Natural Plasmodium falciparum Infection Stimulates Human Antibodies to MSP1 Epitopes Identified in Mice Infection Models upon Non-Natural Modified Peptidomimetic Vaccination
Abstract
1. Introduction
2. Results
2.1. Bioinformatic Analysis and MSP1-Epitope-Peptide Selection
2.2. Functional Assessment of MSP1 Native, -ψ-[CH2–NH]-Peptide-Bond Isosteres and D-Amino Acid Modified Sequences
2.3. Individuals Exposed to Natural Plasmodium spp. Infection
2.4. The Cellular Th1/Th2 Immune Response of Protected Vaccinated Mice against a Malaria Experimental Challenge
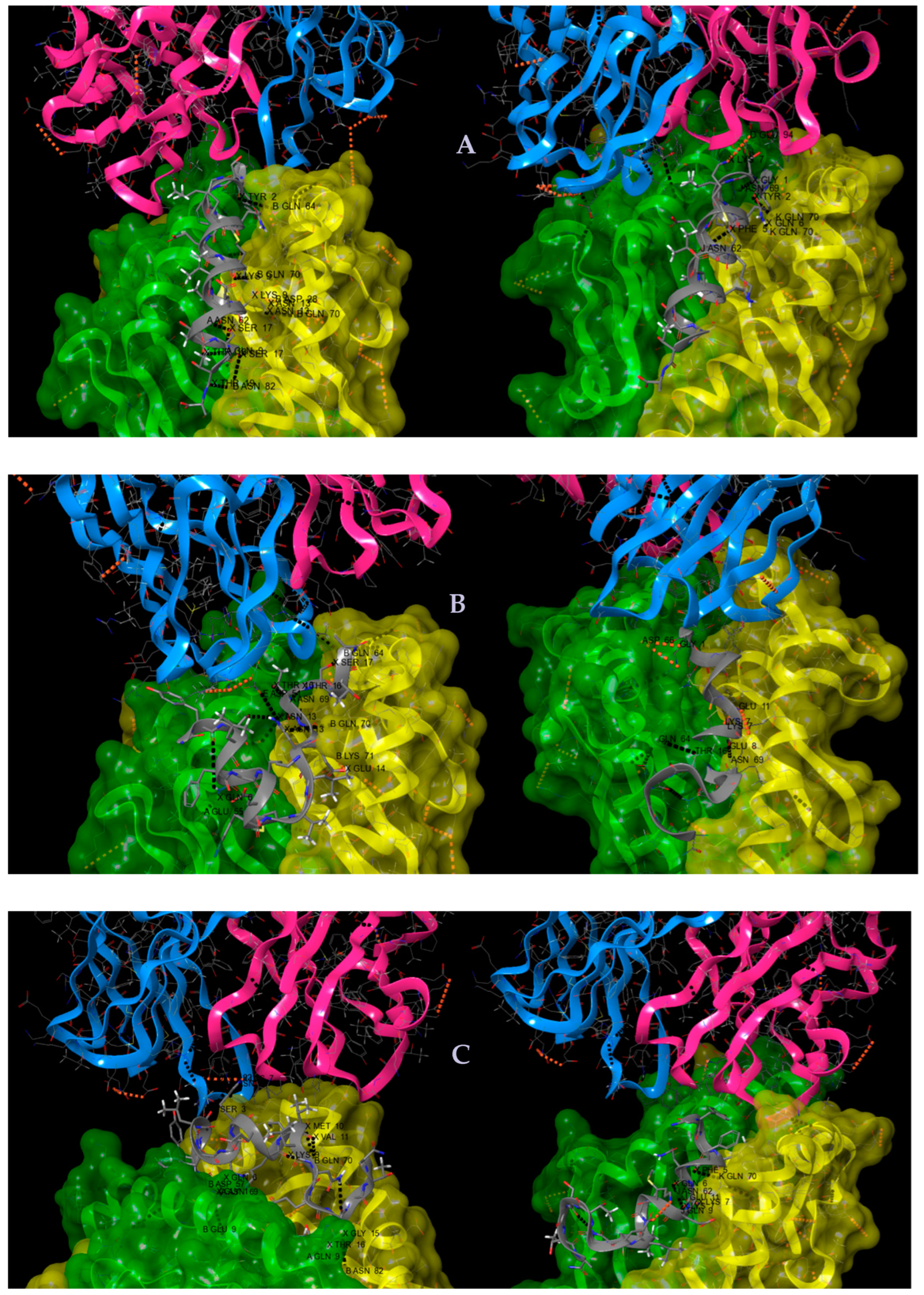
2.5. In Silico Studies for the MSP142–61 Fragment, Its Modified Analogs, and Their HLA-Peptide-TCR Complexes
2.6. RMSD, Radius of Gyration (RG), Root Mean Square Fluctuations (RMSF), Number of H Bonds, Solvent-Accessible Surface Area (SASA), Secondary Structure Changes in Peptides
3. Discussion
4. Materials and Methods
4.1. Predicting MSP1 Epitopes, Bioinformatic Selection
4.2. Solid-Phase Synthesis of Modified-Antigens and Physicochemical Characterization
4.3. Peptide Characterization
4.4. Serological Study and Volunteers
4.5. Functional In Vivo Activity of Proposed Epitopes
4.5.1. Mice Immunization
4.5.2. Immunoreactivity of MSP1 Peptide Analogs
4.5.3. Vaccination of BALB/c Mice and Experimental Challenge with Rodent Malaria Species
4.5.4. CellularTh1/Th2 Immune Responses of Malaria-Protected Animals
4.5.5. In Silico Docking Experiments, Getting-Ready Protocols
4.5.6. Selection of Structures’ Ternary Complexes and Docking Models
4.5.7. Molecular Dynamics Simulations of Highly Immunogenic MSP1 Peptide Analogs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- World Health Organization. World Malaria Report 2021; WHO: Geneva, Switzerland, 2021.
