Development and Recent Advances in Lysine and N-Terminal Bioconjugation for Peptides and Proteins
Abstract
1. Introduction
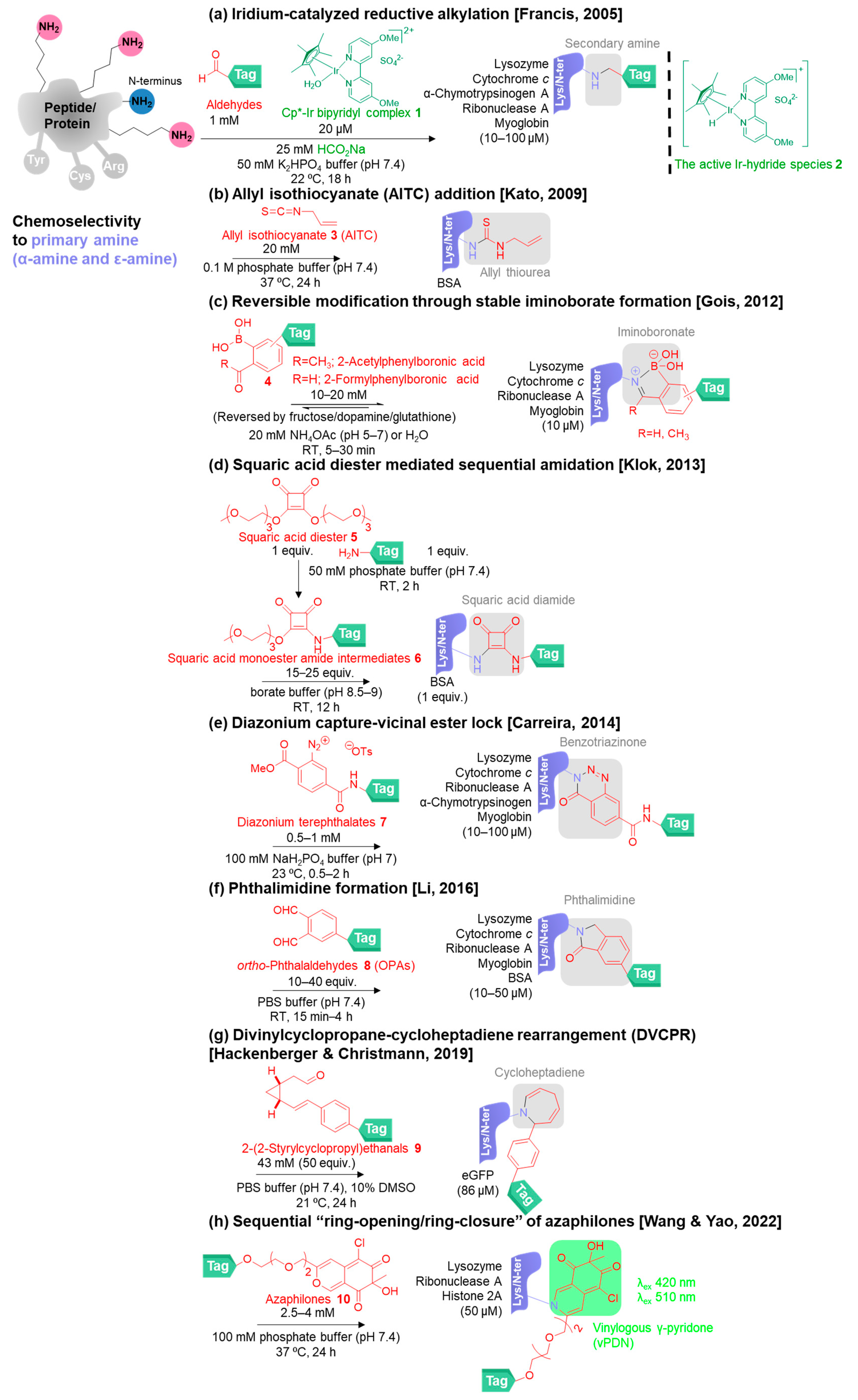
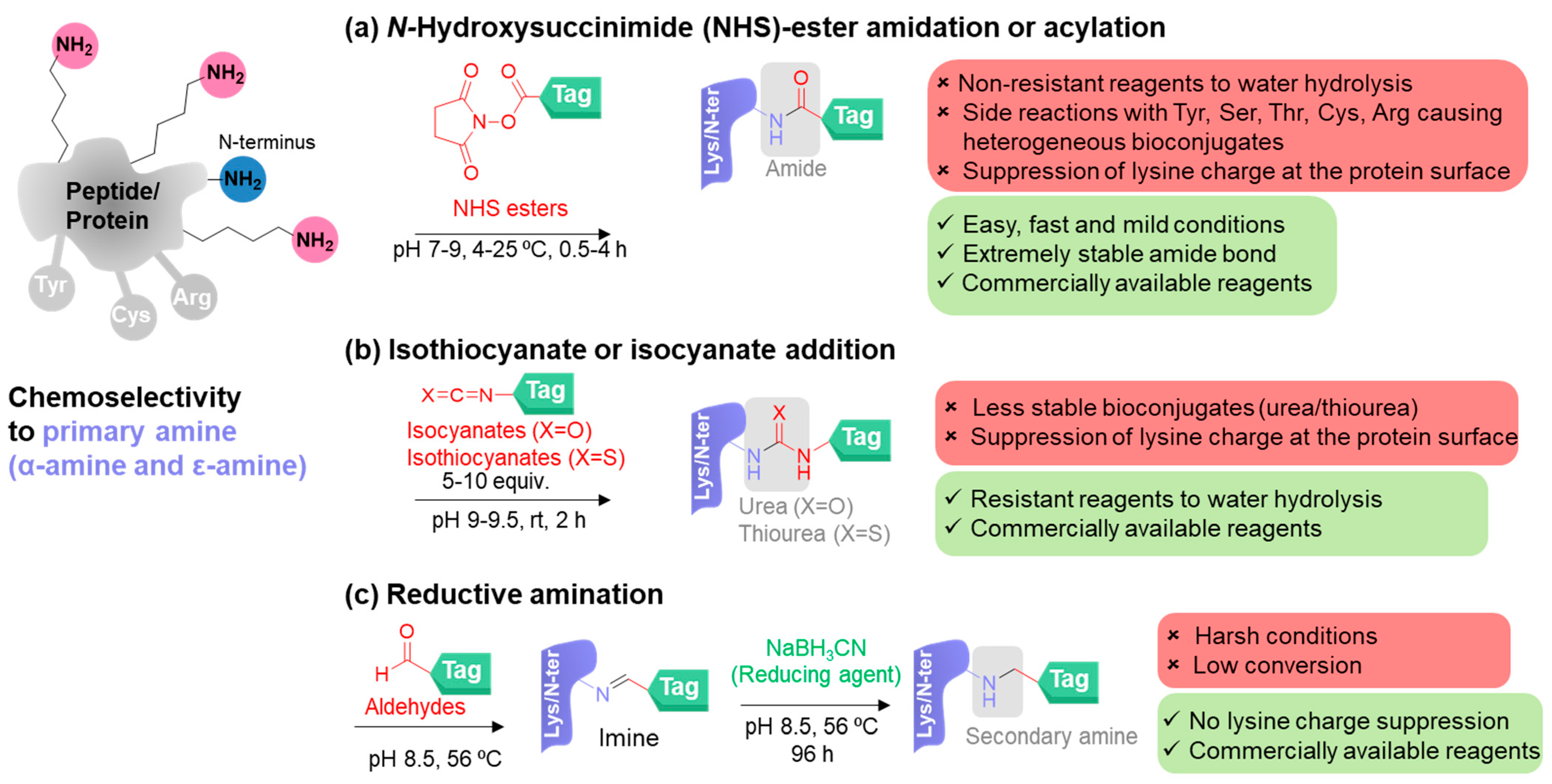
2. Chemoselective Methods for Primary Amine Modification
2.1. Classical Methods for Primary Amine Modification
2.2. New Methods for Primary Amine Modification
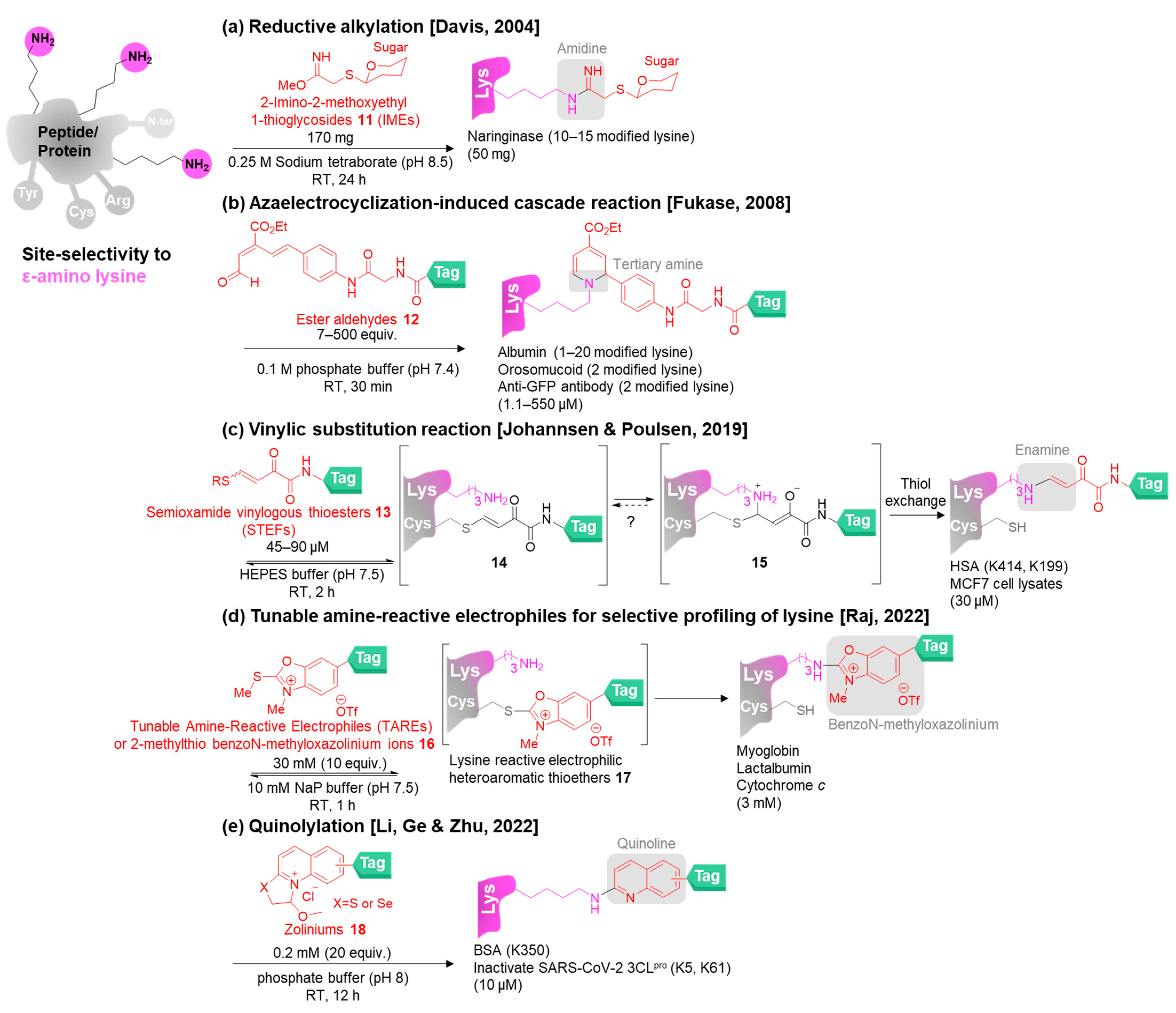
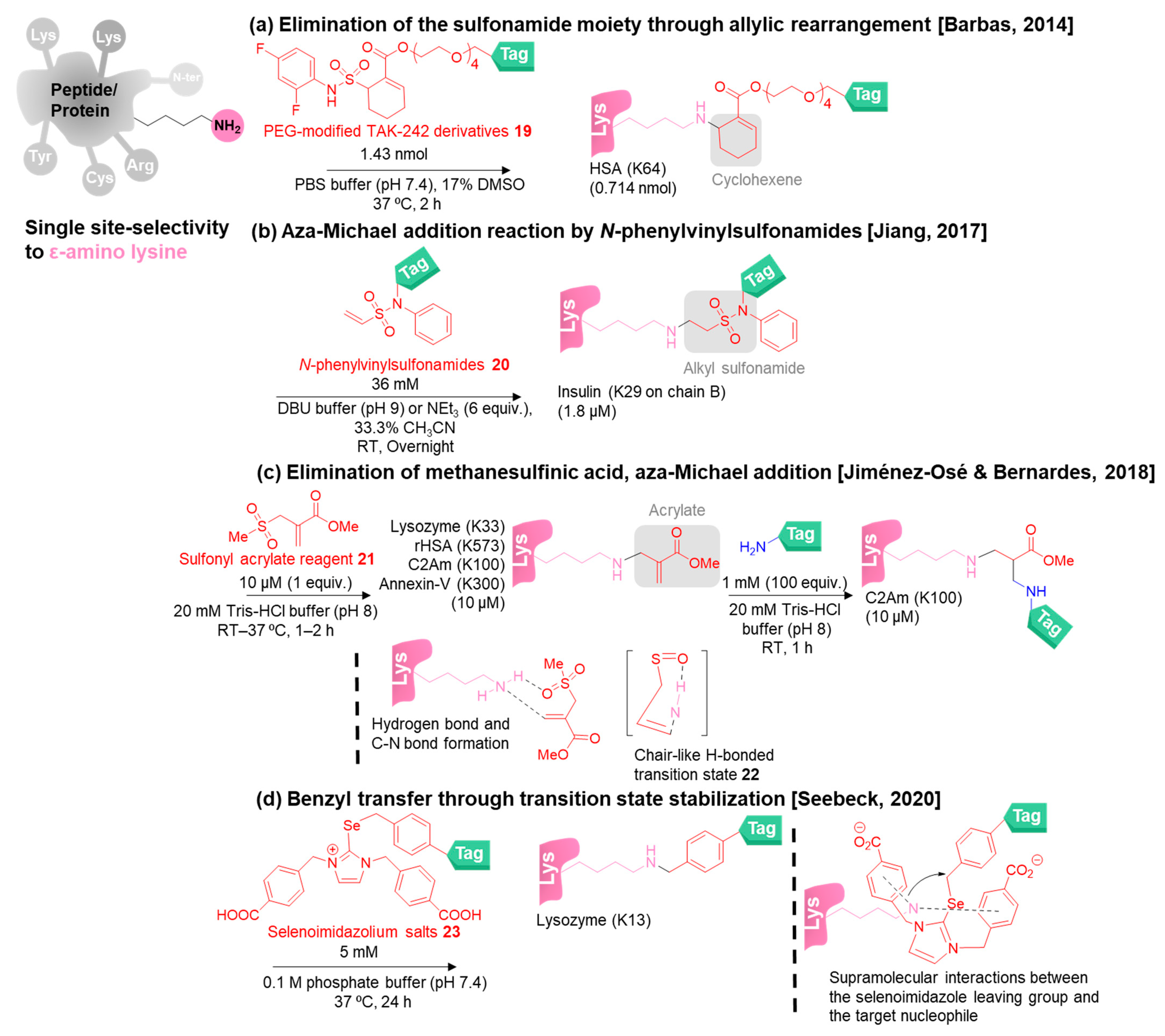
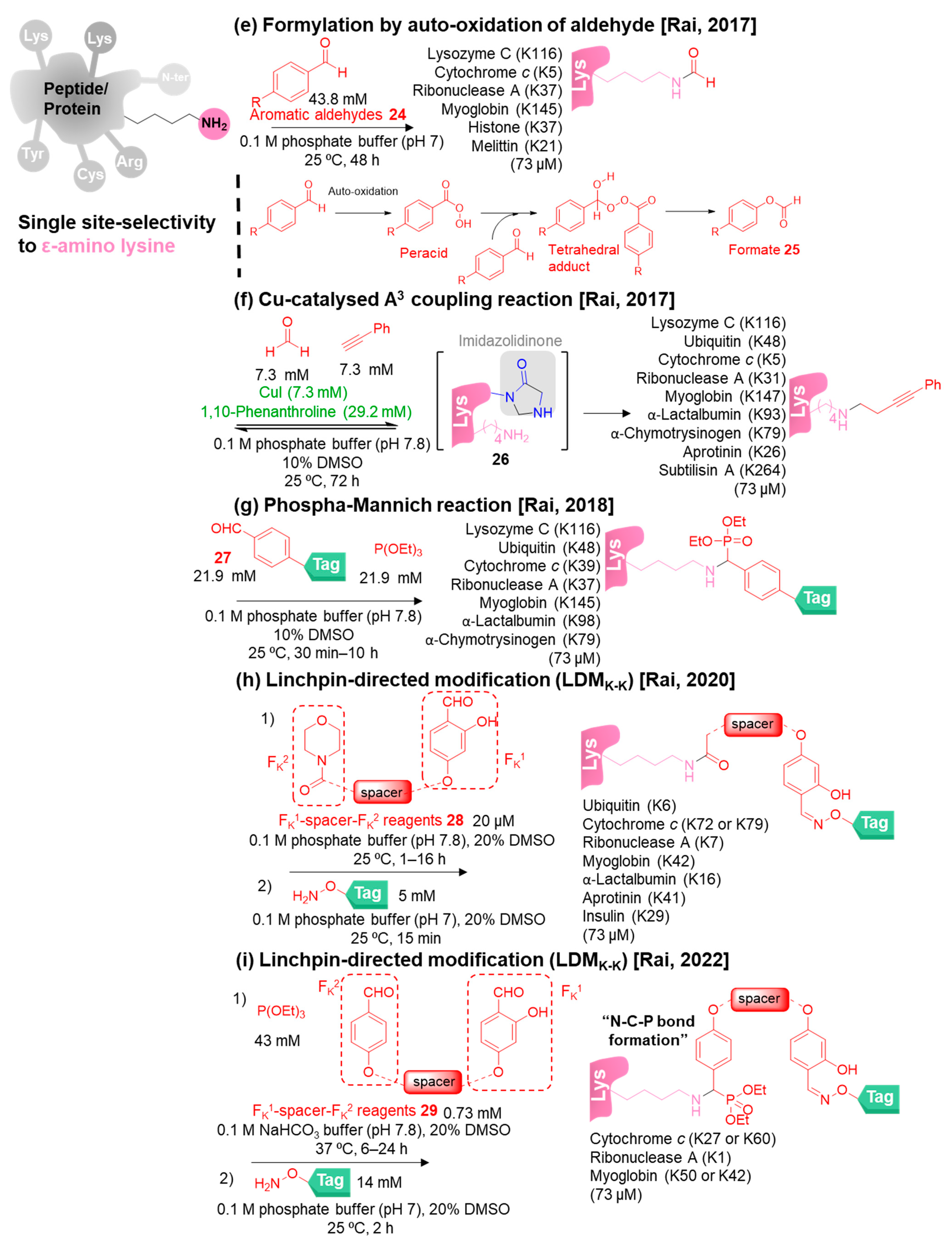
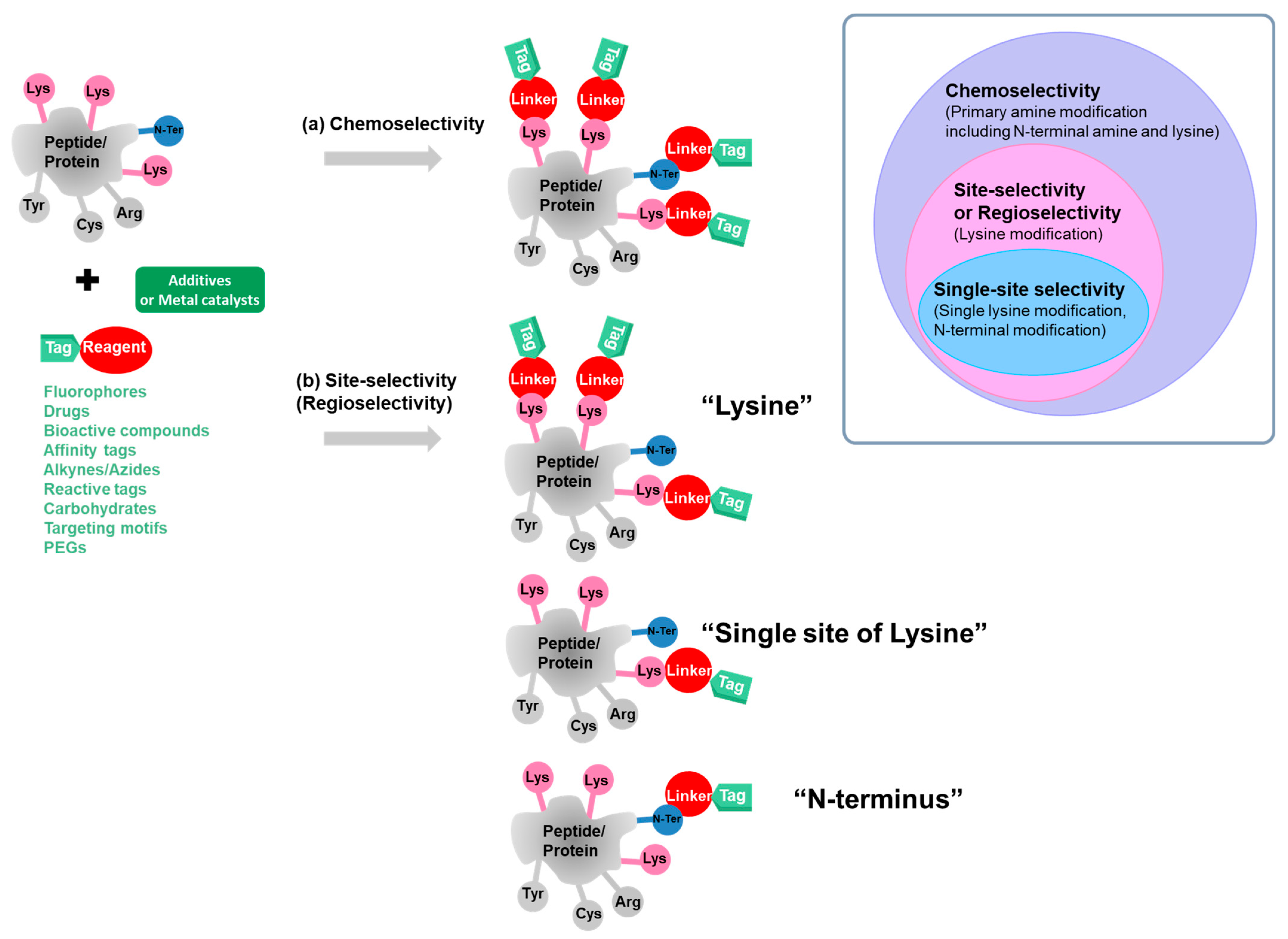
3. Site-Selective Methods for Primary Amine Modification
3.1. Site-Selective Lysine Modification
3.2. Single-Site-Selective Lysine Modification
3.3. Site-Selective N-Terminal Modification
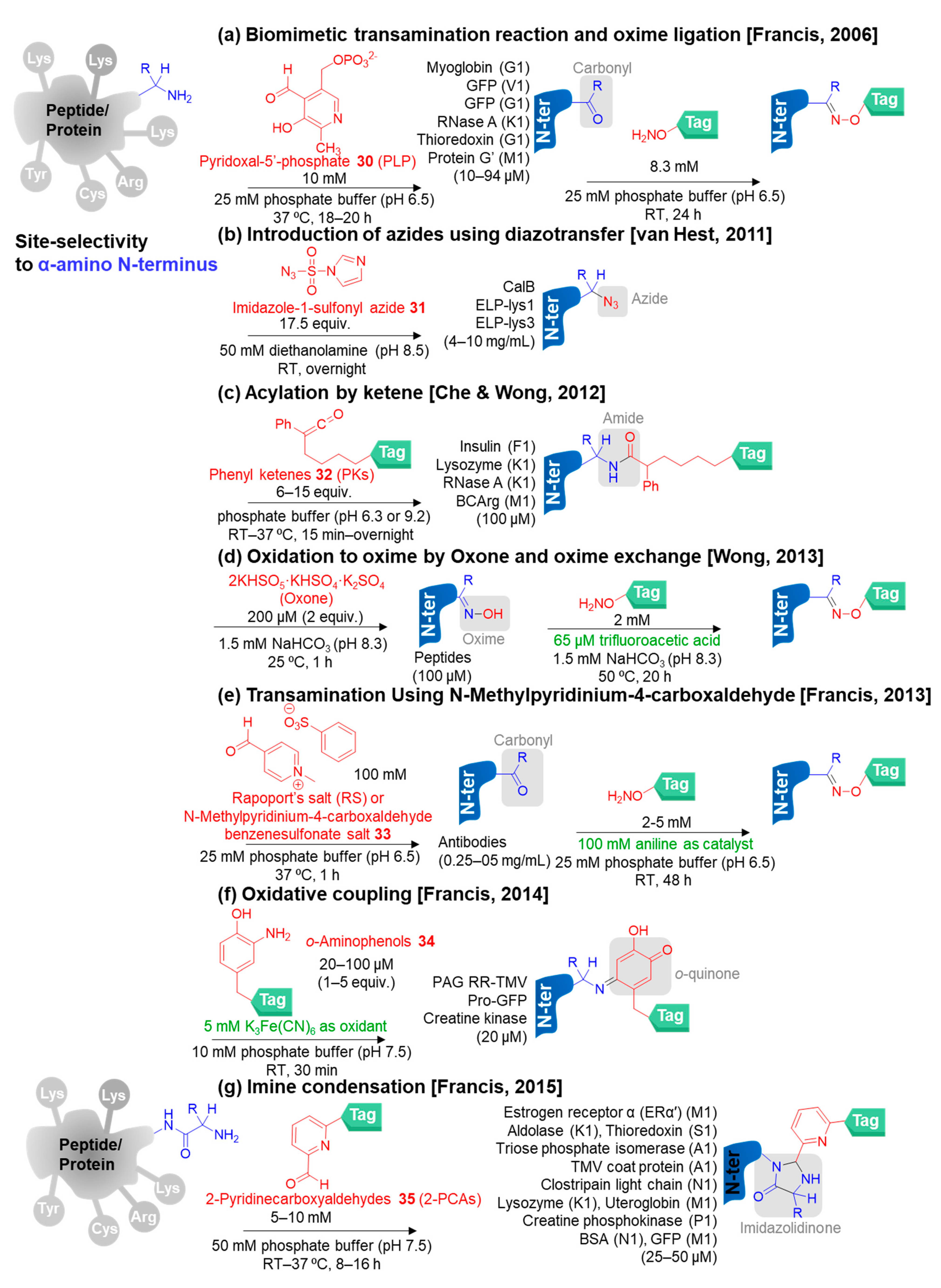
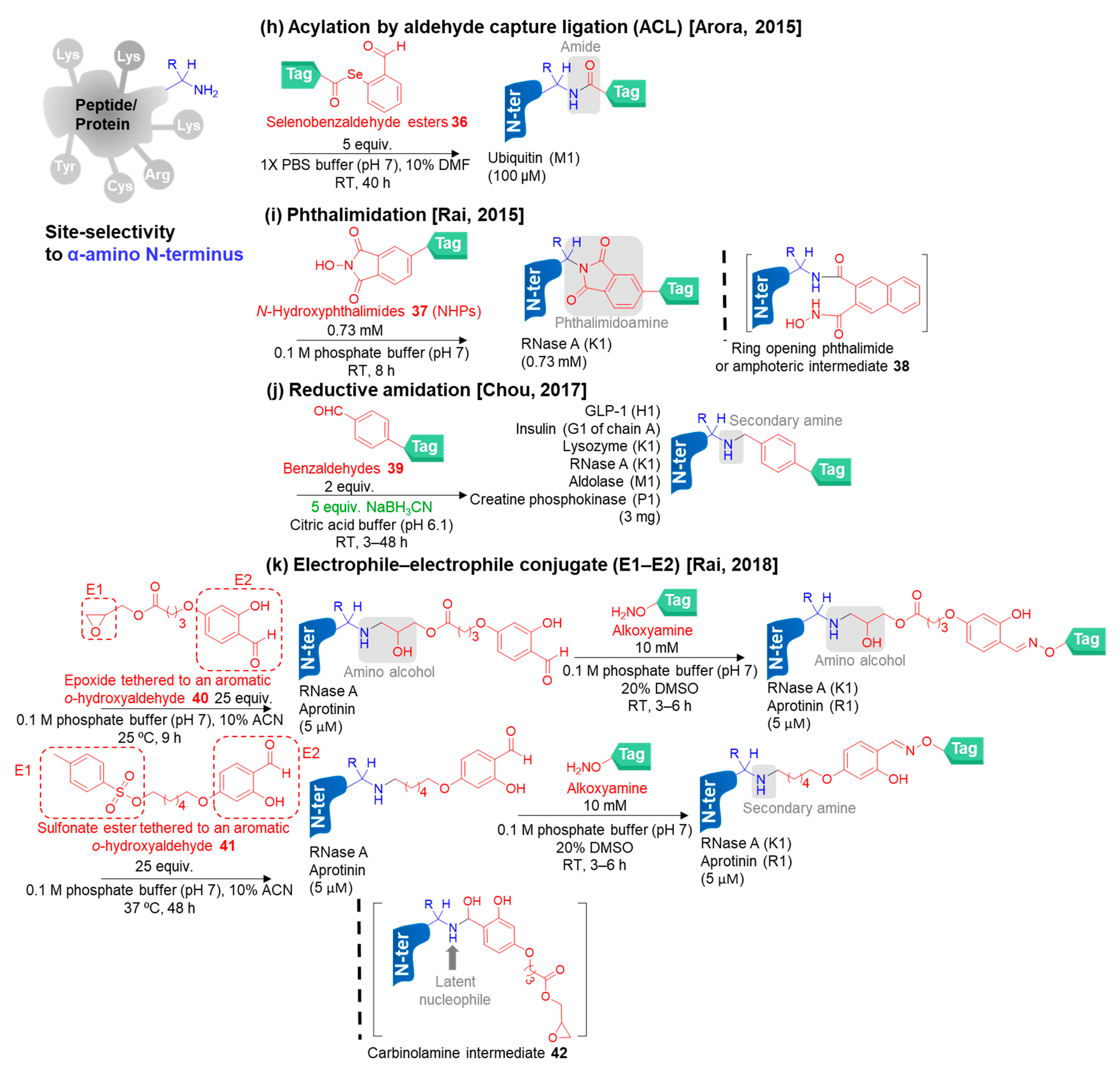
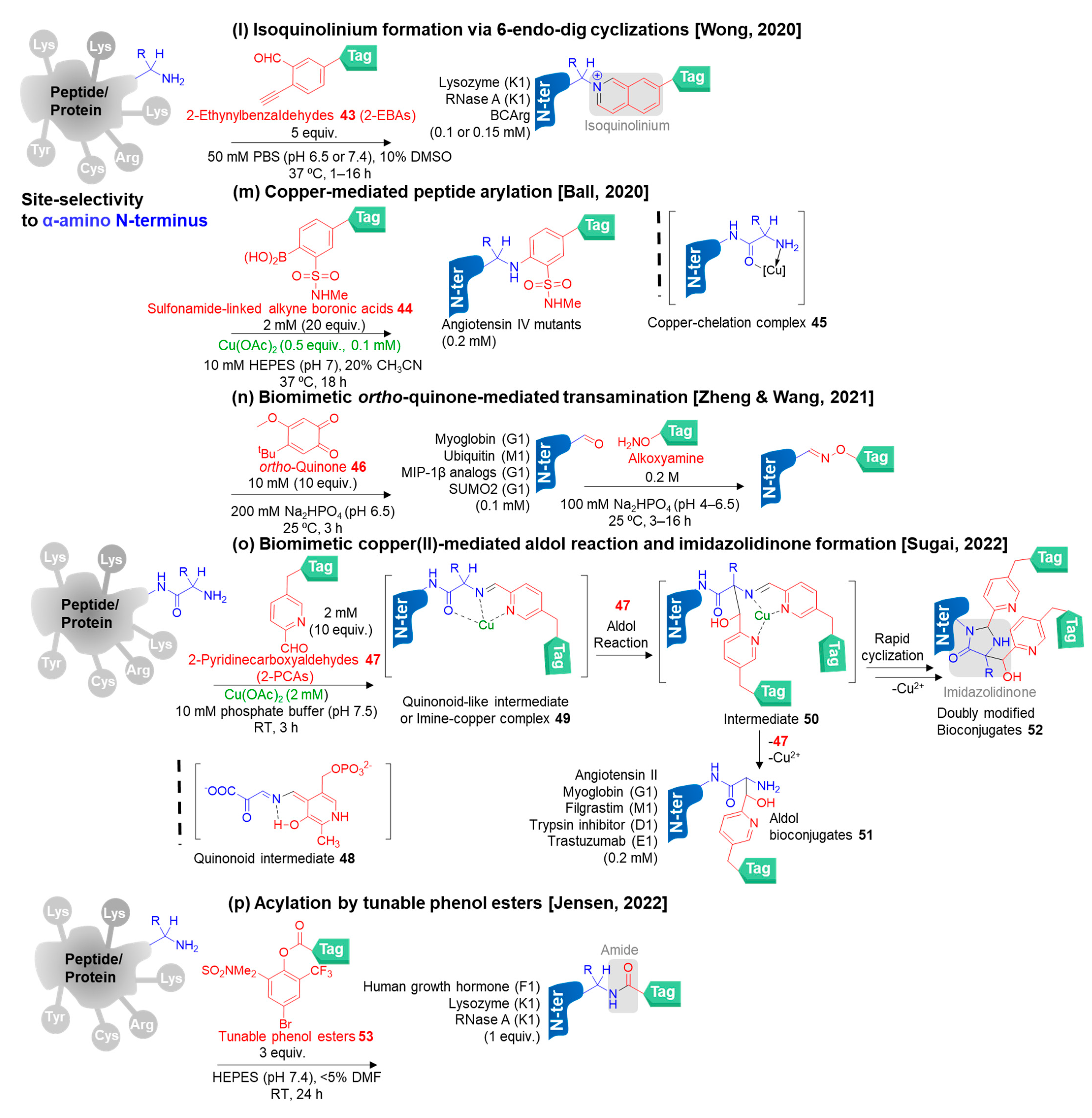
3.3.1. Selective N-Terminal Modification on α-Amino Group
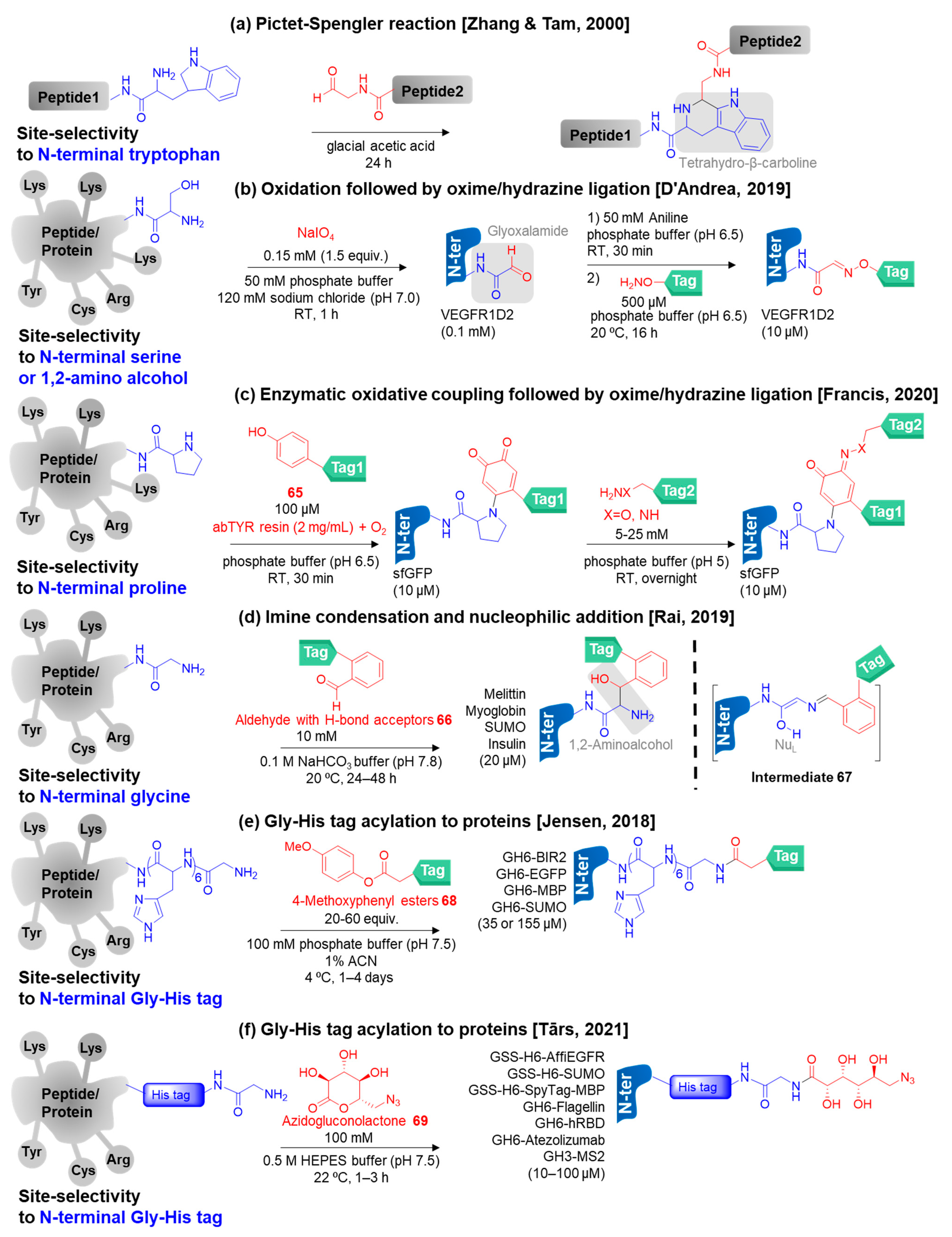
3.3.2. Selective N-Terminal Modification on Certain Amino Acid Residues
- Cysteine
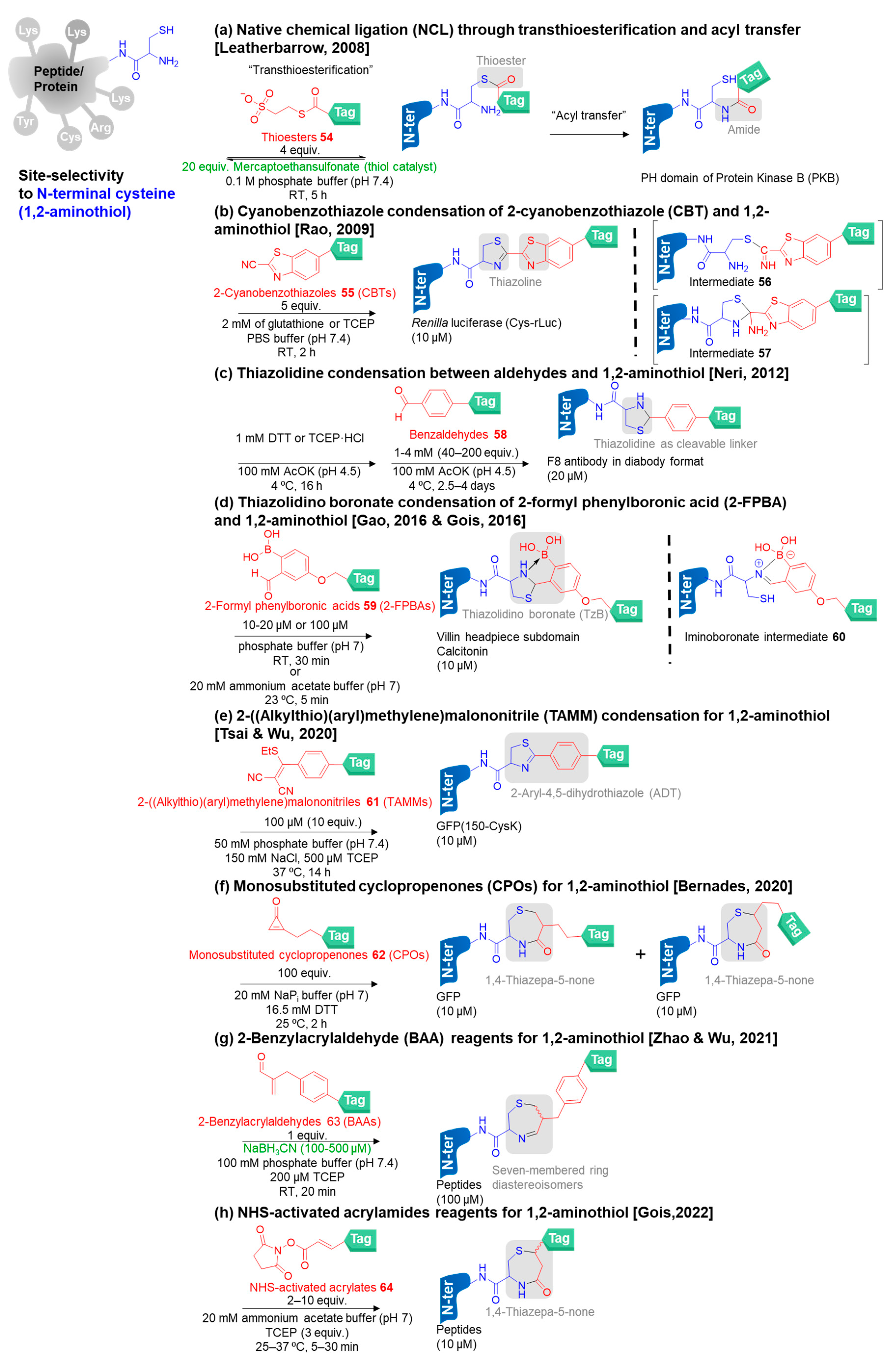
- Other specific N-terminal amino acids and special sequences at the N-terminus
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stephanopoulos, N.; Francis, M.B. Choosing an effective protein bioconjugation strategy. Nat. Chem. Biol. 2011, 7, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Sletten, E.M.; Bertozzi, C.R. Bioorthogonal chemistry: Fishing for selectivity in a sea of functionality. Angew. Chem. Int. Ed. 2009, 48, 6974–6998. [Google Scholar] [CrossRef] [PubMed]
- Rudra, A.; Li, J.; Shakur, R.; Bhagchandani, S.; Langer, R. Trends in therapeutic conjugates: Bench to clinic. Bioconjug. Chem. 2020, 31, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Kalia, J.; Raines, R.T. Advances in bioconjugation. Curr. Org. Chem. 2010, 14, 138–147. [Google Scholar] [CrossRef]
- Kolb, H.C.; Finn, M.; Sharpless, K.B. Click chemistry: Diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. 2001, 40, 2004–2021. [Google Scholar] [CrossRef]
- Zaia, J. The 2022 Nobel Prize in Chemistry for the development of click chemistry and bioorthogonal chemistry. Anal. Bioanal. Chem. 2023, 415, 527–532. [Google Scholar] [CrossRef]
- Available online: https://www.nobelprize.org/prizes/chemistry/2022/prize-announcement/ (accessed on 19 January 2023).
- Ramazi, S.; Zahiri, J. Post-translational modifications in proteins: Resources, tools and prediction methods. Database 2021, 2021. [Google Scholar] [CrossRef]
- Spicer, C.D.; Davis, B.G. Selective chemical protein modification. Nat. Commun. 2014, 5, 1–14. [Google Scholar] [CrossRef]
- Boutureira, O.; Bernardes, G.a.J. Advances in chemical protein modification. Chem. Rev. 2015, 115, 2174–2195. [Google Scholar] [CrossRef]
- Koniev, O.; Wagner, A. Developments and recent advancements in the field of endogenous amino acid selective bond forming reactions for bioconjugation. Chem. Soc. Rev. 2015, 44, 5495–5551. [Google Scholar] [CrossRef]
- Krall, N.; Da Cruz, F.P.; Boutureira, O.; Bernardes, G.J. Site-selective protein-modification chemistry for basic biology and drug development. Nat. Chem. 2016, 8, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Rawale, D.G.; Thakur, K.; Adusumalli, S.R.; Rai, V. Chemical methods for selective labeling of proteins. Eur. J. Org. Chem. 2019, 2019, 6749–6763. [Google Scholar] [CrossRef]
- Hoyt, E.A.; Cal, P.M.; Oliveira, B.L.; Bernardes, G.J. Contemporary approaches to site-selective protein modification. Nat. Rev. Chem. 2019, 3, 147–171. [Google Scholar] [CrossRef]
- Sakamoto, S.; Hamachi, I. Recent progress in chemical modification of proteins. Anal. Sci. 2019, 35, 5–27. [Google Scholar] [CrossRef]
- Reddy, N.C.; Kumar, M.; Molla, R.; Rai, V. Chemical methods for modification of proteins. Org. Biomol. Chem. 2020, 18, 4669–4691. [Google Scholar] [CrossRef]
