-methyl)-β-CD, often referred to as dimethyl-β-cyclodextrin (DM-β-CD, Figure 1b). Compared to β-CD, the primary and one of the two secondary hydroxy (-OH) groups are substituted for methoxy (-OCH3) groups in each of the monomers of DM-β-CD. This macrocycle is therefore ideally suited to assess the methylation in both faces of β-CD, while still one secondary -OH group remains as an internal comparison to β-CD. In this context, the characterization and comparison of the different non-covalent interactions in the CD inclusion complexes is possible, in particular (i) intramolecular interactions within the CD hosts to determine the effect of the -OH and -OCH3 groups, (ii) intermolecular interactions between the CD hosts and the guest (in this study phenol) to compare the complex stability in terms of van der Waals (vdW) interactions, hydrogen bonds (HBs) and binding free energy, and (iii) the formation of host dimers to estimate their contribution to the underestimation of the observed binding constant.
2. Results and Discussion
2.1. Intramolecular Interactions within the β-CD and DM-β-CD Macrocycles
The intramolecular interactions in β-CD and DM-β-CD were evaluated, and since both constitute of seven monomeric units, we investigated the seven intramolecular host interactions occurring between the monomers on both faces. As shown in Figure 2, β-CD exhibits intramolecular HBs on both faces, while DM-β-CD only has HBs on the secondary face. This leads to a higher degree of freedom of the primary face in DM-β-CD, compared to the highly rigid β-CD [45]. The only significant contribution to the interactions in the primary face of DM-β-CD is vdW interactions (green regions, Figure 2b), whose energies were estimated together with those of the HBs in the primary face (Table 1).
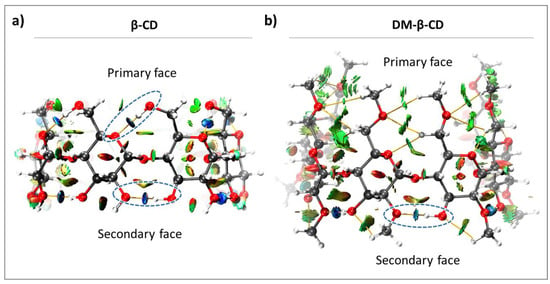
Figure 2.
Color-filled RDG isosurface for intramolecular interactions within (a) β-CD and (b) DM-β-CD. The different types of interaction regions are highlighted, blue: attractive interactions (hydrogen bonds), green: slightly attractive vdW interactions, red: repulsive steric interactions. Hydrogen bonds are additionally marked with blue, dashed ellipsoids.

Table 1.
Intramolecular interaction energies (in kcal/mol) within β-CD (left) and DM-β-CD (right), calculated from the electron density at the critical bond points. The standard error of the mean was calculated for both faces of both hosts.
The individual HB interaction energies, estimated for each face, are largely constant in β-CD, and this can be attributed to its high symmetry and structural rigidity (Table 1). In contrast, the individual intramolecular interactions of DM-β-CD show more fluctuations, particularly in the secondary face (standard error in Table 1). This is due to a substantial loss of symmetry/rigidity between the monomers that constitute DM-β-CD, caused by the absence of HBs at the primary face and the electronic repulsions experienced by the -OCH3 groups. The absolute average energies of the intramolecular interactions were significantly different for both macrocycles, with β-CD having stronger intramolecular interaction on both faces (Table 1). The primary face of β-CD shows a 5.53 kcal/mol higher averaged intramolecular interaction energy than DM-β-CD, while the intramolecular interactions on the secondary face yielded a smaller difference of 1.55 kcal/mol between both hosts.
Taken together, the intramolecular interactions within DM-β-CD are weaker than for β-CD, due to methylation and the absence of intramolecular HBs in the primary face of DM-β-CD. Hence, DM-β-CD is less rigid than β-CD and is expected to be more flexible in encapsulating guest molecules.
2.2. Intermolecular Interactions between β-CD/DM-β-CD and the Model Guest Phenol
After evaluating the non-covalent interactions within both CD hosts, we investigated the intermolecular interactions between β-CD/DM-β-CD and the guest phenol occurring in host-guest complexes of the stoichiometry 1:1 [46]. We chose phenol for this purpose as it is a simple molecule constituted of a hydrophobic group (phenyl moiety) and a hydrogen bond acceptor and donor (hydroxyl group). This makes it possible to reliably evaluate both vdW interactions and hydrogen bonds occurring with the CD hosts [26].
