A Theoretical Analysis of Interaction Energies and Intermolecular Interactions between Amphotericin B and Potential Bioconjugates Used in the Modification of Nanocarriers for Drug Delivery
Abstract
1. Introduction
2. Results and Discussion
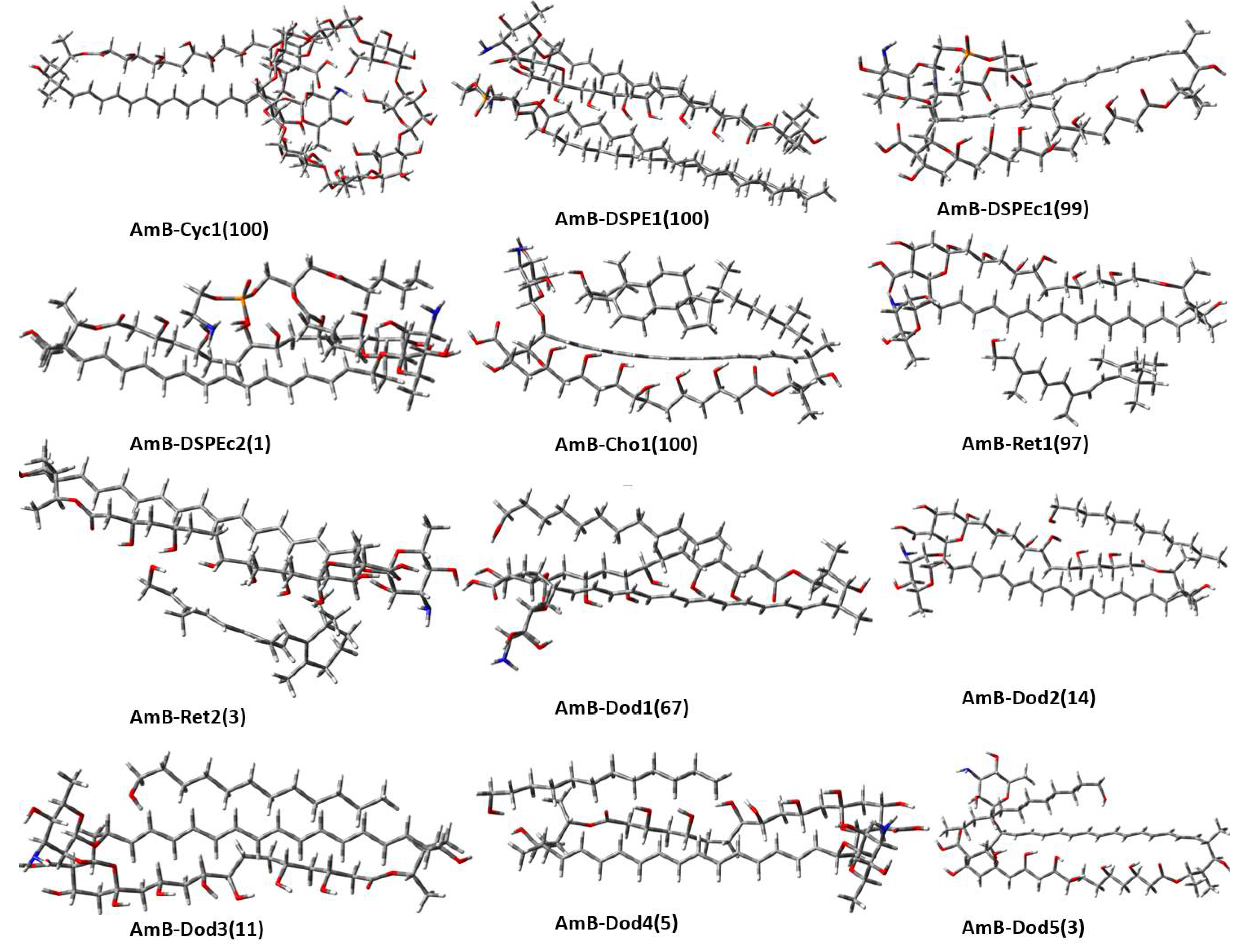
2.1. Potential Energy Surface Exploration
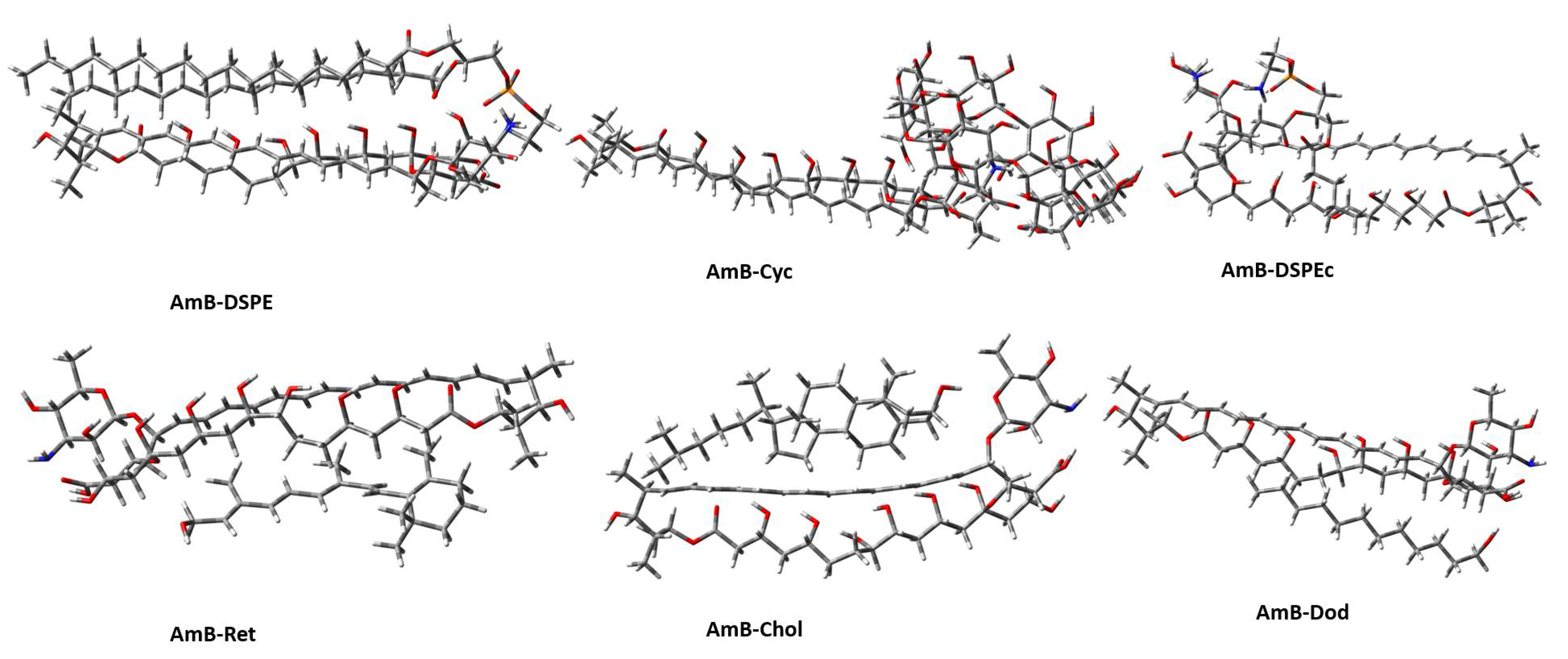
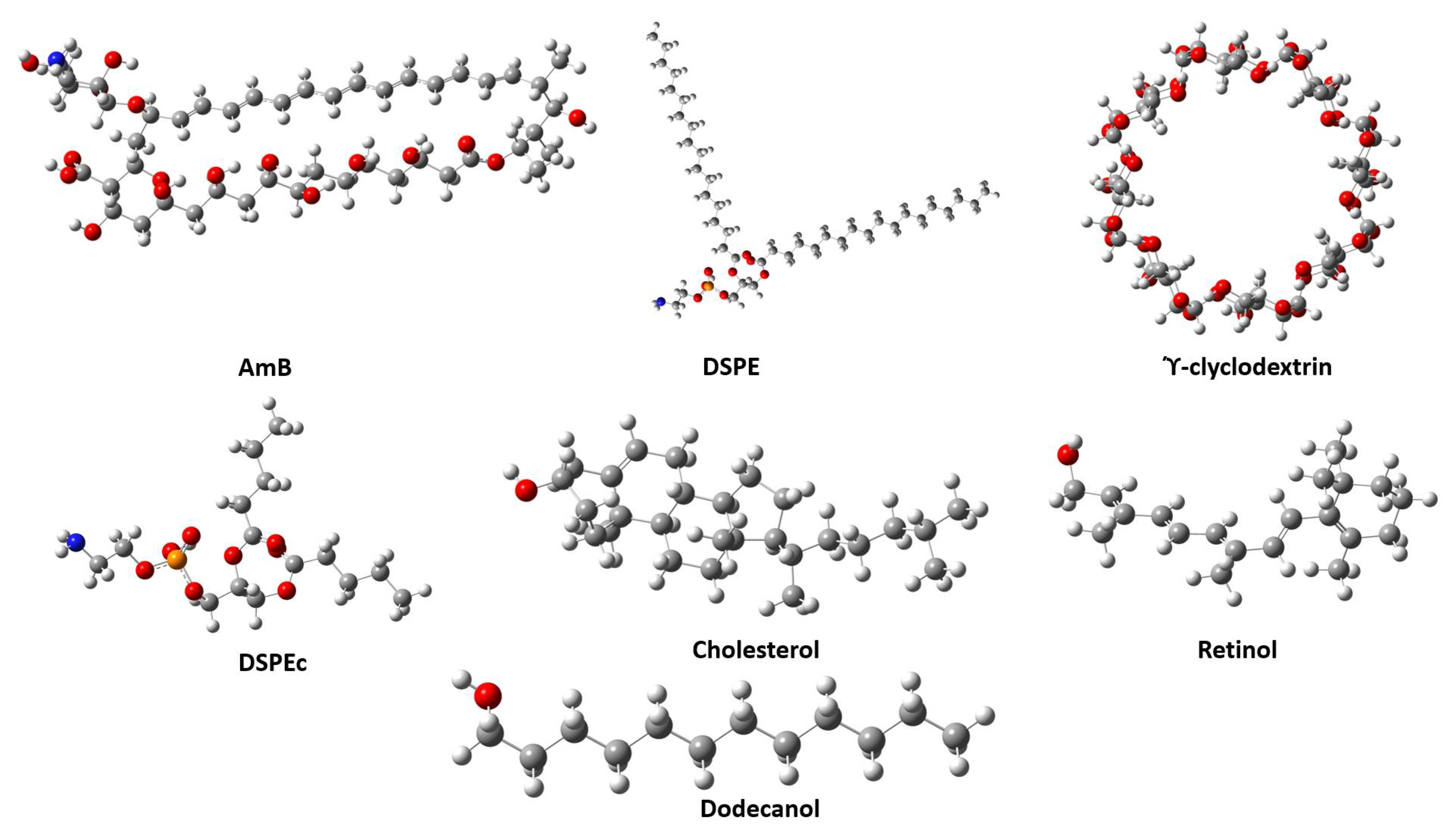
2.2. Geometries and Thermodynamic Properties
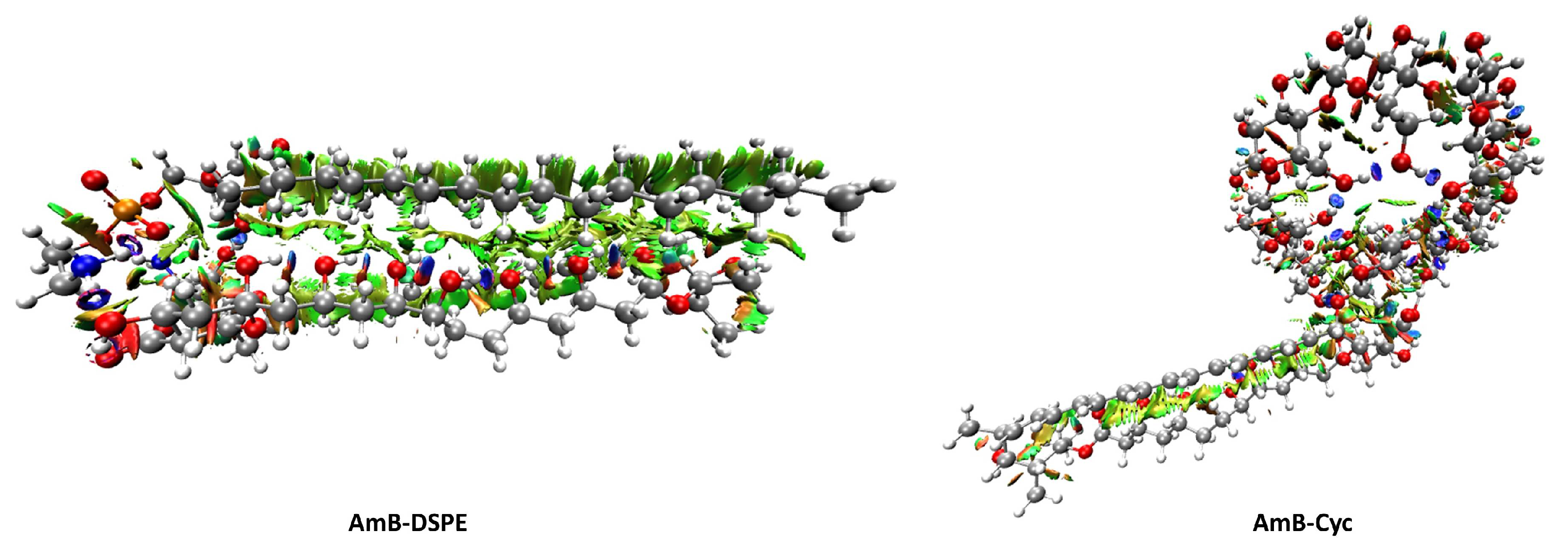
2.3. Non-Covalent Interactions Analysis from Quantum Chemistry Tools
2.3.1. Quantum Theory of Atoms in Molecules Analysis
2.3.2. Non-Covalent Interactions Index Analysis
2.3.3. NBO Analysis
3. Computational Procedure
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arnusch, C.J.; Ulm, H.; Josten, M.; Shadkchan, Y.; Osherov, N.; Sahl, H.G.; Shai, Y. Ultrashort peptide bioconjugates are exclusively antifungal agents and synergize with cyclodextrin and amphotericin B. Antimicrob. Agents Chemother. 2012, 56, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Van Etten, E.W.M.; Van Den Heuvel-de Groot, C.; Bakker-Woudenberg, I.A.J.M. Efficacies of amphotericin b-desoxycholate (fungizone), liposomal amphotericin b (ambisome) and fluconazole in the treatment of systemic candidosis in immunocompetent and leucopenic mice. J. Antimicrob. Chemother. 1993, 32, 723–739. [Google Scholar] [CrossRef] [PubMed]
- Cortijo, G.P.; Espejo, A.C.; Criado, J. Rivero Romn. Infecciones por hongos levaduriformes: Candida sp. y Cryptococcus sp. Med. Programa Form. Médica Contin. Acreditado 2006, 9, 3693–3701. [Google Scholar] [CrossRef]
- Catalán, M.; Montejo, J.C. Systemic antifungals. Pharmacodynamia and pharmacokinetics. Rev. Iberoam. Micol. 2006, 23, 39–49. [Google Scholar] [CrossRef]
- Hamill, R.J. Amphotericin B formulations: A comparative review of efficacy and toxicity. Drugs 2013, 73, 919–934. [Google Scholar] [CrossRef] [PubMed]
- Botero, M.C.; Puentes-Herrera, M.; Cortés, J.A. Lipid formulations of amphotericin. Rev. Chil. Infectol. 2014, 31, 518–527. [Google Scholar] [CrossRef]