- Adakkattil, R.; Thakur, K.; Rai, V. Reactivity and selectivity principles in native protein bioconjugation. Chem. Rec. 2021, 21, 1941–1956. [Google Scholar] [CrossRef]
- Kumar, M.; Reddy, N.C.; Rai, V. Chemical technologies for precise protein bioconjugation interfacing biology and medicine. Chem. Commun. 2021, 57, 7083–7095. [Google Scholar] [CrossRef]
- Sornay, C.; Vaur, V.; Wagner, A.; Chaubet, G. An overview of chemo-and site-selectivity aspects in the chemical conjugation of proteins. R. Soc. Open Sci. 2022, 9, 211563. [Google Scholar] [CrossRef]
- Xie, Y.; Du, S.; Liu, Z.; Liu, M.; Xu, Z.; Wang, X.; Kee, J.X.; Yi, F.; Sun, H.; Yao, S.Q. Chemical biology tools for protein lysine acylation. Angew. Chem. Int. Ed. 2022, e202200303. [Google Scholar]
- Liu, Z.; Chen, X. Simple bioconjugate chemistry serves great clinical advances: Albumin as a versatile platform for diagnosis and precision therapy. Chem. Soc. Rev. 2016, 45, 1432–1456. [Google Scholar] [CrossRef]
- Gomez, S.; Tsung, A.; Hu, Z. Current targets and bioconjugation strategies in photodynamic diagnosis and therapy of cancer. Molecules 2020, 25, 4964. [Google Scholar] [CrossRef] [PubMed]
- Birrer, M.J.; Moore, K.N.; Betella, I.; Bates, R.C. Antibody-drug conjugate-based therapeutics: State of the science. J. Natl. Cancer Inst. 2019, 111, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Walsh, S.J.; Bargh, J.D.; Dannheim, F.M.; Hanby, A.R.; Seki, H.; Counsell, A.J.; Ou, X.; Fowler, E.; Ashman, N.; Takada, Y. Site-selective modification strategies in antibody–drug conjugates. Chem. Soc. Rev. 2021, 50, 1305–1353. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, X.; Chen, X.; Aramsangtienchai, P.; Tong, Z.; Lin, H. Protein lipidation: Occurrence, mechanisms, biological functions, and enabling technologies. Chem. Rev. 2018, 118, 919–988. [Google Scholar] [CrossRef]
- Lim, S.I.; Kwon, I. Bioconjugation of therapeutic proteins and enzymes using the expanded set of genetically encoded amino acids. Crit. Rev. Biotechnol. 2016, 36, 803–815. [Google Scholar] [CrossRef] [PubMed]
- Kintzing, J.R.; Interrante, M.V.F.; Cochran, J.R. Emerging strategies for developing next-generation protein therapeutics for cancer treatment. Trends Pharmacol. Sci. 2016, 37, 993–1008. [Google Scholar] [CrossRef]
- Bentley, M.D.; Bossard, M.J.; Burton, K.W.; Viegas, T.X. Poly(ethylene) glycol conjugates of biopharmaceuticals in drug delivery. In Modern Biopharmaceuticals: Design, Development and Optimization; Wiley: Weinheim, Germany, 2005; pp. 1393–1418. [Google Scholar]
- Meier-Menches, S.M.; Casini, A. Design strategies and medicinal applications of metal-peptidic bioconjugates. Bioconjug. Chem. 2020, 31, 1279–1288. [Google Scholar] [CrossRef]
- Chen, K.; Conti, P.S. Target-specific delivery of peptide-based probes for PET imaging. Adv. Drug Deliv. Rev. 2010, 62, 1005–1022. [Google Scholar] [CrossRef]
- Ibraheem, A.; Campbell, R.E. Designs and applications of fluorescent protein-based biosensors. Curr. Opin. Chem. Biol. 2010, 14, 30–36. [Google Scholar] [CrossRef]
- Dai, Y.; Wang, C.; Chiu, L.-Y.; Abbasi, K.; Tolbert, B.S.; Sauvé, G.; Yen, Y.; Liu, C.-C. Application of bioconjugation chemistry on biosensor fabrication for detection of TAR-DNA binding protein 43. Biosens. Bioelectron. 2018, 117, 60–67. [Google Scholar] [CrossRef]
- Zhang, G.; Zheng, S.; Liu, H.; Chen, P.R. Illuminating biological processes through site-specific protein labeling. Chem. Soc. Rev. 2015, 44, 3405–3417. [Google Scholar] [CrossRef] [PubMed]
- Himiyama, T.; Okamoto, Y. Artificial metalloenzymes: From selective chemical transformations to biochemical applications. Molecules 2020, 25, 2989. [Google Scholar] [CrossRef] [PubMed]
- Pessatti, T.B.; Terenzi, H.; Bertoldo, J.B. Protein modifications: From chemoselective probes to novel biocatalysts. Catalysts 2021, 11, 1466. [Google Scholar] [CrossRef]