- Moreno, A.; Joyner, C. Malaria Vaccine Clinical Trials: What’s on the Horizon. Curr. Opin. Immunol. 2015, 35, 98–106. [Google Scholar] [CrossRef]
- Coelho, C.H.; Doritchamou, J.Y.A.; Zaidi, I.; Duffy, P.E. Advances in Malaria Vaccine Development: Report from the 2017 Malaria Vaccine Symposium. NPJ Vaccines 2017, 2, 34. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Recommends Groundbreaking Malaria Vaccine for Children at Risk. Available online: https://www.who.int/news/item/06-10-2021-who-recommends-groundbreaking-malaria-vaccine-for-children-at-risk (accessed on 31 October 2021).
- Dobaño, C.; Sanz, H.; Sorgho, H.; Dosoo, D.; Mpina, M.; Ubillos, I.; Aguilar, R.; Ford, T.; Díez-Padrisa, N.; Williams, N.; et al. Concentration and Avidity of Antibodies to Different Circumsporozoite Epitopes Correlate with RTS,S/AS01E Malaria Vaccine Efficacy. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Moon, J.E.; Greenleaf, M.E.; Regules, J.A.; Debois, M.; Duncan, E.H.; Sedegah, M.; Chuang, I.; Lee, C.K.; Sikaffy, A.K.; Garver, L.S.; et al. A Phase IIA Extension Study Evaluating the Effect of Booster Vaccination with a Fractional Dose of RTS,S/AS01E in a Controlled Human Malaria Infection Challenge. Vaccine 2021, 39, 6398–6406. [Google Scholar] [CrossRef]
- Patarroyo, M.E.; Bermúdez, A.; Moreno-Vranich, A. Towards the Development of a Fully Protective Plasmodium falciparum Antimalarial Vaccine. Expert. Rev. Vaccines 2012, 11, 1057–1070. [Google Scholar] [CrossRef] [PubMed]
- Gerold, P.; Schofield, L.; Blackman, M.; Holder, A.; Schwarz, R. Structural Analysis of the Glycosyl-Phosphatidylinositol Membrane Anchor of the Merozoite Surface Proteins-1 and -2 of Plasmodium Falciparum. Mol. Biochem. Parasitol. 1996, 75, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Marshall, V.; Tieqiao, W.; Coppel, R.L. Close Linkage of Three Merozoite Surface Protein Genes on Chromosome 2 of Plasmodium Falciparum. Mol. Biochem. Parasitol. 1998, 94, 13–25. [Google Scholar] [CrossRef]
- Tanabe, K.; Mackay, M.; Goman, M.; Scaife, J.G. Allelic Dimorphism in a Surface Antigen Gene of the Malaria Parasite Plasmodium Falciparum. J. Mol. Biol. 1987, 195, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Lyon, J.; Angov, E.; Fay, M.; Sullivan, J.; Girourd, A. Protection Induced by Plasmodium falciparum MSP1 42 Is Strain-Specific, Antigen and Adjuvant Dependent, and Correlates with Antibody Responses. PLoS ONE 2008, 3, 2830. [Google Scholar] [CrossRef]
- Sheehy, S.H.; Duncan, C.J.; Elias, S.C.; Choudhary, P.; Biswas, S.; Halstead, F.D.; Collins, K.A.; Edwards, N.J.; Douglas, A.D.; Anagnostou, N.A.; et al. ChAd63-MVA-Vectored Blood-Stage Malaria Vaccines Targeting MSP1 and AMA1: Assessment of Efficacy against Mosquito Bite Challenge in Humans. Mol. Ther. 2012, 20, 2355–2368. [Google Scholar] [CrossRef]
- Bastian, M.; Lozano, J.M.; Patarroyo, M.E.; Pluschke, G.; Daubenberger, C.A. Characterization of a Reduced Peptide Bond Analogue of a Promiscuous CD4 T Cell Epitope Derived from the Plasmodium falciparum Malaria Vaccine Candidate Merozoite Surface Protein 1. Mol. Immunol. 2004, 41, 775–784. [Google Scholar] [CrossRef]
- Spatola, A. Chemistry and Biochemistry of Amino Acids, Peptides and Proteins; Weinstein, B., Ed.; Marcel Dekker: New York, NY, USA, 1983. [Google Scholar]
- Lozano, J.M.; Espejo, F.; Diaz, D.; Salazar, L.M.; Rodríguez, J.; Pinzón, C.; Calvo, J.; Guzmán, F.; Patarroyo, M.E. Reduced Amide Pseudopeptide Analogues of a Malaria Peptide Possess Secondary Structural Elements Responsible for Induction of Functional Antibodies Which React with Native Proteins Expressed in Plasmodium falciparum Erythrocyte Stages. J. Pept. Res. 1998, 52, 457–469. [Google Scholar] [CrossRef]
- Sanni, L.; Fonseca, L.F.; Langhorne, J. Mouse Models for Erythrocytic-Stage Malaria. Methods Mol. Med. 2002, 72, 57–76. [Google Scholar] [CrossRef]