- Baginski, M.; Sternal, K.; Czub, J.; Borowski, E. Molecular modelling of membrane activity of amphotericin B, a polyene macrolide antifungal antibiotic. Acta Biochim. Pol. 2005, 52, 655–658. [Google Scholar] [CrossRef]
- AL-Quadeib, B.T.; Radwan, M.; Siller, L.; Horrocks, B.; Wright, M.C. Stealth Amphotericin B nanoparticles for oral drug delivery: In vitro optimization. Saudi Pharm. J. 2015, 23, 290–302. [Google Scholar] [CrossRef]
- Gallis, H.A.; Drew, R.; Pickard, W.W. Amphotericin B: 30 years of clinical experience. Rev. Infect. Dis. 1990, 12, 308–329. [Google Scholar] [CrossRef]
- Valadez, T.N.; Norton, J.; Neary, M.C. The Reaction of Cp* (Cl)M(Diene) (M = Ti, Hf) with Isonitriles (Supporting Information). J. Chem. Inf. Model. 2013, 53, 1689–1699. [Google Scholar]
- Voncik, K.S.; Fermino, B.L.; Cardoso, N.C.S.; de Mattos, R.H.; Pinto, E.C.; Ignachewski, J.D.; Carraro, E.; de Freitas, G.B.; da Silva, W.C.F.N.; Bonini, J.S. Difficulties in antifungal therapy with amphotericin B and the continuous search for new formulations: A literature review. Afr. J. Pharm. Pharmacol. 2016, 10, 512–520. [Google Scholar] [CrossRef]
- Alvarez, C.; Shin, D.; Kwon, G.S. Reformulation of Fungizone by PEG-DSPE Micelles: Deaggregation and Detoxification of Amphotericin B. Pharm. Res. 2016, 33, 2098–2106. [Google Scholar] [CrossRef] [PubMed]
- Villamil, J.C.; Parra-Giraldo, C.; Pérez, L.D. Enhancing the performance of PEG-b-PCL copolymers as precursors of micellar vehicles for amphotericin B through its conjugation with cholesterol. Colloids Surf. A Physicochem. Eng. Asp. 2019, 572, 79–87. [Google Scholar] [CrossRef]
- Diaz, I.L.; Parra, C.; Linarez, M.; Perez, L.D. Design of Micelle Nanocontainers Based on PDMAEMA-b-PCL-b-PDMAEMA Triblock Copolymers for the Encapsulation of Amphotericin B. AAPS PharmSciTech 2015, 16, 1069–1078. [Google Scholar] [CrossRef] [PubMed]
- Azanza, J.R.; Sádada, B.; Reis, J. Liposomal formulations of amphotericin B: Differences according to the scientific evidence. Rev. Esp. Quim. 2015, 28, 275–281. [Google Scholar]