The DFT optimized structure of the β-CD/phenol inclusion complex shows a docked host-guest conformation, with most of the phenol being included within the cavity. The hydroxyl group interacts favourably with the portals of the primary face of β-CD (via OH-OH), and this is in agreement with previous results obtained from NMR spectroscopy [46]. Similar results were found for the DM-β-CD/phenol complex, where the guest hydroxyl group interacts with the primary face of DM-β-CD (via OH-OCH3). Because the hydroxyl group of the phenol interacts with the same side of both CDs (Figure 3), the effect of the CD methylation at the primary face on the stability of the inclusion complex can be directly compared (Table 2). The results show that the intermolecular HB energy of β-CD (between the host and guest hydroxyl groups) was −4.99 kcal/mol, while the OH-OCH3 interaction in the DM-β-CD/phenol complex was stronger (−7.31 kcal/mol). This difference can be rationalized with the intramolecular interactions in the CD hosts, specifically the intramolecular interaction of the host oxygen that interacts with the hydroxyl group of phenol. Here, the β-CD/phenol complex presents with a strong HB of −5.38 kcal/mol, which is highly similar to the free β-CD as discussed above (Table 1). For the DM-β-CD/phenol complex, the strong and attractive intramolecular interaction does not occur (+0.17 kcal/mol, Table 2), and this allows the -OCH3 groups to form strong OH-OCH3 HBs with phenol.
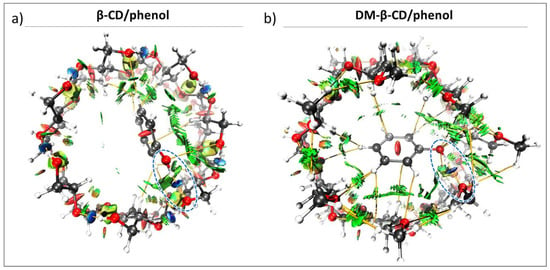
Figure 3.
Color-filled RDG isosurface for intermolecular host-guest interactions between (a) β-CD/phenol and (b) DM-β-CD/phenol. The different types of interaction regions are highlighted in different colors; blue: attractive interactions (hydrogen bonds), green: slightly attractive vdW interactions, red: repulsive steric interactions. Hydrogen bonds between the host and phenol are additionally marked with blue, dashed ellipsoids.

Table 2.
Binding parameters of CD/phenol host-guest complexes (in kcal/mol).
For the β-CD/phenol and DM-β-CD/phenol complexes, the interaction energies (−14.96 kcal/mol and −18.27 kcal/mol, respectively) are mainly governed by vdW interactions (ca. 70%, Table 2), as previously reported for similar complexes [28,34,47]. The estimated ΔΕvdW for the β-CD/phenol complex is −11.40 kcal/mol, while for the DM-β-CD/phenol complex, the energy is −14.67 kcal/mol. This difference can be explained by the greater flexibility that DM-β-CD/phenol possesses by having a weaker network of intramolecular interactions compared to β-CD (Table 1) [33,48]. This is in contrast to the rationalization suggested by Wenz, who assumed the absence of an intramolecular hydrogen network in the primary face of β-CD due to the too long distance between the hydroxyl groups, and hence that methylation there had no major influence on the rigidity of the host [33]. In our work, we found intramolecular HBs in the primary face of β-CD (discussed above) and a higher flexibility when these were absent in DM-β-CD. This apparently allows DM-β-CD to organize itself and adapt a favorable cavity for phenol, changing the position of the guest within the complex to a conformation with stronger vdW interactions (Figure 3, green).
Finally, ΔGcalc was calculated for both complexes (Table 2). The DM-β-CD/phenol complex has a binding energy of −5.23 kcal/mol, whereas the one of the β-CD/phenol complex is −2.62 kcal/mol. The latter is in excellent agreement with calorimetric data previously reported (−2.69 kcal/mol) [26], and this validates the level of theory used in this work. The absolute difference in binding energy (2.61 kcal/mol, ≈50%) between both complexes supports the points discussed above, and suggests that DM-β-CD is a better host than β-CD for the guest phenol. For the same reasons, we also anticipate that DM-β-CD, compared to β-CD, is better suited to form stable host-guest complexes with alcohols in general.
2.3. Dimers of the Macrocycles β-CD and DM-β-CD
A detailed knowledge of the species present in assays with macrocycles such as CDs is important for experimentally determining host-guest binding constants of a given complex. The latter depends on the concentration of the free host and hence the formation of dimers (or even oligomers) leads to an absolute concentration decrease, resulting in an underestimation of the binding constant. Thus, the stability of the β-CD and DM-β-CD-based dimers is linked to the differences in their observed host-guest binding constants and should hence be considered when discussing host-guest complex stability.