- Cubillos, M.; Salazar, L.M.; Torres, L.; Patarroyo, M. Protection against Experimental P. falciparum Malaria Is Associated with Short AMA-1 Peptide Analogue α-Helical Structures. Biochimie 2002, 84, 1181–1188. [Google Scholar] [CrossRef]
- Daubenberger, C.A.; Nickel, B.; Ciatto, C.; Grütter, M.G.; Pöltl-Frank, F.; Rossi, L.; Siegler, U.; Robinson, J.; Kashala, O.; Patarroyo, M.E.; et al. Amino Acid Dimorphism and Parasite Immune Evasion: Cellular Immune Responses to a Promiscuous Epitope of Plasmodium falciparum Merozoite Surface Protein 1 Displaying Dimorphic Amino Acid Polymorphism Are Highly Constrained. Eur. J. Immunol. 2002, 32, 3667–3677. [Google Scholar] [CrossRef]
- Urquiza, M.; Rodríguez, L.E.; Suarez, J.; Guzmán, F.; Ocampo, M.; Curtidor, H.; Segura, C.; Trujillo, E.; Patarroyo, M.E. Identification of Plasmodium falciparum MSP-1 Peptides Able to Bind to Human Red Blood Cells. Parasite Immunol. 1996, 18, 515–526. [Google Scholar] [CrossRef]
- Lozano, J.M.; Espejo, F.; Ocampo, M.; Salazar, L.M.; Tovar, D.; Barrera, N.; Guzmán, F.; Patarroyo, M.E. Mapping the Anatomy of a Plasmodium falciparum MSP-1 Epitope Using Pseudopeptide-Induced Mono- and Polyclonal Antibodies and CD and NMR Conformation Analysis. J. Struct. Biol. 2004, 148, 110–122. [Google Scholar] [CrossRef]
- Lozano, J.M.; Rivera, Z.; Patarroyo, M.E. What Is Hidden Behind Peptide Bond Restriction and Alfa-Carbon Asymmetry of Conserved Antigens? Peptide Bond Isosters and Chirally Transformed Pseudopeptides as Novel Elements for Synthetic Vaccines and Therapeutic Agents Against Malaria. Curr. Org. Chem. 2006, 10, 433–456. [Google Scholar] [CrossRef]
- Patarroyo, M.E.; Moreno-Vranich, A.; Bermúdez, A. Phi (Φ) and Psi (Ψ) Angles Involved in Malarial Peptide Bonds Determine Sterile Protective Immunity. Biochem. Biophys. Res. Commun. 2012, 429, 75–80. [Google Scholar] [CrossRef]
- Vanegas, M.; Bermúdez, A.; Guerrero, Y.; Cortes-Vecino, J.A.; Curtidor, H.; Patarroyo, M.E.; Lozano, J.M. Protecting Capacity against Malaria of Chemically Defined Tetramer Forms Based on the Plasmodium falciparum Apical Sushi Protein as Potential Vaccine Components. Biochem. Biophys. Res. Commun. 2014, 451, 15–23. [Google Scholar] [CrossRef]
- Lozano, J.M.; Guerrero, Y.; Alba, M.; Lesmes, L.; Escobar, J.; Patarroyo, M.E. Redefining an Epitope of a Malaria Vaccine Candidate, with Antibodies against the N-Terminal MSA-2 Antigen of Plasmodium Harboring Non-Natural Peptide Bonds. Amino Acids 2013, 45, 913–935. [Google Scholar] [CrossRef]
- Instituto Nacional de Salud. Informe Quincenal Epidemiológico Nacional: Informe de Evento Malaria 2019. Bogotá: INS; c2018 10 P. Available online: https//:www.ins.gov.co (accessed on 14 January 2019).
- Aucan, C.; Traoré, Y.; Tall, F.; Nacro, B.; Traoré-Leroux, T.; Fumoux, F.; Rihet, P. High Immunoglobulin G2 (IgG2) and Low IgG4 Levels Are Associated with Human Resistance to Plasmodium falciparum Malaria. Infect. Immun. 2000, 68, 1252. [Google Scholar] [CrossRef]
- Garraud, O.; Mahanty, S.; Perraut, R. Malaria-Specific Antibody Subclasses in Immune Individuals: A Key Source of Information for Vaccine Design. Trends Immunol. 2003, 24, 30–35. [Google Scholar] [CrossRef]
- Perez-Mazliah, D.; Langhorne, J.; Krzych, U.; Army, W.R.; Rodrigues, M.M. CD4 T-Cell Subsets in Malaria: TH1/TH2 Revisited. Front. Immunol. 2015, 5, 1–8. [Google Scholar] [CrossRef]
- Popa, G.L.; Popa, M.I. Recent Advances in Understanding the Inflammatory Response in Malaria: A Review of the Dual Role of Cytokines. J. Immunol. Res. 2021, 2021, 7785180. [Google Scholar] [CrossRef]
- Riley, E.M.; Stewart, V.A. Immune Mechanisms in Malaria: New Insights in Vaccine Development. Nat. Med. 2013, 19, 168–178. [Google Scholar] [CrossRef]
- O’Garra, A.; Murphy, K. Role of Cytokines in Determining T-Lymphocyte Function. Curr. Opin. Immunol. 1994, 6, 458–466. [Google Scholar] [CrossRef]