- Liu, X.; Sun, J.; Gao, W. Site-selective protein modification with polymers for advanced biomedical applications. Biomaterials 2018, 178, 413–434. [Google Scholar] [CrossRef] [PubMed]
- Foubert, A.; Beloglazova, N.V.; Rajkovic, A.; Sas, B.; Madder, A.; Goryacheva, I.Y.; De Saeger, S. Bioconjugation of quantum dots: Review & impact on future application. TrAC—Trends Anal. Chem. 2016, 83, 31–48. [Google Scholar]
- Geyik, C.; Guler, E.; Gumus, Z.P.; Barlas, F.B.; Akbulut, H.; Demirkol, D.O.; Timur, S.; Yagci, Y. Bioconjugation and applications of amino functional fluorescence polymers. Macromol. Biosci. 2017, 17, 1600232. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, Z.; Gu, Z. Bioinspired and biomimetic nanomedicines. Acc. Chem. Res. 2019, 52, 1255–1264. [Google Scholar] [CrossRef]
- Shadish, J.A.; DeForest, C.A. Site-selective protein modification: From functionalized proteins to functional biomaterials. Matter 2020, 2, 50–77. [Google Scholar] [CrossRef]
- Spassov, V.Z.; Karshikov, A.D.; Atanasov, B.P. Electrostatic interactions in proteins. A theoretical analysis of lysozyme ionization. Biochim. Biophys. Acta (BBA)—Protein Struct. Mol. Enzymol. 1989, 999, 1–6. [Google Scholar] [CrossRef]
- Chen, D.; Disotuar, M.M.; Xiong, X.; Wang, Y.; Chou, D.H.-C. Selective N-terminal functionalization of native peptides and proteins. Chem. Sci. 2017, 8, 2717. [Google Scholar] [CrossRef]
- Hermanson, G.T. Bioconjugate Techniques; Academic Press: Boston, MA, USA, 2013. [Google Scholar]
- Anderson, G.W.; Zimmerman, J.E.; Callahan, F.M. The use of esters of N-hydroxysuccinimide in peptide synthesis. J. Am. Chem. Soc. 1964, 86, 1839–1842. [Google Scholar] [CrossRef]
- Anderson, G.W.; Zimmerman, J.E.; Callahan, F.M. N-hydroxysuccinimide esters in peptide synthesis. J. Am. Chem. Soc. 1963, 85, 3039. [Google Scholar] [CrossRef]
- Todrick, A.; Walker, E. A note on the combination of cysteine with allyl isothiocyanate. Biochem. J. 1937, 31, 297. [Google Scholar] [CrossRef] [PubMed]
- Riggs, J.; Seiwald, R.; Burckhalter, J.; Downs, C.M.; Metcalf, T. Isothiocyanate compounds as fluorescent labeling agents for immune serum. Am. J. Clin. Pathol. 1958, 34, 1081. [Google Scholar]
- Jentoft, N.; DG, D. Labeling of proteins by reductive methylation using sodium cyanoborohydride. J. Biol. Chem. 1979, 254, 4359–4365. [Google Scholar] [CrossRef]
- McFarland, J.M.; Francis, M.B. Reductive alkylation of proteins using iridium catalyzed transfer hydrogenation. J. Am. Chem. Soc. 2005, 127, 13490–13491. [Google Scholar] [CrossRef]
- Nakamura, T.; Kawai, Y.; Kitamoto, N.; Osawa, T.; Kato, Y. Covalent modification of lysine residues by allyl isothiocyanate in physiological conditions: Plausible transformation of isothiocyanate from thiol to amine. Chem. Res. Toxicol. 2009, 22, 536–542. [Google Scholar] [CrossRef]
- Cal, P.M.; Vicente, J.B.; Pires, E.; Coelho, A.V.; Veiros, L.s.F.; Cordeiro, C.; Gois, P.M. Iminoboronates: A new strategy for reversible protein modification. J. Am. Chem. Soc. 2012, 134, 10299–10305. [Google Scholar] [CrossRef]
- António, J.P.; Russo, R.; Carvalho, C.P.; Cal, P.M.; Gois, P.M. Boronic acids as building blocks for the construction of therapeutically useful bioconjugates. Chem. Soc. Rev. 2019, 48, 3513–3536. [Google Scholar] [CrossRef]
- Bandyopadhyay, A.; McCarthy, K.A.; Kelly, M.A.; Gao, J. Targeting bacteria via iminoboronate chemistry of amine-presenting lipids. Nat. Commun. 2015, 6, 1–9. [Google Scholar] [CrossRef]
- Bandyopadhyay, A.; Gao, J. Iminoboronate-based peptide cyclization that responds to pH, oxidation, and small molecule modulators. J. Am. Chem. Soc. 2016, 138, 2098–2101. [Google Scholar] [CrossRef] [PubMed]
- Wurm, F.; Steinbach, T.; Klok, H.-A. One-pot squaric acid diester mediated aqueous protein conjugation. Chem. Commun. 2013, 49, 7815–7817. [Google Scholar] [CrossRef] [PubMed]