The dimer formation of CDs was previously reported in the literature, and the favoured host-host conformation of β-CD is head-to-head (Figure 4a) [49,50]. For both macrocycles, inter- and intramolecular HBs as well as vdW interactions were quantified (Table 3). The β-CD dimer was found to be more stable (ΔΕint = −53.88 kcal/mol) than the DM-β-CD dimer (ΔΕint = −41.65 kcal/mol), and the comparison of ΔGcalc for both dimers was not possible, as the value for DM-β-CD could not be determined. The large ΔΕint difference can be rationalized with the different dimer conformations (head-to-head for β-CD and head to main body for DM-β-CD, Figure 4a,b) and the occurrence of intermolecular hydrogen bonds for β-CD (−5.33 kcal/mol averaged over each monomer, Table 3, Figure 4c). In contrast, the head to main body dimer of DM-β-CD lacks intermolecular HB interactions (Figure 4d). This is also quantitatively reflected in the electrostatic interaction energies of both dimers, where the β-CD dimer yields ΔΕele = −46.40 kcal/mol and the DM-β-CD dimer ΔΕele = −13.28 kcal/mol (Table 3). Noteworthy is also that the averaged individual intramolecular interaction energy in the β-CD dimer (−5.22 kcal/mol, Table 3) is significantly weaker compared to the one of the secondary face of the β-CD monomer (−5.86 kcal/mol, Table 1), which once more highlights the interplay between intramolecular and intermolecular interactions.
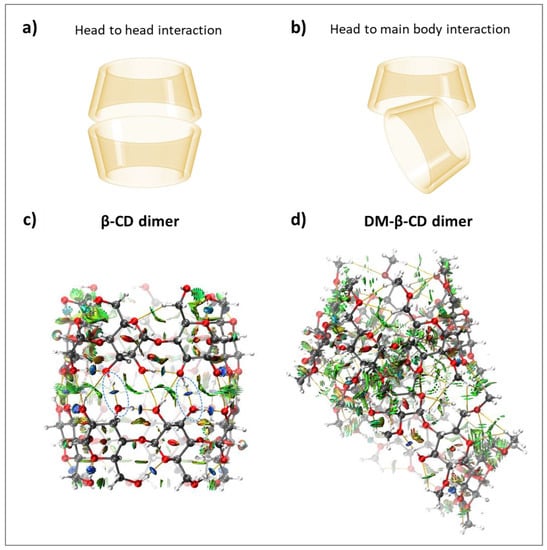
Figure 4.
Schematic of (a) head-to-head interactions and (b) head to main body interactions of β-CD and DM-β-CD dimers, respectively. Color-filled RDG isosurface for intermolecular host-host interactions between (c) β-CD dimers and (d) DM-β-CD dimers. The different types of interaction regions are highlighted in different colors, blue: attractive interactions (hydrogen bonds), green: slightly attractive vdW interactions, red: repulsive steric interactions. Hydrogen bonds are additionally marked with blue, dashed ellipsoids.

Table 3.
Binding parameters for the host-host dimers of β-CD and DM-β-CD in kcal/mol.
3. Methods
3.1. Preparation of Initial Structures and Geometry Optimization
The initial structures of β-CD and DM-β-CD were obtained from X-ray crystallography data [51,52]. These were used as inputs for electronic structure optimization using density functional theory (method-basis set B97-3c) [53,54], under the TightSCF convergence protocol established by default in the ORCA program package (Program Version 5.0.3) [55]. The method-basis set B97-3c was chosen because it is a low-cost and reliable level of theory [54], suitable for evaluating large systems such as cyclodextrin dimers. The accuracy was validated with the comparison of the free binding energy of β-CD/phenol with the experimental literature data (Table 2), and hence other levels of theory were not tested. The local energy minimum for each electronic structure was confirmed as all vibrational modes presented positive magnitudes.
The initial interaction conformation of the host-host and host-guest complexes were docked from the Autodock program (Version 4.2) [56] using Merz-Kollmann (MK) charges [57], for a 1:1 complex stoichiometry ratio. The MK charges were calculated from the optimized structures of the monomers using the Multiwfn program (Version 3.8) [58]. Docked complexes were optimized as described above for β-CD and DM-β-CD. Optimized coordinates and minimum energies of β-CD, DM-β-CD and phenol, as well as the corresponding host-guest complexes and host-host dimers, can be found in the supplementary dataset for both gas phase and aqueous phase calculations.
3.2. Noncovalent Interaction (NCI) Analysis
The NCI analysis was carried out by the reduced density gradient (RDG) method as described by Yang et al. [59], and as implemented in the Multiwfn program [58]. The electron densities (ρ) of the monomers and complexes were mapped according to Equation (1), as a function of the sign of the second largest eigenvalue of the Hessian matrix of electron density (sign(λ2)) multiplied by ρ, to distinguish between different types of noncovalent interactions at the critical bond points obtained from Bader’s AIM theory (Figure S1 for β-CD/DM-β-CD including the description of the color scale and Figure S2 for the β-CD/DM-β-CD complexes with phenol as well as the dimers of β-CD and DM-β-CD, respectively) [60]. For all cases, the chosen isovalue was 0.6, adequate for constructing the RDG isosurface and to reveal all the weak interactions involved in a 3D space.
3.3. Calculation of the Individual Non-Covalent Interactions Energies
The energies of the non-covalent interactions involved in the monomers and the different dimers/host-guest complexes were calculated as described by Lu et al. [21]. Each of the structures optimized with the B97-3c level of theory underwent a single point calculation with b3lyp-D3BJ/ma-TZVPP, corresponding to the method and basis set used by Lu et al. in their predictive model (Equation (2)) [21].