- Lyke, K.E.; Fernández-Viňa, M.A.; Cao, K.; Hollenbach, J.; Coulibaly, D.; Kone, A.K.; Guindo, A.; Burdett, L.A.; Hartzman, R.J.; Wahl, A.R.; et al. Association of HLA alleles with Plasmodium falciparum severity in Malian children. Tissue Antigens 2011, 77, 562–571. [Google Scholar] [CrossRef]
- Garamszegi, L.Z. Global distribution of malaria-resistant MHC-HLA alleles: The number and frequencies of alleles and malaria risk. Malar. J. 2014, 13, 349. [Google Scholar] [CrossRef]
- Stern, L.J.; Brown, J.H.; Jardetzky, T.S.; Gorga, J.C.; Urban, R.G.; Strominger, J.L.; Wiley, D.C. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature 1994, 368, 215–221. [Google Scholar] [CrossRef]
- Hennecke, J.; Wiley, D.C. Structure of a complex of the human alpha/beta T cell receptor (TCR) HA1.7, influenza hemagglutinin peptide, and major histocompatibility complex class II molecule, HLA-DR4 (DRA*0101 and DRB1*0401): Insight into TCR cross-restriction and alloreactivity. J. Exp. Med. 2002, 195, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Espejo, F.; Bermúdez, A.; Torres, E.; Urquiza, M.; Rodríguez, R.; López, Y.; Patarroyo, M.E. Shortening and modifying the 1513 MSP-1 peptide’s alpha-helical region induces protection against malaria. Biochem. Biophys. Res. Commun. 2004, 315, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Gardner, M.; Shallom, S.; Carlton, J.; Salzberg, S.; Nene, V.; Shoaibi, A.; Ciecko, A. Sequence of Plasmodium falciparum Chromosomes 2, 10, 11 and 14. Nature 2002, 419, 527–531. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2-A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Verspurten, J.; Gevaert, K.; Declercq, W.; Vandenabeele, P. SitePredicting the Cleavage of Proteinase Substrates. Trends Biochem. Sci. 2009, 34, 319–323. [Google Scholar] [CrossRef]
- Saha, S.; Bhasin, M.; Raghava, G. Bcipep: A Database of B-Cell Epitopes. BMC Genom. 2005, 6, 1–7. [Google Scholar] [CrossRef]
- Vita, R.; Overton, J.; Greenbaum, J.A.; Ponomarenko, J.; Clark, J.D.; Cantrell, J.R.; Wheeler, D.K.; Gabbard, J.L.; Hix, D.; Sette, A.; et al. The Immune Epitope Database (IEDB) 3.0. Nucleic Acids Res. 2015, 43, D405–D412. [Google Scholar] [CrossRef]
- Fleri, W.; Paul, S.; Dhanda, S.K.; Mahajan, S.; Xu, X.; Peters, B.; Sette, A. The Immune Epitope Database and Analysis Resource in Epitope Discovery and Synthetic Vaccine Design. Front. Immunol. 2017, 8, 278. [Google Scholar] [CrossRef]
- Chen, J.S.; Liu, H.; Yang, J.; Chou, K. Prediction of Linear B-Cell Epitopes Using Amino Acid Pair Antigenicity Scale. Amino Acids 2007, 33, 423–428. [Google Scholar] [CrossRef]
- El-Manzalawy, Y.; Dobbs, D.; Honavar, V. Predicting Linear B-Cell Epitopes Using String Kernels. J. Mol. Recognit. 2008, 21, 243–255. [Google Scholar] [CrossRef]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving Sequence-Based B-Cell Epitope Prediction Using Conformational Epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef]
- Jensen, K.K.; Andreatta, M.; Marcatili, P.; Buus, S.; Greenbaum, J.A.; Yan, Z.; Sette, A.; Peters, B.; Nielsen, M. Improved Methods for Predicting Peptide Binding Affinity to MHC Class II Molecules. Immunology 2018, 154, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Lozano, J.M.; Varela, Y.; Silva, Y.; Ardila, K.; Forero, M.; Guasca, L.; Guerrero, Y.; Bermudez, A.; Alba, P.; Vanegas, M.; et al. A Large Size Chimeric Highly Immunogenic Peptide Presents Multistage Plasmodium Antigens as a Vaccine Candidate System against Malaria. Molecules 2017, 22, 1837. [Google Scholar] [CrossRef] [PubMed]
- Lozano, J.M.; Montoya-Fajardo, F.J.; Hoebecke, J.; Cifuentes, G.; Forero, M.; Patarroyo, M.E. Antibodies Induced by Plasmodium falciparum Merozoite Surface Antigen-2-Designed Pseudopeptides Possess Neutralizing Properties of the in vitro Malarial Infection. Peptides 2007, 28, 1954–1965. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Guidance on Regulations for the Transport of Infectious Substances 2019–2020; World Health Organization: Geneva, Switzerland, 2019.