- Tevyashova, A.N.; Korolev, A.M.; Trenin, A.S.; Dezhenkova, L.G.; Shtil, A.A.; Polshakov, V.I.; Savelyev, O.Y.; Olsufyeva, E.N. New conjugates of polyene macrolide amphotericin B with benzoxaboroles: Synthesis and properties. J. Antibiot. 2016, 69, 549–560. [Google Scholar] [CrossRef]
- Tang, X.; Jiao, R.; Xie, C.; Xu, L.; Huo, Z.; Dai, J.; Qian, Y.; Xu, W.; Hou, W.; Wang, J.; et al. Improved antifungal activity of amphotericin B-loaded TPGS-b-(PCL-ran-PGA) nanoparticles. Int. J. Clin. Exp. Med. 2015, 8, 5150–5162. [Google Scholar]
- Abdellatif, A.A.H.; Ibrahim, M.A.; Amin, M.A.; Maswadeh, H.; Alwehaibi, M.N.; Al-Harbi, S.N.; Alharbi, Z.A.; Mohammed, H.A.; Mehany, A.B.M.; Saleem, I. Cetuximab Conjugated with Octreotide and Entrapped Calcium Alginate-beads for Targeting Somatostatin Receptors. Sci. Rep. 2020, 10, 4736. [Google Scholar] [CrossRef]
- Xu, L.; Li, Y. Responsive Guest Encapsulation of Dynamic Conjugated Microporous Polymers. Sci. Rep. 2016, 6, 28784. [Google Scholar] [CrossRef]
- Vikmon, M.; Stadler-szoke, A.; Szejtli, J.; Chinoin, C. Solubilization of amphotericin B with γ-cyclodextrin. J. Antibiot. 1985, 38, 1822–1824. [Google Scholar] [CrossRef]
- Moribe, K.; Maruyama, S.; Inoue, Y.; Suzuki, T.; Fukami, T.; Tomono, K.; Higashi, K.; Tozuka, Y.; Yamamoto, K. Ascorbyl dipalmitate/PEG-lipid nanoparticles as a novel carrier for hydrophobic drugs. Int. J. Pharm. 2010, 387, 236–243. [Google Scholar] [CrossRef]
- Yañez, O.; Báez-Grez, R.; Inostroza, D.; Pino-Rios, R.; Rabanal-León, W.A.; Contreras-García, J.; Cardenas, C.; Tiznado, W. Kick-Fukui: A Fukui Function-Guided Method for Molecular Structure Prediction. J. Chem. Inf. Model. 2021, 61, 3955–3963. [Google Scholar] [CrossRef] [PubMed]
- Wilcock, B.C.; Endo, M.; Uno, B.; Burke, M.D. C2′-OH of amphotericin B plays an important role in binding the primary sterol of human cells but not yeast cells. J. Am. Chem. Soc. 2013, 135, 8488–8491. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, E.; Souhassou, M.; Lachekar, H.; Lecomte, C. Topological analysis of the electron density in hydrogen bonds. 1999. Acta Crystallogr. Sect. B Struct. Sci. 1999, 55, 563–572. [Google Scholar] [CrossRef]