Here, the mean absolute percentage error of prediction (MAPE) is 14.7% and is the electron density at the critical bond point of the corresponding interaction, determined by the Multiwfn program (Version 3.8) [58].
3.4. Calculation of the Complex Formation Interaction Energies (ΔΕint)
The interaction energies of the host-guest complexes and host-host dimers were calculated by the total electronic energy difference between the complex and the isolated constituting building blocks. The electronic structure of each building block was the same individually as in the supramolecular complex. The basis set superposition error (BSSE) of the complexes was calculated and considered in the correction of the total electronic energy. All energies described were determined using the B97-3c level of theory [54], implemented in the ORCA program package (Program Version 5.0.3) [55].
3.5. Calculation of the Complex Formation Interaction Energies Based on Molecular Forcefield (ΔΕint-FF)
The ΔΕint-FF was calculated as the sum of the electrostatic interaction energy (ΔΕele) and the vdW interaction energy (ΔΕvdW) using the potential of pairwise non-covalent interactions (Equations (3) and (4)) [61].
and are the partial charges centred on the individual atoms of the building blocks A and B, respectively. is the separation distance and the vacuum permittivity. The MK charges were calculated [57] and used as partial charges in each building block, with the exact conformation that they individually possess in the supramolecular complex through a single point energy calculation at the B97-3c level of theory.
The parameters and [62], defined as and , were obtained using GAFF and AMBER99 force fields, using the molecular structure of the complex obtained by DFT (as discussed above). Equations (3) and (4) were calculated by the Multiwfn program [58].
3.6. Calculation of the Complex Formation Free Binding Energies (ΔGcalc)
The free binding energy for the formation of the complex was estimated from Equation (5) [63].
The free binding energies of the building blocks A and B, as well as of the complex AB were calculated in the aqueous phase (ΔGaq) using the B97-3c level of theory at 298.15 K and 1 atm from the ORCA program, with previous optimization of their structures assuming implicit solvent (water). The solvation was simulated using the conductor-like polarizable continuum model with the COSMO epsilon function (CPCMC) [64]. Vibrational entropy was computed according to the quasi-rigid-rotor harmonic oscillator (QRRHO) approximation [65].
4. Conclusions
With this work, we characterize the non-covalent interactions of β-CD and DM-β-CD host-guest complexes, and more precisely the intramolecular interactions within the hosts, the intermolecular host-guest interactions with phenol as well as the host dimer interactions. In support of experimental data, our results suggest significantly more stable host-guest complexes of the methylated host DM-β-CD with phenol compared to β-CD. We attribute this behaviour to the disruption of the HB network in the primary face of β-CD upon methylation, which leads to a greater flexibility, and hence a larger stabilization of the guest through both vdW interactions and stronger intermolecular HBs.
For the intramolecular interactions in the CDs, free β-CD presented a network of strong hydrogen bonds with an average energy of −5.66 kcal/mol on the primary face and −5.86 kcal/mol on the secondary face. In contrast, the free DM-β-CD yielded lower and more fluctuating energies, attributable to the loss of symmetry in the CD portals (due to electronic repulsions upon methylation) and the absence of intramolecular HBs in the primary face.
The lack of the latter in DM-β-CD allows on the one hand -OCH3 groups to form stronger intermolecular interactions with hydrogen bond donor groups such as the -OH group of phenol (OH-OCH3, ≈1.5-fold relative to the hydroxyl groups of β-CD), and on the other hand leads to a greater flexibility with respect to the guest, where stronger intermolecular vdW interactions occur (≈1.3-fold).
When investigating the dimerization of both CD hosts, we found a greater stability of the β-CD dimer compared to DM-β-CD. Although this is not directly related to the observed stability of the CD-phenol complexes on a molecular level as discussed above, the greater stability of the β-CD dimer over the DM-β-CD dimer suggests that if these dimers are formed in such host-guest assays, the binding constant in the presence of β-CD will be underestimated to a greater extent than the constant obtained in the presence of DM-β-CD.
Taken together, the applied methodology presents a complementary way of assessing non-covalent interactions in host-guest complexes, and we propose this theoretical framework as a suitable addition to the analytical toolkit of such supramolecular structures.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules28062625/s1. Color-filled scatter plots of the reduced density gradient versus the sign of the second Hessian eigenvalue multiplied by the electron density for β-CD, DM-β-CD, the β-CD dimer, the DM-β-CD dimer, the β-CD/phenol complex and the DM-β-CD/phenol complex; including the description of the color scale.