- Singh, B.; Cabrera-Mora, M.; Jiang, J.; Galinski, M.; Moreno, A. Genetic Linkage of Autologous T Cell Epitopes in a Chimeric Recombinant Construct Improves Anti-Parasite and Anti-Disease Protective Effect of a Malaria Vaccine Candidate. Vaccine 2010, 28, 2580. [Google Scholar] [CrossRef] [PubMed]
- National Research Council (US). Guide for the Care and Use of Laboratory Animals; National Academies Press: Cambridge, MA, USA, 2011. [Google Scholar] [CrossRef]
- Florez, M.M.; Rodríguez, R.; Cabrera, J.A.; Robledo, S.M.; Delgado, G.; Pavanelli, W.; Pal, C.; Palmisano, G. Leishmania Spp Epitopes in Humans Naturally Resistant to the Disease: Working Toward a Synthetic Vaccine. Front. Cell Infect. Microbiol. 2021, 11, 631019. [Google Scholar] [CrossRef]
- Lozano, J.M.; Muller, S. Monkeypox: Potential vaccine development strategies. Trends Pharmacol. Sci. 2023, 44, 15–19. [Google Scholar] [CrossRef]

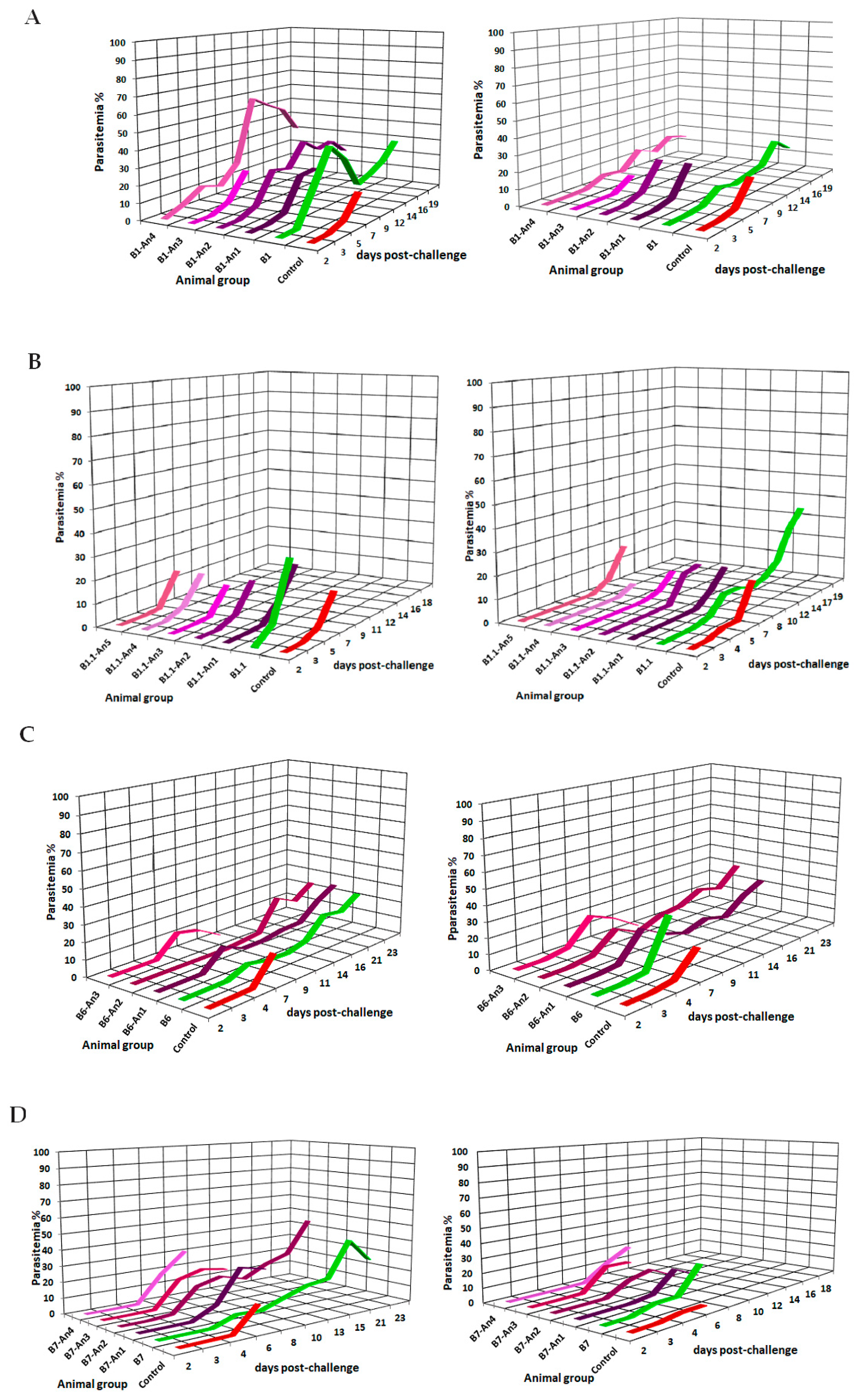

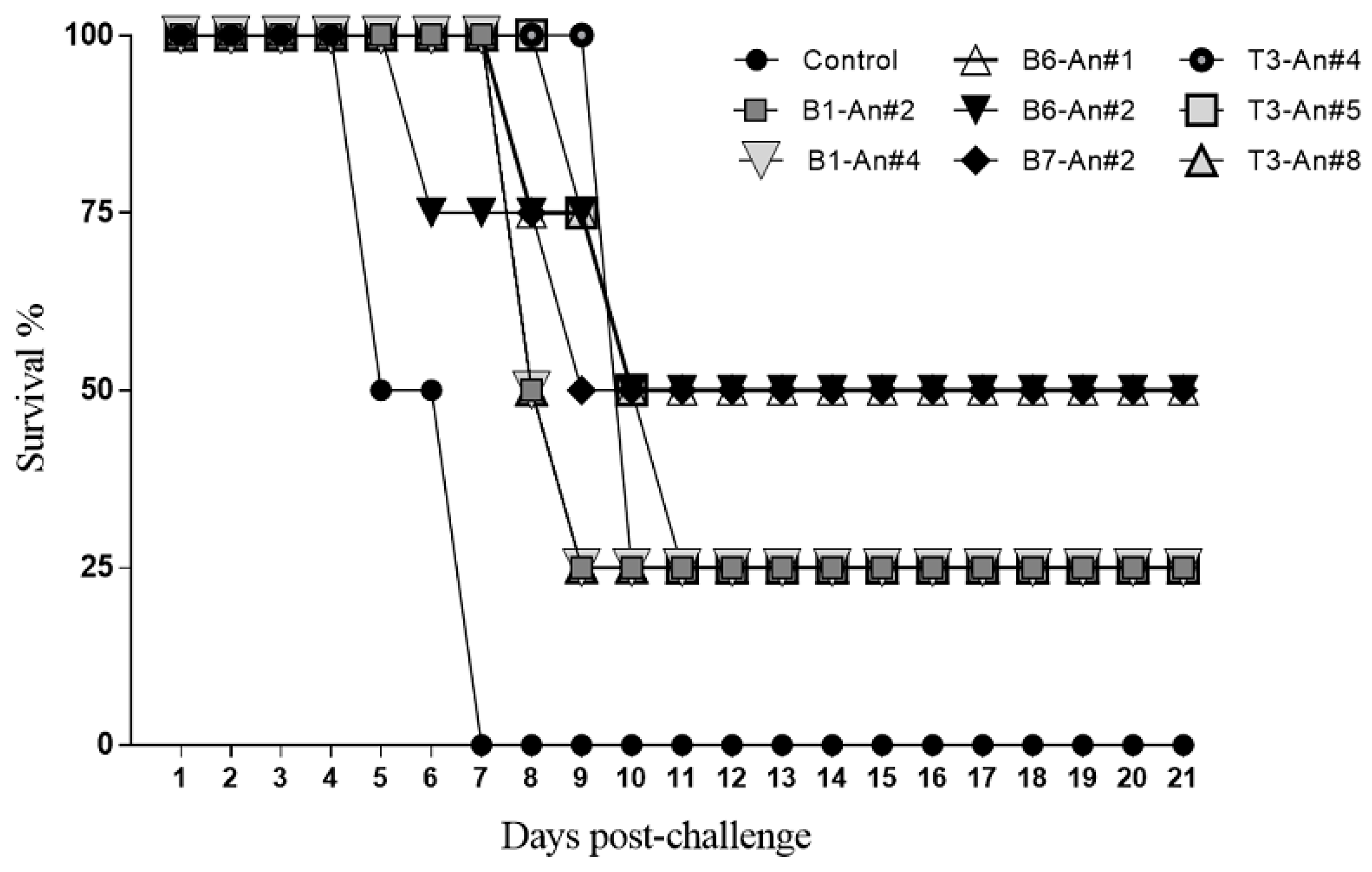
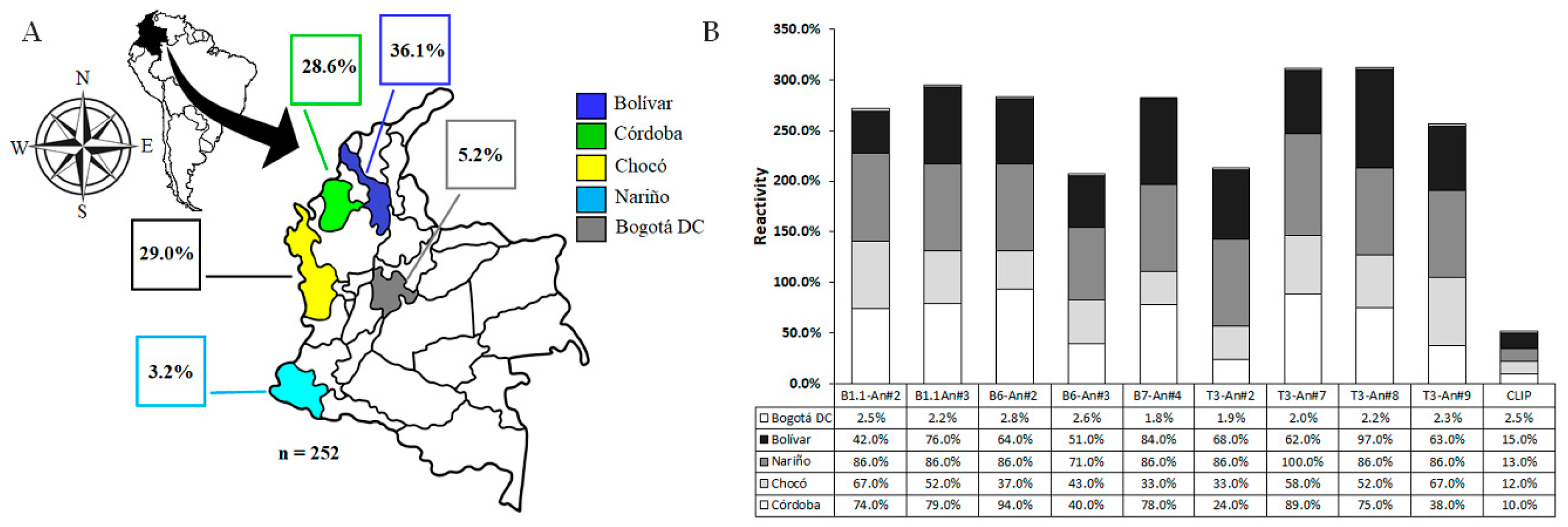
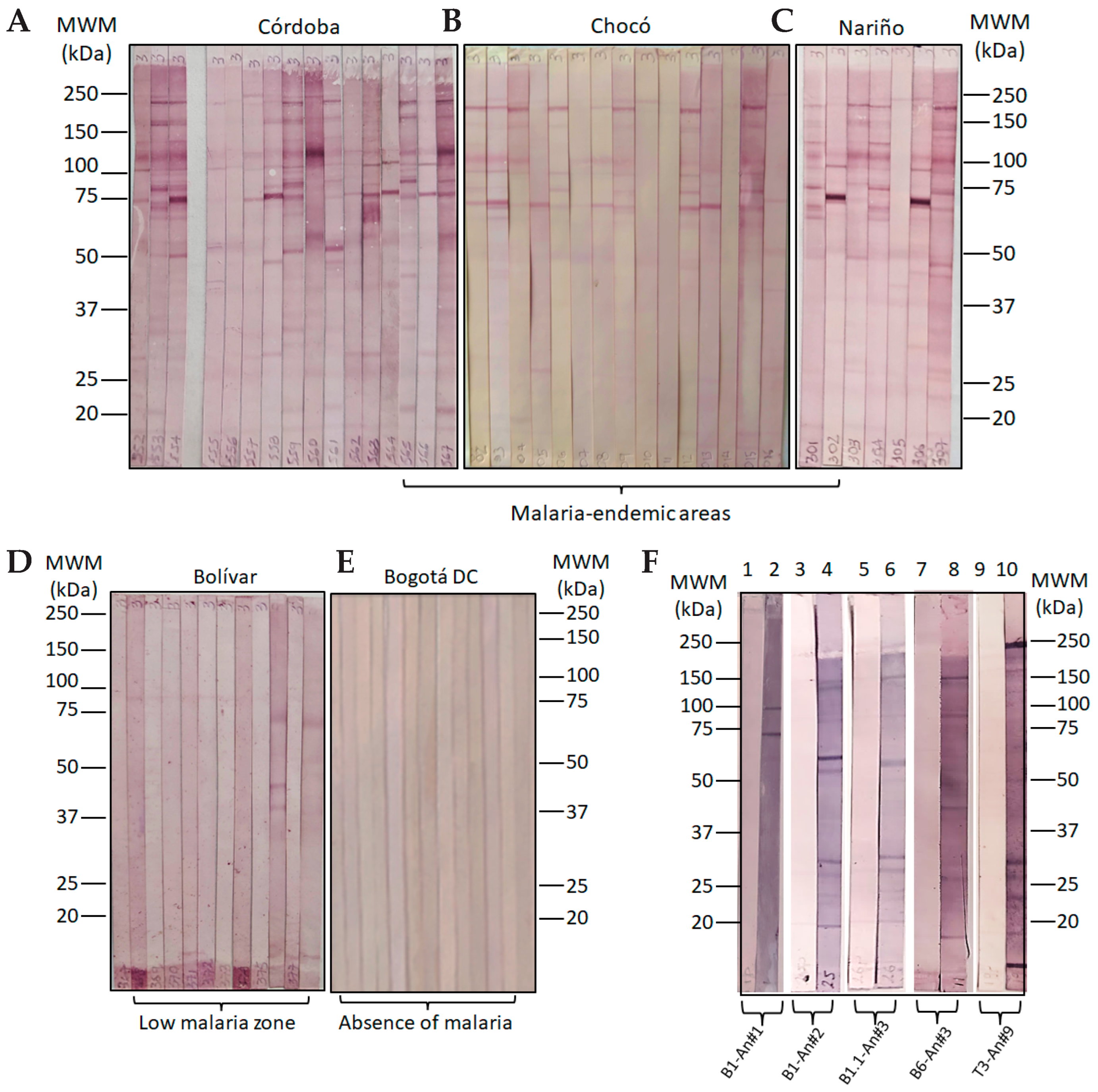
| Epitope-Peptide | Modification Position | Antigenicity a | Anti-Molecule Ig Titers | |
|---|---|---|---|---|
| B1 | B1 | Pf.3D7.MSP-142–61 | ** | 1:400 |
| B1-An1 | Pf.3D7.MSP-1-G42-ψ[CH2-NH]-Y43- | ** | 1:800 | |
| B1-An2 | Pf.3D7.MSP-1-Y43-ψ[CH2-NH]-S44- | ** | 1:51,200 | |
| B1-An3 | Pf.3D7.MSP-1-L45-ψ[CH2-NH]-F46- | NS | ND | |