- Sławomir, J.; Sokalski, W.A.; Dyguda, E.; Leszczyński, J. Quantitative classification of covalent and noncovalent H-bonds. J. Phys. Chem. B 2006, 110, 6444–6446. [Google Scholar] [CrossRef]
- Chacón, K.N.; Espinal, J.; Montero-Campillo, M.; Yáñez, M.; Mejiá, S.M. Looking for the Azeotrope: A Computational Study of (Ethanol)6-Water, (Methanol)6-Water, (Ethanol)7, and (Methanol)7Heptamers. J. Phys. Chem. A 2020, 124, 7080–7087. [Google Scholar] [CrossRef] [PubMed]
- Mejía, S.M.; Espinal, J.; Mills, M.L.; Mondragón, F. The role of OH…O and CH…O hydrogen bonds and H…H interactions in ethanol/methanol–water heterohexamers. J. Mol. Model. 2016, 22, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Matta, C.F.; Boyd, R.J. An Introduction to the Quantum Theory of Atoms in Molecules; Wiley-VCH: Hoboken, NY, USA, 2007. [Google Scholar] [CrossRef]
- Rozas, I.; Alkorta, I.; Elguero, J. Behavior of ylides containing N, O, and C atoms as hydrogen bond acceptors. J. Am. Chem. Soc. 2000, 122, 11154–11161. [Google Scholar] [CrossRef]
- Contreras-Garcia, J.; Johnson, E.R.; Keinan, S.; Chaudret, R.; Piquemal, J.-P.; Beratan, D.; Yang, W. NCIPLOT: A program for plotting noncovalent interaction regions. J. Chem. Theory Comput. 2011, 7, 625–632. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Hettne, K.M.; Williams, A.; Van Mulligen, E.; Kleinjans, J.; Tkachenko, V.; Kors, J.A. Automatic vs. manual curation of a multi-source chemical dictionary: The impact on text mining. J. Cheminform. 2010, 2, 10–11. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.; Lonie, D.; Vandermeerschd, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Singh, I.; El-Emam, A.A.; Pathak, S.K.; Srivastava, R.; Shukla, V.K.; Prasad, O.; Sinha, L. Experimental and theoretical DFT (B3LYP, X3LYP, CAM-B3LYP and M06-2X) study on electronic structure, spectral features, hydrogen bonding and solvent effects of 4-methylthiadiazole-5-carboxylic acid. Mol. Simul. 2019, 45, 1029–1043. [Google Scholar] [CrossRef]