Author Contributions
Conceptualization, J.J.A.; Formal analysis, J.J.A.; Funding acquisition, P.R.C.; Investigation, N.G. and J.J.A.; Methodology, J.J.A.; Software, J.J.A.; Visualization, J.J.A.; Writing—original draft, N.G. and J.J.A.; Writing—review & editing, N.G., J.J.A. and P.R.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The supporting data referred to in this manuscript is contained within a Supplementary Information Document and in a Supplementary Dataset available on Figshare 10.6084/m9.figshare.22010837.
Acknowledgments
J.J.A. thanks the Vicerectoria de Investigacion y Doctorado (VID), Universidad de Desarrollo, for a postdoctoral fellowship. The authors acknowledge FONDEQUIP EQM150093 for computational resources, Eliecer Pérez for designing the table of contents graphic and the Instituto de Ciencias e Innovación en Medicina (ICIM), Facultad de Medicina, Universidad de Desarrollo, for general support.
Conflicts of Interest
There are no conflict to declare.
References
- Li, S.; Purdy, W.C. Cyclodextrins and Their Applications in Analytical Chemistry. Chem. Rev. 1992, 92, 1457–1470. [Google Scholar] [CrossRef]
- Dodziuk, H. Cyclodextrins and Their Complexes: Chemistry, Analytical Methods, Applications; Wiley-VCH: Weinheim, Germany, 2006; ISBN 3527312803. [Google Scholar]
- Crini, G. Review: A History of Cyclodextrins. Chem. Rev. 2014, 114, 10940–10975. [Google Scholar] [CrossRef] [PubMed]
- Crini, G.; Fourmentin, S.; Fenyvesi, É.; Torri, G.; Fourmentin, M.; Morin-Crini, N. Cyclodextrins, from Molecules to Applications. Environ. Chem. Lett. 2018, 16, 1361–1375. [Google Scholar] [CrossRef]
- Brewster, M.E.; Loftsson, T. Cyclodextrins as Pharmaceutical Solubilizers. Adv. Drug Deliv. Rev. 2007, 59, 645–666. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.D.; Tang, G.P.; Chu, P.K. Cyclodextrin-Based Host-Guest Supramolecular Nanoparticles for Delivery: From Design to Applications. Acc. Chem. Res. 2014, 47, 2017–2025. [Google Scholar] [CrossRef]
- Braga, S.S. Cyclodextrins: Emerging Medicines of the New Millennium. Biomolecules 2019, 9, 801. [Google Scholar] [CrossRef]
- Tian, B.; Hua, S.; Liu, J. Cyclodextrin-Based Delivery Systems for Chemotherapeutic Anticancer Drugs: A Review. Carbohydr. Polym. 2020, 232, 115805. [Google Scholar] [CrossRef]
- Astray, G.; Mejuto, J.C.; Simal-Gandara, J. Latest Developments in the Application of Cyclodextrin Host-Guest Complexes in Beverage Technology Processes. Food Hydrocoll. 2020, 106, 105882. [Google Scholar] [CrossRef]
- Matencio, A.; Navarro-Orcajada, S.; García-Carmona, F.; López-Nicolás, J.M. Applications of Cyclodextrins in Food Science. A Review. Trends Food Sci. Technol. 2020, 104, 132–143. [Google Scholar] [CrossRef]
- Ogoshi, T.; Harada, A. Chemical Sensors Based on Cyclodextrin Derivatives. Sensors 2008, 8, 4961–4982. [Google Scholar] [CrossRef]
- Zhu, G.; Yi, Y.; Chen, J. Recent Advances for Cyclodextrin-Based Materials in Electrochemical Sensing. Trends Anal. Chem. 2016, 80, 232–241. [Google Scholar] [CrossRef]
- Tang, W.; Zou, C.; Da, C.; Cao, Y.; Peng, H. A Review on the Recent Development of Cyclodextrin-Based Materials Used in Oilfield Applications. Carbohydr. Polym. 2020, 240, 116321. [Google Scholar] [CrossRef]
- Liu, Q.; Zhou, Y.; Lu, J.; Zhou, Y. Novel Cyclodextrin-Based Adsorbents for Removing Pollutants from Wastewater: A Critical Review. Chemosphere 2020, 241, 125043. [Google Scholar] [CrossRef]
- Tian, B.; Hua, S.; Tian, Y.; Liu, J. Cyclodextrin-Based Adsorbents for the Removal of Pollutants from Wastewater: A Review. Environ. Sci. Pollut. Res. 2021, 28, 1317–1340. [Google Scholar] [CrossRef]
- Mura, P. Analytical Techniques for Characterization of Cyclodextrin Complexes in Aqueous Solution: A Review. J. Pharm. Biomed. Anal. 2014, 101, 238–250. [Google Scholar] [CrossRef]