- Diethelm, S.; Schafroth, M.A.; Carreira, E.M. Amine-selective bioconjugation using arene diazonium salts. Org. Lett. 2014, 16, 3908–3911. [Google Scholar] [CrossRef] [PubMed]
- Tung, C.L.; Wong, C.T.; Fung, E.Y.M.; Li, X. Traceless and chemoselective amine bioconjugation via phthalimidine formation in native protein modification. Org. Lett. 2016, 18, 2600–2603. [Google Scholar] [CrossRef]
- Apel, C.; Kasper, M.-A.; Stieger, C.E.; Hackenberger, C.P.; Christmann, M. Protein modification of lysine with 2-(2-styrylcyclopropyl)ethanal. Org. Lett. 2019, 21, 10043–10047. [Google Scholar] [CrossRef]
- Yi, S.; Wei, S.; Wu, Q.; Wang, H.; Yao, Z.J. Azaphilones as activation-free primary-amine-specific bioconjugation reagents for peptides, proteins and lipids. Angew. Chem. Int. Ed. 2022, 61, e202111783. [Google Scholar] [CrossRef]
- Hamblett, K.J.; Senter, P.D.; Chace, D.F.; Sun, M.M.; Lenox, J.; Cerveny, C.G.; Kissler, K.M.; Bernhardt, S.X.; Kopcha, A.K.; Zabinski, R.F. Effects of drug loading on the antitumor activity of a monoclonal antibody drug conjugate. Clin. Cancer Res. 2004, 10, 7063–7070. [Google Scholar] [CrossRef]
- Panowski, S.; Bhakta, S.; Raab, H.; Polakis, P.; Junutula, J.R. Site-specific antibody drug conjugates for cancer therapy. mAbs; 2014; 6, pp. 34–45. [Google Scholar]
- Yamada, K.; Shikida, N.; Shimbo, K.; Ito, Y.; Khedri, Z.; Matsuda, Y.; Mendelsohn, B.A. AJICAP: Affinity peptide mediated regiodivergent functionalization of native antibodies. Angew. Chem. Int. Ed. 2019, 131, 5648–5653. [Google Scholar] [CrossRef]
- Sadiki, A.; Vaidya, S.R.; Abdollahi, M.; Bhardwaj, G.; Dolan, M.E.; Turna, H.; Arora, V.; Sanjeev, A.; Robinson, T.D.; Koid, A. Site-specific conjugation of native antibody. Antib. Ther. 2020, 3, 271–284. [Google Scholar] [CrossRef]
- Hacker, S.M.; Backus, K.M.; Lazear, M.R.; Forli, S.; Correia, B.E.; Cravatt, B.F. Global profiling of lysine reactivity and ligandability in the human proteome. Nat. Chem. 2017, 9, 1181–1190. [Google Scholar] [CrossRef]
- Zhao, S.; Allis, C.D.; Wang, G.G. The language of chromatin modification in human cancers. Nat. Rev. Cancer 2021, 21, 413–430. [Google Scholar] [CrossRef] [PubMed]
- Jacob, E.; Unger, R. A tale of two tails: Why are terminal residues of proteins exposed? Bioinformatics 2007, 23, e225–e230. [Google Scholar] [CrossRef] [PubMed]
- Westheimer, F.H.; Schmidt Jr, D.E. pK of the lysine amino group at the active site of acetoacetate decarboxylase. Biochemistry 1971, 10, 1249–1253. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.C.; Stowell, C.P.; Krantz, M.J. 2-Imino-2-methoxyethyl 1-thioglycosides: New reagents for attaching sugars to proteins. Biochemistry 1976, 15, 3956–3963. [Google Scholar] [CrossRef]
- Robinson, M.A.; Charlton, S.T.; Garnier, P.; Wang, X.-t.; Davis, S.S.; Perkins, A.C.; Frier, M.; Duncan, R.; Savage, T.J.; Wyatt, D.A. LEAPT: Lectin-directed enzyme-activated prodrug therapy. Proc. Natl. Acad. Sci. USA 2004, 101, 14527–14532. [Google Scholar] [CrossRef]
- Tanaka, K.; Fujii, Y.; Fukase, K. Site-selective and nondestructive protein labeling through azaelectrocyclization-induced cascade reactions. ChemBioChem 2008, 9, 2392–2397. [Google Scholar] [CrossRef]
- Tanaka, K.; Fukase, K.; Katsumura, S. Exploring a unique reactivity of 6π-azaelectrocyclization to enzyme inhibition, natural products synthesis, and molecular imaging: An approach to chemical biology by synthetic chemists. Synlett 2011, 2011, 2115–2139. [Google Scholar] [CrossRef]
- Tanaka, K.; Fukase, K. PET (positron emission tomography) imaging of biomolecules using metal–DOTA complexes: A new collaborative challenge by chemists, biologists, and physicians for future diagnostics and exploration of in vivo dynamics. Org. Biomol. Chem. 2008, 6, 815–828. [Google Scholar] [CrossRef]