- Sebastian, S.; Sundaraganesan, N. The spectroscopic (FT-IR, FT-IR gas phase, FT-Raman and UV) and NBO analysis of 4-Hydroxypiperidine by density functional method. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2010, 75, 941–952. [Google Scholar] [CrossRef] [PubMed]
- Gómez, S.A.; Rojas-Valencia, N.; Gómez, S.; Egidi, F.; Cappelli, C.; Restrepo, A. Binding of SARS-CoV-2 to Cell Receptors: A Tale of Molecular Evolution. ChemBioChem 2021, 22, 724–732. [Google Scholar] [CrossRef]
- Gómez, S.; Rojas-Valencia, N.; Gómez, S.; Cappelli, C.; Merino, G.; Restrepo, A. A molecular twist on hydrophobicity. Chem. Sci. 2021, 12, 9233–9245. [Google Scholar] [CrossRef]
- Ruiz, H.K.; Serrano, D.R.; Dea-Ayuela, M.A.; Bilbao-Ramos, P.E.; Bolás-Fernández, F.; Torrado, J.J.; Molero, G. New amphotericin B-gamma cyclodextrin formulation for topical use with synergistic activity against diverse fungal species and Leishmania spp. Int. J. Pharm. 2014, 473, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Oqmhula, K.; Hongo, K.; Maezono, R.; Ichibha, T. Ab Initio Evaluation of Complexation Energies for Cyclodextrin-Drug Inclusion Complexes. ACS Omega 2020, 5, 19371–19376. [Google Scholar] [CrossRef] [PubMed]
- Korth, M. Third-Generation Hydrogen-Bonding Corrections for Semiempirical QM Methods and Force Fields. J. Chem. Theory Comput. 2010, 12, 3808–3816. [Google Scholar] [CrossRef]
- Stewart, J.J.P. MOPAC: A semiempirical molecular orbital program. J. Comput. Aided Mol. Des. 1990, 4, 1–103. [Google Scholar] [CrossRef]
- Petersson, G.A.; Zhong, S.; Montgomery, J.; Frisch, M.J. On the optimization of gaussian basis sets. J. Chem. Phys. 2003, 118, 1101–1109. [Google Scholar] [CrossRef]
- Kar, T.; Scheiner, S. Comparison between hydrogen and dihydrogen bonds among H3BNH3, H2BNH2, and NH3. J. Chem. Phys. 2003, 119, 1473–1482. [Google Scholar] [CrossRef]
- Liu, B.; Mclean, A.D. Accurate calculation of the attractive interaction of two ground state helium atoms. J. Chem. Phys. 1973, 59, 4557–4558. [Google Scholar] [CrossRef]