| B1-An4 | Pf.3D7.MSP-1-M51-ψ[CH2-NH]-V52- | **** | 1:400 | |
| B1.1 | B1.1 | Pf.3D7.MSP-138–58 | ** | 1:400 |
| B1.1-An1 | Pf.3D7.MSP-1-V52-ψ[CH2-NH]-L53- | * | 1:200 | |
| B1.1-An2 | Pf.3D7.MSP-1-M51-ψ[CH2-NH]-V52- | ** | 1:200 | |
| B1.1-An3 | Pf.3D7.MSP-1-K50-ψ[CH2-NH]-M51- | ** | 1:12,800 | |
| B1.1-An4 | Pf.3D7.MSP-1-E49-ψ[CH2-NH]-K50- | *** | 1:200 | |
| B1.1-An5 | Pf.3D7.MSP-1-K48-ψ[CH2-NH]-E49- | ** | 1:400 | |
| B6 | B6 | Pf.3D7.MSP-11545-1560 | ** | 1:200 |
| B6-An1 | Pf.3D7.MSP-1-Y-dL1546-K- | ** | 1:6400 | |
| B6-An2 | Pf.3D7.MSP-1-Y-dK1547-P- | ** | 1:800 | |
| B6-An3 | Pf.3D7.MSP-1-K-dP1548-L- | ** | 1:200 | |
| B7 | B7 | Pf.3D7.MSP-11803–1821 | ** | 1:400 |
| B7-An1 | Pf.3D7.MSP-1-M1803-ψ[CH2-NH]-L1804- | ** | 1:400 | |
| B7-An2 | Pf.3D7.MSP-1-L1804-ψ[CH2-NH]-N1805- | **** | 1:400 | |
| B7-An3 | Pf.3D7.MSP-1-K1815-ψ[CH2-NH]-Q1816- | * | 1:200 | |
| B7-An4 | Pf.3D7.MSP-1-C1817-ψ[CH2-NH]-P1818- | ** | 1:200 | |
| T3 | T3 | Pf.3D7.MSP-1217–236 | *** | 1:200 |
| T3-An1 | Pf.3D7.MSP-1-L222-ψ[CH2-NH]-K223- | **** | 1:1600 | |
| T3-An2 | Pf.3D7.MSP-1-K223-ψ[CH2-NH]-I224- | NS | ND | |
| T3-An3 | Pf.3D7.MSP-1-R225-ψ[CH2-NH]-A226- | *** | 1:1600 | |
| T3-An4 | Pf.3D7.MSP-1-N227-ψ[CH2-NH]-E228- | * | 1:3200 | |
| T3-An5 | Pf.3D7.MSP-1-L229-ψ[CH2-NH]-D230- | * | 1:1600 | |
| T3-An6 | Pf.3D7.MSP-1-D230-ψ[CH2-NH]-V231- | ** | 1:800 | |
| T3-An7 | Pf.3D7.MSP-1-V231-ψ[CH2-NH]-L232- | **** | 1:51,200 | |