- Tanaka, K.; Masuyama, T.; Hasegawa, K.; Tahara, T.; Mizuma, H.; Wada, Y.; Watanabe, Y.; Fukase, K. A submicrogram-scale protocol for biomolecule-based PET imaging by rapid 6π-azaelectrocyclization: Visualization of sialic acid dependent circulatory residence of glycoproteins. Angew. Chem. Int. Ed. 2008, 120, 108–111. [Google Scholar] [CrossRef]
- Hansen, B.K.; Loveridge, C.J.; Thyssen, S.; Wørmer, G.J.; Nielsen, A.D.; Palmfeldt, J.; Johannsen, M.; Poulsen, T.B. STEFs: Activated vinylogous protein-reactive electrophiles. Angew. Chem. Int. Ed. 2019, 58, 3533–3537. [Google Scholar] [CrossRef]
- Tang, K.C.; Cao, J.; Boatner, L.M.; Li, L.; Farhi, J.; Houk, K.N.; Spangle, J.; Backus, K.M.; Raj, M. Tunable amine-reactive electrophiles for selective profiling of lysine. Angew. Chem. Int. Ed. 2022, 134, e202112107. [Google Scholar]
- Sun, H.; Xi, M.; Jin, Q.; Zhu, Z.; Zhang, Y.; Jia, G.; Zhu, G.; Sun, M.; Zhang, H.; Ren, X. Chemo-and site-selective lysine modification of peptides and proteins under native conditions using the water-soluble zolinium. J. Med. Chem. 2022. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Lee, J.; Brooks, B.R. Origin of pKa shifts of internal lysine residues in SNase studied via equal-molar VMMS simulations in explicit water. J. Phys. Chem. B 2017, 121, 3318–3330. [Google Scholar] [CrossRef]
- Asano, S.; Patterson, J.T.; Gaj, T.; Barbas III, C.F. Site-selective labeling of a lysine residue in human serum albumin. Angew. Chem. Int. Ed. 2014, 53, 11783–11786. [Google Scholar] [CrossRef] [PubMed]
- Patterson, J.T.; Wilson, H.D.; Asano, S.; Nilchan, N.; Fuller, R.P.; Roush, W.R.; Rader, C.; Barbas III, C.F. Human serum albumin domain I fusion protein for antibody conjugation. Bioconjug. Chem. 2016, 27, 2271–2275. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Huang, R.; Li, Z.; Zhu, W.; Chen, J.; Zhan, Y.; Jiang, B. Selective lysine modification of native peptides via aza-Michael addition. Org. Biomol. Chem. 2017, 15, 7339–7345. [Google Scholar] [CrossRef]
- Matos, M.J.; Oliveira, B.L.; Martínez-Sáez, N.; Guerreiro, A.; Cal, P.M.; Bertoldo, J.; Maneiro, M.; Perkins, E.; Howard, J.; Deery, M.J. Chemo-and regioselective lysine modification on native proteins. J. Am. Chem. Soc. 2018, 140, 4004–4017. [Google Scholar] [CrossRef]
- Lim, D.; Wen, X.; Seebeck, F.P. Selenoimidazolium salts as supramolecular reagents for protein alkylation. ChemBioChem 2020, 21, 3515–3520. [Google Scholar] [CrossRef]
- Purushottam, L.; Adusumalli, S.R.; Chilamari, M.; Rai, V. Chemoselective and site-selective peptide and native protein modification enabled by aldehyde auto-oxidation. Chem. Commun. 2017, 53, 959–962. [Google Scholar] [CrossRef]
- Chilamari, M.; Purushottam, L.; Rai, V. Site-selective labeling of native proteins by a multicomponent approach. Chem. Eur. J. 2017, 23, 3819–3823. [Google Scholar] [CrossRef]
- Chilamari, M.; Kalra, N.; Shukla, S.; Rai, V. Single-site labeling of lysine in proteins through a metal-free multicomponent approach. Chem. Commun. 2018, 54, 7302–7305. [Google Scholar] [CrossRef] [PubMed]
- Adusumalli, S.R.; Rawale, D.G.; Thakur, K.; Purushottam, L.; Reddy, N.C.; Kalra, N.; Shukla, S.; Rai, V. Chemoselective and site-selective lysine-directed lysine modification enables single-site labeling of native proteins. Angew. Chem. Int. Ed. 2020, 132, 10418–10422. [Google Scholar] [CrossRef]
- Sahu, T.; Chilamari, M.; Rai, V. Protein inspired chemically orthogonal imines for linchpin directed precise and modular labeling of lysine in proteins. Chem. Commun. 2022, 58, 1768–1771. [Google Scholar] [CrossRef] [PubMed]
- Rosen, C.B.; Francis, M.B. Targeting the N terminus for site-selective protein modification. Nat. Chem. Biol. 2017, 13, 697–705. [Google Scholar] [CrossRef]
- De Rosa, L.; Di Stasi, R.; Romanelli, A.; D’Andrea, L.D. Exploiting protein N-terminus for site-specific bioconjugation. Molecules 2021, 26, 3521. [Google Scholar] [CrossRef]