- Arias, E.R.; Angarita-Villamizar, V.; Baena, Y.; Parra-Giraldo, C.; Perez, L.D. Phospholipid-conjugated peg-b-pcl copolymers as precursors of micellar vehicles for amphotericin b. Polymers 2021, 13, 1747. [Google Scholar] [CrossRef]
- Grabowski, S.J. Non-covalent interactions—QTAIM and NBO analysis. J. Mol. Model. 2013, 19, 4713–4721. [Google Scholar] [CrossRef]
- Cruz, J.C.; Hernández-Esparza, R.; Vázquez-Mayagoitia, Á.; Vargas, R.; Garza, J. Implementation of the Molecular Electrostatic Potential over Graphics Processing Units. J. Chem. Inf. Model. 2019, 59, 3120–3127. [Google Scholar] [CrossRef]
- Bader, R.F.W. A Quantum Theory of Molecular Structure and Its Applications. Chem. Rev. 1991, 91, 893–928. [Google Scholar] [CrossRef]
- Analyzer, A.M.W.; Lu, T. Multiwfn; Sobereva: Beijing, China, 2021; Volume 8. [Google Scholar]
- VMD. VMD User’s Guide Version 1.9.3.; Beckman Institute for Advanced Science and Technology: Urbana, IL, USA, 2016; Volume Manual. [Google Scholar]


| Abbreviation | Definition | Abbreviation | Definition |
|---|---|---|---|
| AmB | Amphotericin B | ZPE | Zero point energy |
| DSPE | 1,2-distearoyl-sn-glycerol-3-phosphorylethanolamine | UFF | Universal force field |
| Cyc | Cyclodextrin | HBs | Hydrogen bond |
| β-Cyc | Beta-cyclodextrin | BCP | Bond critical points |
| γ-Cyc | alpha-cyclodextrin | ρ(rCP) | Electron density |
| DSPEc | Homologous DSPE with a shorter aliphatic chain | ∇2ρ(rCP) | Laplacian of the electron density |
| Ret | Retinol | H(rCP) | Total energy density |
| Chol | Cholesterol | (lV(rCP)l/G(rCP)) | Relation between the Virial field and kinetic energy density |
| Dod | Dodecanol | IE | Interaction energy |
| AmB-DSPE | Dimer formed between Amphotericin B and 1,2-distearoyl-sn-glycerol-3-phosphorylethanolamine | Donor–acceptor second-order interaction energies | |
| AmB_γ-cyclodextrin or AmB-Cyc | Dimer formed between Amphotericin B and alpha-cyclodextrin | ΔE | Dimerization energy |
| AmB_DSPEc | Dimer formed between Amphotericin B and DSPE homologous with a shorter aliphatic chain | ΔH | Dimerization enthalpy |