| T3-An8 | Pf.3D7.MSP-1-L232-ψ[CH2-NH]-K233- | **** | 1:51,200 | |
| T3-An9 | Pf.3D7.MSP-1-K234-ψ[CH2-NH]-L235- | ** | 1:51,200 | |
| T3-An10 | Pf.3D7.MSP-1-L235-ψ[CH2-NH]-V236- | ** | 1:3200 |
| Molecule | TNF | IFNγ | IL-2 | IL-4 | IL-5 | Associated Immune Pattern | |
|---|---|---|---|---|---|---|---|
| [pg/mL] | [pg/mL] | [pg/mL] | [pg/mL] | [pg/mL] | |||
| PHA | 6.35 | 139.86 | 366.31 | 17.98 | 221.93 | ||
| Control | 3.81 | 1.18 | 17.01 | 4.73 | ND | ||
| Cell stimulation at 48 h | B1 | 15.16 | 2.32 | 202.04 | 9.29 | 33.86 | Th1 |
| B1An4 | ND | 1.12 | ND | 3.31 | ND | Th1/Th2 | |
| B1An2 | 3.59 | 1.22 | 79.29 | 8.08 | 1.05 | Th1 | |
| B1.1 | 25.07 | 1.25 | 108.78 | 5.65 | 3.25 | Th1 | |
| B6An1 | 26.11 | 1.58 | 110.59 | 2.87 | 6.54 | Th1 | |
| B6An2 | 7.75 | 1.28 | 7.17 | 5.11 | 11.12 | Th1/Th2 | |
| T3 | 14.82 | 1.35 | 94.90 | 4.64 | ND | Th1 | |
| T3An5 | ND | 1.16 | 10.09 | 6.66 | 3.54 | Th1/Th2 | |
| Control | 3.61 | 1.06 | ND | ND | ND | ||
| Cell stimulation at 72 h | B1 | 22.44 | 1.39 | 38.44 | 3.86 | 2.68 | Th1 |
| B1An4 | ND | 1.05 | ND | 3.73 | ND | Th1/Th2 | |
| B1An2 | ND | 1.26 | 118.65 | 6.01 | 3.41 | Th1 | |
| B1.1 | 9.97 | 1.14 | 15.6 | 6.07 | 6.34 | Th1/Th2 | |
| B6An1 | 97.25 | 2.38 | 324.1 | 13.37 | 21.99 | Th1 | |
| B6An2 | 47.22 | 1.46 | 43.36 | 8.06 | 9.36 | Th1 | |
| T3 | 6.18 | 1.29 | 74.65 | 4.72 | 3.32 | Th1 | |