- Asiimwe, N.; Al Mazid, M.F.; Murale, D.P.; Kim, Y.K.; Lee, J.S. Recent advances in protein modifications techniques for the targeting N-terminal cysteine. Pept. Sci. 2022, 114, e24235. [Google Scholar] [CrossRef]
- Jiang, H.; Chen, W.; Wang, J.; Zhang, R. Selective N-terminal modification of peptides and proteins: Recent progresses and applications. Chin. Chem. Lett. 2021. [Google Scholar] [CrossRef]
- Gilmore, J.M.; Scheck, R.A.; Esser-Kahn, A.P.; Joshi, N.S.; Francis, M.B. N-terminal protein modification through a biomimetic transamination reaction. Angew. Chem. Int. Ed. 2006, 45, 5307–5311. [Google Scholar] [CrossRef]
- Schoffelen, S.; van Eldijk, M.B.; Rooijakkers, B.; Raijmakers, R.; Heck, A.J.; van Hest, J.C. Metal-free and pH-controlled introduction of azides in proteins. Chem. Sci. 2011, 2, 701–705. [Google Scholar] [CrossRef]
- Chan, A.O.-Y.; Ho, C.-M.; Chong, H.-C.; Leung, Y.-C.; Huang, J.-S.; Wong, M.-K.; Che, C.-M. Modification of N-terminal α-amino groups of peptides and proteins using ketenes. J. Am. Chem. Soc. 2012, 134, 2589–2598. [Google Scholar] [CrossRef]
- Kung, K.K.-Y.; Wong, K.-F.; Leung, K.-C.; Wong, M.-K. N-terminal α-amino group modification of peptides by an oxime formation–exchange reaction sequence. Chem. Commun. 2013, 49, 6888–6890. [Google Scholar] [CrossRef] [PubMed]
- Witus, L.S.; Netirojjanakul, C.; Palla, K.S.; Muehl, E.M.; Weng, C.-H.; Iavarone, A.T.; Francis, M.B. Site-specific protein transamination using N-methylpyridinium-4-carboxaldehyde. J. Am. Chem. Soc. 2013, 135, 17223–17229. [Google Scholar] [CrossRef] [PubMed]
- Scheck, R.A.; Francis, M.B. Regioselective labeling of antibodies through N-terminal transamination. ACS Chem. Biol. 2007, 2, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Witus, L.S.; Moore, T.; Thuronyi, B.W.; Esser-Kahn, A.P.; Scheck, R.A.; Iavarone, A.T.; Francis, M.B. Identification of highly reactive sequences for PLP-mediated bioconjugation using a combinatorial peptide library. J. Am. Chem. Soc. 2010, 132, 16812–16817. [Google Scholar] [CrossRef] [PubMed]
- Obermeyer, A.C.; Jarman, J.B.; Francis, M.B. N-terminal modification of proteins with o-aminophenols. J. Am. Chem. Soc. 2014, 136, 9572–9579. [Google Scholar] [CrossRef]
- MacDonald, J.I.; Munch, H.K.; Moore, T.; Francis, M.B. One-step site-specific modification of native proteins with 2-pyridinecarboxyaldehydes. Nat. Chem. Biol. 2015, 11, 326–331. [Google Scholar] [CrossRef]
- Raj, M.; Wu, H.; Blosser, S.L.; Vittoria, M.A.; Arora, P.S. Aldehyde capture ligation for synthesis of native peptide bonds. J. Am. Chem. Soc. 2015, 137, 6932–6940. [Google Scholar] [CrossRef]
- Singudas, R.; Adusumalli, S.R.; Joshi, P.N.; Rai, V. A phthalimidation protocol that follows protein defined parameters. Chem. Commun. 2015, 51, 473–476. [Google Scholar] [CrossRef]
- Adusumalli, S.R.; Rawale, D.G.; Rai, V. Aldehydes can switch the chemoselectivity of electrophiles in protein labeling. Org. Biomol. Chem. 2018, 16, 9377–9381. [Google Scholar] [CrossRef]
- Deng, J.-R.; Lai, N.C.-H.; Kung, K.K.-Y.; Yang, B.; Chung, S.-F.; Leung, A.S.-L.; Choi, M.-C.; Leung, Y.-C.; Wong, M.-K. N-Terminal selective modification of peptides and proteins using 2-ethynylbenzaldehydes. Commun. Chem. 2020, 3, 1–9. [Google Scholar] [CrossRef]