| AmB_retinol or AmB-Ret | Dimer formed between Amphotericin B and retinol | ΔG | Dimerization Gibbs free energy |
| AmB_cholesterol or AmB-Chol | Dimer formed between Amphotericin B and cholesterol | ΔS | Dimerization entropy |
| AmB_dodecanol or AmB-Dod | Dimer formed between Amphotericin B and dodecanol | SI | Supporting Information |
| DFT | Density Functional Theory | PES | Potential Energy Surface |
| NCI | Non-Covalent interactions Index | QTAIM | Quantum theory of atoms in molecules |
| Dimers | ΔE | ΔH | ΔG | ΔS |
|---|---|---|---|---|
| AmB-DSPE | −10.66 | −141.46 | −101.65 | −133.53 |
| AmB-Cyc | −64.79 | −95.30 | −69.69 | −85.88 |
| AmB-DSPEc | −56.01 | −77.74 | −50.95 | −89.84 |
| AmB-Ret | −22.32 | −34.86 | −16.09 | −62.94 |
| AmB-Cho | −17.82 | −31.51 | −12.28 | −64.49 |
| AmB-Dod | −16.01 | −26.11 | −9.50 | −55.69 |
| IT a | Length b [Å] | ρ(rCP) c 10−2 | ∇2ρ(rCP) d 10−2 | H(rCP) e 10−3 | lV(rCP)l/ G(rCP)) f | IE g 10−2 [kcal/mol] | NI h |
|---|---|---|---|---|---|---|---|
| AmB-DSPE | |||||||
| O-H---O | 1.70 | 4.88 | 13.33 | −4.2 | 1.08 | −2.09 | 2 |
| C-H---O | 2.68 | 0.75 | 2.71 | 0.86 | 0.82 | −0.25 | 25 |
| H---H | 2.51 | 0.48 | 1.71 | 1.00 | 0.69 | −0.11 | 18 |
| C-H---C | 3.04 | 0.43 | 1.51 | 0.88 | 0.71 | −0.10 | 3 |
| N-H---O | 1.89 | 3.68 | 10.71 | −1.06 | 1.01 | −1.44 | 3 |
| AmB-Cyc | |||||||
| O-H---O | 1.89 | 4.16 | 7.87 | −10.64 | 1.11 | −2.05 | 6 |
| C-H---O | 2.63 | 0.85 | 2.99 | 0.75 | 0.83 | −0.30 | 1 |
| H---H | 2.06 | 1.01 | 3.66 | 1.6 | 0.79 | −0.30 | 12 |
| N-H---O | 2.01 | 3.07 | 8.81 | −0.90 | 1.01 | −1.19 | 1 |
| C-H---N | 3.39 | 0.19 | 6.90 | 0.50 | 0.63 | −0.04 | 1 |
| AmB-DSPEc | |||||||
| O-H---O | 1.75 | 3.99 | 11.94 | −1.03 | 1.03 | −1.60 | 3 |
| C-H---O | 2.54 | 0.91 | 3.13 | 0.81 | 0.87 | −0.31 | 11 |
| H---H | 2.43 | 0.56 | 2.09 | 1.10 | 0.70 | −0.15 | 8 |
| C-H---C | 3.13 | 0.45 | 1.55 | 0.80 | 0.72 | −0.12 | 2 |
| O---O | 3.10 | 0.66 | 2.74 | 0.90 | 0.86 | −0.26 | 1 |
| N-H---O | 1.88 | 3.01 | 8.66 | −0.80 | 1.04 | −1.17 | 1 |
| N---O | 2.77 | 1.47 | 6.04 | 1.80 | 0.86 | −0.58 | 1 |
| AmB-Ret | |||||||