- Geue, N.; Winpenny, R.E.P.; Barran, P.E. Structural Characterisation Methods for Supramolecular Chemistry That Go beyond Crystallography. Chem. Soc. Rev. 2022, 51, 8–27. [Google Scholar] [CrossRef]
- Sayede, A.; Monflier, E. Molecular Modeling of Cyclodextrin Inclusion Complexes. In Chemical Modelling; Springbord, M., Joswig, J.-O., Eds.; Royal Society of Chemistry: London, UK, 2021; Volume 16, pp. 72–99. ISBN 9781839161704. [Google Scholar]
- Mazurek, A.H.; Szeleszczuk, Ł.; Gubica, T. Application of Molecular Dynamics Simulations in the Analysis of Cyclodextrin Complexes. Int. J. Mol. Sci. 2021, 22, 9422. [Google Scholar] [CrossRef]
- Mazurek, A.H.; Szeleszczuk, Ł. Current Status of Quantum Chemical Studies of Cyclodextrin Host–Guest Complexes. Molecules 2022, 27, 3874. [Google Scholar] [CrossRef]
- Emamian, S.; Lu, T.; Kruse, H.; Emamian, H. Exploring Nature and Predicting Strength of Hydrogen Bonds: A Correlation Analysis Between Atoms-in-Molecules Descriptors, Binding Energies, and Energy Components of Symmetry-Adapted Perturbation Theory. J. Comput. Chem. 2019, 40, 2868–2881. [Google Scholar] [CrossRef]
- Coleman, A.W.; Nicolis, I.; Keller, N.; Dalbiez, J.P. Aggregation of Cyclodextrins: An Explanation of the Abnormal Solubility of β-Cyclodextrin. J. Incl. Phenom. Mol. Recognit. Chem. 1992, 13, 139–143. [Google Scholar] [CrossRef]
- Saokham, P.; Muankaew, C.; Jansook, P.; Loftsson, T. Solubility of Cyclodextrins and Drug/Cyclodextrin Complexes. Molecules 2018, 23, 1161. [Google Scholar] [CrossRef] [PubMed]
- Fenyvesi, É.; Szemán, J.; Csabai, K.; Malanga, M.; Szente, L. Methyl-Beta-Cyclodextrins: The Role of Number and Types of Substituents in Solubilizing Power. J. Pharm. Sci. 2014, 103, 1443–1452. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, W.; Nguyen, T.A.; Zhao, J. Empirical, Thermodynamic and Quantum-Chemical Investigations of Inclusion Complexation between Flavanones and (2-Hydroxypropyl)-Cyclodextrins. Food Chem. 2012, 134, 926–932. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, G.L.; Faulkner, J.R.; Han, S.M.; Armstrong, D.W. Substituent Effects on the Binding of Phenols to Cyclodextrins in Aqueous Solution. J. Phys. Chem. 1989, 93, 6863–6867. [Google Scholar] [CrossRef]
- Acuña-Rougier, C.; Olea-Azar, C. Thermodynamic and Geometric Study of Diasteroisomeric Complexes Formed by Racemic Flavanones and Three Cyclodextrins through NMR. J. Incl. Phenom. Macrocycl. Chem. 2013, 75, 119–136. [Google Scholar] [CrossRef]
- Sangpheak, W.; Kicuntod, J.; Schuster, R.; Rungrotmongkol, T.; Wolschann, P.; Kungwan, N.; Viernstein, H.; Mueller, M.; Pongsawasdi, P. Physical Properties and Biological Activities of Hesperetin and Naringenin in Complex with Methylated P-Cyclodextrin. Beilstein J. Org. Chem. 2015, 11, 2763–2773. [Google Scholar] [CrossRef]
- Pandey, S.; Xiang, Y.; Walpita Kankanamalage, D.V.D.; Jayawickramarajah, J.; Leng, Y.; Mao, H. Measurement of Single-Molecule Forces in Cholesterol and Cyclodextrin Host-Guest Complexes. J. Phys. Chem. B 2021, 125, 11112–11121. [Google Scholar] [CrossRef]
- Kim, H.; Kim, H.W.; Jung, S. Aqueous Solubility Enhancement of Some Flavones by Complexation with Cyclodextrins. Bull. Korean Chem. Soc. 2008, 29, 590–594. [Google Scholar] [CrossRef]
- Gelb, R.I.; Schwartz, L.M. Complexation of Adamantane-Ammonium Substrates by Beta-Cyclodextrin and Its O-Methylated Derivatives. J. Incl. Phenom. Mol. Recognit. Chem. 1989, 7, 537–543. [Google Scholar] [CrossRef]
- Pereira-Vilar, A.; Martin-Pastor, M.; Pessêgo, M.; García-Río, L. Supramolecular Recognition Induces Nonsynchronous Change of Dye Fluorescence Properties. J. Org. Chem. 2016, 81, 6587–6595. [Google Scholar] [CrossRef]