| T3An5 | ND | 1.12 | 12.34 | 5.21 | 5.64 | Th1/Th2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez, Z.J.; Melo, F.L.; Torres, A.; Agrawal, N.; Cortés-Vecino, J.A.; Lozano, J.M. Natural Plasmodium falciparum Infection Stimulates Human Antibodies to MSP1 Epitopes Identified in Mice Infection Models upon Non-Natural Modified Peptidomimetic Vaccination. Molecules 2023, 28, 2527. https://doi.org/10.3390/molecules28062527
Rodríguez ZJ, Melo FL, Torres A, Agrawal N, Cortés-Vecino JA, Lozano JM. Natural Plasmodium falciparum Infection Stimulates Human Antibodies to MSP1 Epitopes Identified in Mice Infection Models upon Non-Natural Modified Peptidomimetic Vaccination. Molecules. 2023; 28(6):2527. https://doi.org/10.3390/molecules28062527
Chicago/Turabian StyleRodríguez, Zully Johana, Fredy Leonardo Melo, Angela Torres, Nikhil Agrawal, Jesús Alfredo Cortés-Vecino, and José Manuel Lozano. 2023. "Natural Plasmodium falciparum Infection Stimulates Human Antibodies to MSP1 Epitopes Identified in Mice Infection Models upon Non-Natural Modified Peptidomimetic Vaccination" Molecules 28, no. 6: 2527. https://doi.org/10.3390/molecules28062527
APA StyleRodríguez, Z. J., Melo, F. L., Torres, A., Agrawal, N., Cortés-Vecino, J. A., & Lozano, J. M. (2023). Natural Plasmodium falciparum Infection Stimulates Human Antibodies to MSP1 Epitopes Identified in Mice Infection Models upon Non-Natural Modified Peptidomimetic Vaccination. Molecules, 28(6), 2527. https://doi.org/10.3390/molecules28062527