| O-H---O | 1.98 | 2.55 | 6.21 | −1.80 | 1.10 | −0.96 | 1 |
| C-H---O | 2.39 | 1.29 | 3.95 | 0.26 | 0.96 | −0.47 | 5 |
| H---H | 2.39 | 0.58 | 2.07 | 1.14 | 0.70 | −0.14 | 10 |
| C-H---C | 2.97 | 0.53 | 1.68 | 0.83 | 0.75 | −0.13 | 11 |
| AmB-Chol | |||||||
| O-H---O | 1.96 | 2.52 | 7.11 | −1.00 | 1.05 | −0.99 | 1 |
| C-H---O | 2.82 | 0.59 | 2.25 | 9.29 | 0.77 | −0.19 | 7 |
| H---H | 2.32 | 0.64 | 2.30 | 12.20 | 0.72 | −0.17 | 13 |
| C-H---C | 2.95 | 0.59 | 1.80 | 9.00 | 0.74 | −0.13 | 5 |
| AmB-Dod | |||||||
| O-H---O | 1.75 | 4.23 | 11.43 | −1.80 | 1.06 | −1.61 | 1 |
| C-H---O | 2.73 | 0.72 | 2.61 | 0.80 | 0.80 | −0.24 | 4 |
| H---H | 2.47 | 0.49 | 1.69 | 0.95 | 0.70 | −0.12 | 13 |
| C-H---C | 3.06 | 0.49 | 1.62 | 0.82 | 0.73 | −0.12 | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuellar, J.; Parada-Díaz, L.; Garza, J.; Mejía, S.M. A Theoretical Analysis of Interaction Energies and Intermolecular Interactions between Amphotericin B and Potential Bioconjugates Used in the Modification of Nanocarriers for Drug Delivery. Molecules 2023, 28, 2674. https://doi.org/10.3390/molecules28062674
Cuellar J, Parada-Díaz L, Garza J, Mejía SM. A Theoretical Analysis of Interaction Energies and Intermolecular Interactions between Amphotericin B and Potential Bioconjugates Used in the Modification of Nanocarriers for Drug Delivery. Molecules. 2023; 28(6):2674. https://doi.org/10.3390/molecules28062674
Chicago/Turabian StyleCuellar, Jennifer, Lorena Parada-Díaz, Jorge Garza, and Sol M. Mejía. 2023. "A Theoretical Analysis of Interaction Energies and Intermolecular Interactions between Amphotericin B and Potential Bioconjugates Used in the Modification of Nanocarriers for Drug Delivery" Molecules 28, no. 6: 2674. https://doi.org/10.3390/molecules28062674
APA StyleCuellar, J., Parada-Díaz, L., Garza, J., & Mejía, S. M. (2023). A Theoretical Analysis of Interaction Energies and Intermolecular Interactions between Amphotericin B and Potential Bioconjugates Used in the Modification of Nanocarriers for Drug Delivery. Molecules, 28(6), 2674. https://doi.org/10.3390/molecules28062674