- Wenz, G. Influence of Intramolecular Hydrogen Bonds on the Binding Potential of Methylated β-Cyclodextrin Derivatives. Beilstein J. Org. Chem. 2012, 8, 1890–1895. [Google Scholar] [CrossRef]
- Sangpheak, W.; Khuntawee, W.; Wolschann, P.; Pongsawasdi, P.; Rungrotmongkol, T. Enhanced Stability of a Naringenin/2,6-Dimethyl β-Cyclodextrin Inclusion Complex: Molecular Dynamics and Free Energy Calculations Based on MM- and QM-PBSA/GBSA. J. Mol. Graph. Model. 2014, 50, 10–15. [Google Scholar] [CrossRef]
- Jeromin, J.; Noll, O.; Ritter, H. Cyclodextrins in Polymer Synthesis: Free Radical Polymerization of Cyclodextrin Complexes with N-Methacryloyl-11-Aminoundecanoic Acid or N-Methacryloyl-1-Aminononane as Guest Monomers. Macromol. Chem. Phys. 1998, 199, 2641–2645. [Google Scholar] [CrossRef]
- Choi, S.W.; Ritter, H. Towards Green Routes for Polymer Synthesis: Polymerization of Cyclodextrin Host-Guest Complexed Diethyl Fumarate and Copolymerization with Complexed Styrene in Homogenous Aqueous Solution. Macromol. Rapid Commun. 2004, 25, 716–719. [Google Scholar] [CrossRef]
- Köllisch, H.; Barner-Kowollik, C.; Ritter, H. Living Free Radical Polymerization of Cyclodextrin Host-Guest Complexes of Styrene via the Reversible Addition Fragmentation Chain Transfer (RAFT) Process in Aqueous Solution. Macromol. Rapid Commun. 2006, 27, 848–853. [Google Scholar] [CrossRef]
- Fava, F.; Ciccotosto, V.F. Effects of Randomly Methylated-β-Cyclodextrins (RAMEB) on the Bioavailability and Aerobic Biodegradation of Polychlorinated Biphenyls in Three Pristine Soils Spiked with a Transformer Oil. Appl. Microbiol. Biotechnol. 2002, 58, 393–399. [Google Scholar] [CrossRef]
- Fava, F.; Di Gioia, D.; Marchetti, L.; Fenyvesi, E.; Szejtli, J. Randomly Methylated β-Cyclodextrins (RAMEB) Enhance the Aerobic Biodegradation of Polychlorinated Biphenyl in Aged-Contaminated Soils. J. Incl. Phenom. 2002, 44, 417–421. [Google Scholar] [CrossRef]
- Jeschke, S.; Jankowski, P.; Best, A.S.; Johansson, P. Catching TFSI: A Computational–Experimental Approach to Β-Cyclodextrin-Based Host–Guest Systems as Electrolytes for Li-Ion Batteries. ChemSusChem 2018, 11, 1942–1949. [Google Scholar] [CrossRef]
- Dhiman, P.; Bhatia, M. Pharmaceutical Applications of Cyclodextrins and Their Derivatives. J. Incl. Phenom. Macrocycl. Chem. 2020, 98, 171–186. [Google Scholar] [CrossRef]
- Elbashir, A.A.; Suliman, F.E.O.; Saad, B.; Aboul-Enein, H.Y. Determination of Aminoglutethimide Enantiomers in Pharmaceutical Formulations by Capillary Electrophoresis Using Methylated-β-Cyclodextrin as a Chiral Selector and Computational Calculation for Their Respective Inclusion Complexes. Talanta 2009, 77, 1388–1393. [Google Scholar] [CrossRef]
- Varga, E.; Benkovics, G.; Darcsi, A.; Várnai, B.; Sohajda, T.; Malanga, M.; Béni, S. Comparative Analysis of the Full Set of Methylated β-Cyclodextrins as Chiral Selectors in Capillary Electrophoresis. Electrophoresis 2019, 40, 2789–2798. [Google Scholar] [CrossRef] [PubMed]
- Höfler, T.; Wenz, G. Determination of Binding Energies between Cyclodextrins and Aromatic Guest Molecules by Microcalorimetry. J. Incl. Phenom. Mol. Recognit. Chem. 1996, 25, 81–84. [Google Scholar] [CrossRef]
- Zabel, V.; Mason, S.A.; Saenger, W. Neutron Diffraction Study of the Hydrogen Bonding in 0-Cyclodextrin Undecahydrate at 120 K: From Dynamic Flip-Flops to Static Homodromic Chains. J. Am. Chem. Soc. 1986, 108, 3664–3673. [Google Scholar] [CrossRef]
- Chelli, S.; Majdoub, M.; Jouini, M.; Aelyach, S.; Maurel, F.; Chane-Ching, K.I.; Lacaze, P.C. Host-Guest Complexes of Phenol Derivatives with β-Cyclodextrin: An Experimental and Theoretical Investigation. J. Phys. Org. Chem. 2007, 20, 30–43. [Google Scholar] [CrossRef]