- Miller, M.K.; Wang, H.; Hanaya, K.; Zhang, O.; Berlaga, A.; Ball, Z.T. Copper-mediated peptide arylation selective for the N-terminus. Chem. Sci. 2020, 11, 10501–10505. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhou, Q.; Chen, X.; Luo, R.-H.; Li, Y.; Liu, X.; Yang, L.-M.; Zheng, Y.-T.; Wang, P. Modification of N-terminal α-amine of proteins via biomimetic ortho-quinone-mediated oxidation. Nat. Commun. 2021, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hanaya, K.; Yamoto, K.; Taguchi, K.; Matsumoto, K.; Higashibayashi, S.; Sugai, T. Single-step N-terminal modification of proteins via a bio-inspired copper(II)-mediated aldol reaction. Chem. Eur. J. 2022, 28, e202201677. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, J.H.; Gustafsson, M.B.; Skrydstrup, T.; Jensen, K.B. Selective N-terminal acylation of peptides and proteins with tunable phenol esters. Bioconjug. Chem. 2022, 33, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.B.; Mikkelsen, J.H.; Jensen, S.P.; Kidal, S.; Friberg, G.; Skrydstrup, T.; Gustafsson, M.B. New phenol esters for efficient pH-controlled amine acylation of peptides, proteins, and sepharose beads in aqueous media. Bioconjug. Chem. 2022, 33, 172–179. [Google Scholar] [CrossRef]
- Wieland, T.; Bokelmann, E.; Bauer, L.; Lang, H.; Lau, H.; Schafer, W. Polypeptide syntheses. VIII. Formation of sulfur containing peptides by the intramolecular migration of aminoacyl groups. Liebigs Ann. Chem 1953, 583, 129–149. [Google Scholar] [CrossRef]
- Dawson, P.E.; Muir, T.W.; Clark-Lewis, I.; Kent, S.B. Synthesis of proteins by native chemical ligation. Science 1994, 266, 776–779. [Google Scholar] [CrossRef]
- Tam, J.P.; Lu, Y.-A.; Liu, C.-F.; Shao, J. Peptide synthesis using unprotected peptides through orthogonal coupling methods. Proc. Natl. Acad. Sci. USA 1995, 92, 12485–12489. [Google Scholar] [CrossRef]
- Agouridas, V.; El Mahdi, O.; Diemer, V.; Cargoet, M.; Monbaliu, J.-C.M.; Melnyk, O. Native chemical ligation and extended methods: Mechanisms, catalysis, scope, and limitations. Chem. Rev. 2019, 119, 7328–7443. [Google Scholar] [CrossRef]
- Berrade, L.; Camarero, J.A. Expressed protein ligation: A resourceful tool to study protein structure and function. Cell. Mol. Life Sci. 2009, 66, 3909–3922. [Google Scholar] [CrossRef]
- Malins, L.R.; Payne, R.J. Recent extensions to native chemical ligation for the chemical synthesis of peptides and proteins. Curr. Opin. Chem. Biol. 2014, 22, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Bang, D.; Chopra, N.; Kent, S.B. Total chemical synthesis of crambin. J. Am. Chem. Soc. 2004, 126, 1377–1383. [Google Scholar] [CrossRef] [PubMed]
- Ollivier, N.; Vicogne, J.; Vallin, A.; Drobecq, H.; Desmet, R.; El Mahdi, O.; Leclercq, B.; Goormachtigh, G.; Fafeur, V.; Melnyk, O. A one-pot three-segment ligation strategy for protein chemical synthesis. Angew. Chem. Int. Ed. 2012, 124, 213–217. [Google Scholar] [CrossRef]
- Thompson, R.E.; Chan, B.; Radom, L.; Jolliffe, K.A.; Payne, R.J. Chemoselective peptide ligation–desulfurization at aspartate. Angew. Chem. Int. Ed. 2013, 125, 9905–9909. [Google Scholar] [CrossRef]
- Hermanson, G.T. Chapter 3—The reactions of bioconjugation. In Bioconjugate Techniques, 3rd ed.; Hermanson, G.T., Ed.; Academic Press: Boston, MA, USA, 2013; pp. 229–258. [Google Scholar]
- Conibear, A.C.; Watson, E.E.; Payne, R.J.; Becker, C.F. Native chemical ligation in protein synthesis and semi-synthesis. Chem. Soc. Rev. 2018, 47, 9046–9068. [Google Scholar] [CrossRef]
- Hermanson, G.T. Chapter 17—Chemoselective ligation; Bioorthogonal reagents. In Bioconjugate Techniques, 3rd ed.; Hermanson, G.T., Ed.; Academic Press: Boston, MA, USA, 2013; pp. 757–785. [Google Scholar]
- Busch, G.K.; Tate, E.W.; Gaffney, P.R.; Rosivatz, E.; Woscholski, R.; Leatherbarrow, R.J. Specific N-terminal protein labelling: Use of FMDV 3C pro protease and native chemical ligation. Chem. Commun. 2008, 3369–3371. [Google Scholar] [CrossRef]
- Dempsey, D.R.; Jiang, H.; Kalin, J.H.; Chen, Z.; Cole, P.A. Site-specific protein labeling with N-hydroxysuccinimide-esters and the analysis of ubiquitin ligase mechanisms. J. Am. Chem. Soc. 2018, 140, 9374–9378. [Google Scholar] [CrossRef]
- Ren, H.; Xiao, F.; Zhan, K.; Kim, Y.P.; Xie, H.; Xia, Z.; Rao, J. A biocompatible condensation reaction for the labeling of terminal cysteine residues on proteins. Angew. Chem. Int. Ed. 2009, 48, 9658–9662. [Google Scholar] [CrossRef]