- Mahalapbutr, P.; Nutho, B.; Wolschann, P.; Chavasiri, W.; Kungwan, N.; Rungrotmongkol, T. Molecular Insights into Inclusion Complexes of Mansonone E and H Enantiomers with Various β-Cyclodextrins. J. Mol. Graph. Model. 2018, 79, 72–80. [Google Scholar] [CrossRef]
- Creasy, W.R.; Farrar, J.M. Reactive Scattering from Double Minimum Potentials: Energetics and Mechanism of the Gas Phase Dehydration Reaction of Lithium Ion with Tert-Butyl Alcohol. J. Chem. Phys. 1986, 85, 162–178. [Google Scholar] [CrossRef]
- Bonnet, P.; Jaime, C.; Morin-Allory, L. α- β-, and γ-Cyclodextrin Dimers. Molecular Modeling Studies by Molecular Mechanics and Molecular Dynamics Simulations. J. Org. Chem. 2001, 66, 689–692. [Google Scholar] [CrossRef]
- Zhang, H.; Tan, T.; Feng, W.; Van Der Spoel, D. Molecular Recognition in Different Environments: β-Cyclodextrin Dimer Formation in Organic Solvents. J. Phys. Chem. B 2012, 116, 12684–12693. [Google Scholar] [CrossRef]
- Aree, T.; Saenger, W.; Leibnitz, P.; Hoier, H. Crystal Structure of Heptakis(2,6-Di-O-Methyl)-β-Cyclodextrin Dihydrate: A Water Molecule in an Apolar Cavity. Carbohydr. Res. 1999, 315, 199–205. [Google Scholar] [CrossRef]
- Ramos, A.I.; Braga, T.M.; Silva, P.; Fernandes, J.A.; Ribeiro-Claro, P.; De Fátima Silva Lopes, M.; Paz, F.A.A.; Braga, S.S. Chloramphenicol·cyclodextrin Inclusion Compounds: Co-Dissolution and Mechanochemical Preparations and Antibacterial Action. CrystEngComm 2013, 15, 2822–2834. [Google Scholar] [CrossRef]
- Assaba, I.M.; Rahali, S.; Belhocine, Y.; Allal, H. Inclusion Complexation of Chloroquine with α and β-Cyclodextrin: Theoretical Insights from the New B97-3c Composite Method. J. Mol. Struct. 2021, 1227, 129696. [Google Scholar] [CrossRef]
- Brandenburg, J.G.; Bannwarth, C.; Hansen, A.; Grimme, S. B97-3c: A Revised Low-Cost Variant of the B97-D Density Functional Method. J. Chem. Phys. 2018, 148, 064104. [Google Scholar] [CrossRef]
- Neese, F.; Wennmohs, F.; Becker, U.; Riplinger, C. The ORCA Quantum Chemistry Program Package. J. Chem. Phys. 2020, 152, 224108. [Google Scholar] [CrossRef]
- Allouche, A.R. Gabedita—A Graphical User Interface for Computational Chemistry Softwares. J. Comput. Chem. 2011, 32, 174–182. [Google Scholar] [CrossRef]
- Zhang, J. Libreta: Computerized Optimization and Code Synthesis for Electron Repulsion Integral Evaluation. J. Chem. Theory Comput. 2018, 14, 572–587. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Chen, F. Multiwfn: A Multifunctional Wavefunction Analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.R.; Keinan, S.; Mori-Sánchez, P.; Contreras-García, J.; Cohen, A.J.; Yang, W. Revealing Noncovalent Interactions. J. Am. Chem. Soc. 2010, 132, 6498–6506. [Google Scholar] [CrossRef] [PubMed]
- Bader, R.F.W. Atoms in Molecules. Acc. Chem. Res. 1985, 18, 9–15. [Google Scholar] [CrossRef]
- Anchique, L.; Alcázar, J.J.; Ramos-Hernandez, A.; Méndez-López, M.; Mora, J.R.; Rangel, N.; Paz, J.L.; Márquez, E. Predicting the Adsorption of Amoxicillin and Ibuprofen on Chitosan and Graphene Oxide Materials: A Density Functional Theory Study. Polymers 2021, 13, 1620. [Google Scholar] [CrossRef]
- Johnson, J.K.; Zollweg, J.A.; Gubbins, K.E. The Lennard-Jones Equation of State Revisited. Mol. Phys. 1993, 78, 591–618. [Google Scholar] [CrossRef]
- Jensen, J.H. Predicting Accurate Absolute Binding Energies in Aqueous Solution: Thermodynamic Considerations for Electronic Structure Methods. Phys. Chem. Chem. Phys. 2015, 17, 12441–12451. [Google Scholar] [CrossRef]
- Barone, V.; Cossi, M. Quantum Calculation of Molecular Energies and Energy Gradients in Solution by a Conductor Solvent Model. J. Phys. Chem. A 1998, 102, 1995–2001. [Google Scholar] [CrossRef]
- Grimme, S. Supramolecular Binding Thermodynamics by Dispersion-Corrected Density Functional Theory. Chem. Eur. J. 2012, 18, 9955–9964. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).