- Zheng, Z.; Chen, P.; Li, G.; Zhu, Y.; Shi, Z.; Luo, Y.; Zhao, C.; Fu, Z.; Cui, X.; Ji, C. Mechanistic study of CBT-Cys click reaction and its application for identifying bioactive N-terminal cysteine peptides in amniotic fluid. Chem. Sci. 2017, 8, 214–222. [Google Scholar] [CrossRef]
- Yuan, Y.; Liang, G. A biocompatible, highly efficient click reaction and its applications. Org. Biomol. Chem. 2014, 12, 865–871. [Google Scholar] [CrossRef]
- Friedman Ohana, R.; Hurst, R.; Rosenblatt, M.; Levin, S.; Machleidt, T.; Kirkland, T.A.; Encell, L.P.; Robers, M.B.; Wood, K.V. Utilizing a simple method for stoichiometric protein labeling to quantify antibody blockade. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Gao, J. N, S-double labeling of N-terminal cysteines via an alternative conjugation pathway with 2-cyanobenzothiazole. J. Org. Chem. 2020, 85, 1756–1763. [Google Scholar] [CrossRef] [PubMed]
- Casi, G.; Huguenin-Dezot, N.; Zuberbühler, K.; Scheuermann, J.r.; Neri, D. Site-specific traceless coupling of potent cytotoxic drugs to recombinant antibodies for pharmacodelivery. J. Am. Chem. Soc. 2012, 134, 5887–5892. [Google Scholar] [CrossRef] [PubMed]
- Bermejo-Velasco, D.; Nawale, G.N.; Oommen, O.P.; Hilborn, J.; Varghese, O.P. Thiazolidine chemistry revisited: A fast, efficient and stable click-type reaction at physiological pH. Chem. Commun. 2018, 54, 12507–12510. [Google Scholar] [CrossRef] [PubMed]
- Murale, D.P.; Hong, S.C.; Jang, S.y.; Lee, J.S. Reinvestigation of an o-salicylaldehyde ester functional group in aqueous buffer and discovery of a coumarin scaffold probe for selective N-terminal cysteine labeling. ChemBioChem 2018, 19, 2545–2549. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Li, J.; Jin, K.; Liu, J.; Chen, Z.; Yang, J.; Li, X. Cysteine/penicillamine ligation independent of terminal steric demands for chemical protein synthesis. Angew. Chem. Int. Ed. 2020, 132, 12841–12845. [Google Scholar] [CrossRef]
- Bandyopadhyay, A.; Cambray, S.; Gao, J. Fast and selective labeling of N-terminal cysteines at neutral pH via thiazolidino boronate formation. Chem. Sci. 2016, 7, 4589–4593. [Google Scholar] [CrossRef]
- Faustino, H.; Silva, M.J.; Veiros, L.F.; Bernardes, G.J.; Gois, P.M. Iminoboronates are efficient intermediates for selective, rapid and reversible N-terminal cysteine functionalisation. Chem. Sci. 2016, 7, 5052–5058. [Google Scholar] [CrossRef]
- Li, K.; Wang, W.; Gao, J. Fast and stable N-Terminal cysteine modification through thiazolidino boronate mediated acyl transfer. Angew. Chem. Int. Ed. 2020, 59, 14246–14250. [Google Scholar] [CrossRef]
- Zheng, X.; Li, Z.; Gao, W.; Meng, X.; Li, X.; Luk, L.Y.; Zhao, Y.; Tsai, Y.-H.; Wu, C. Condensation of 2-((alkylthio)(aryl) methylene) malononitrile with 1,2-aminothiol as a novel bioorthogonal reaction for site-specific protein modification and peptide cyclization. J. Am. Chem. Soc. 2020, 142, 5097–5103. [Google Scholar] [CrossRef]
- Istrate, A.; Navo, C.D.; Sousa, B.B.; Marques, M.C.; Deery, M.; Bond, A.; Corzana, F.; Jiménez-Osés, G.; Bernardes, G. Selective N-terminal cysteine protein modification with cyclopropenones. 2020. J. Am. Chem. Soc. 2020, 144, 10396–10406. [Google Scholar] [CrossRef] [PubMed]
- Istrate, A.; Geeson, M.B.; Navo, C.D.; Sousa, B.B.; Marques, M.C.; Taylor, R.J.; Journeaux, T.; Oehler, S.R.; Mortensen, M.R.; Deery, M.J. Platform for orthogonal N-cysteine-specific protein modification enabled by cyclopropenone reagents. J. Am. Chem. Soc. 2022. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, C.; Fan, S.; Zhao, Y.; Wu, C. Fast and selective reaction of 2-benzylacrylaldehyde with 1,2-aminothiol for stable N-terminal cysteine modification and peptide cyclization. Bioconjug. Chem. 2021, 32, 2065–2072. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.J.; Faustino, H.; Coelho, J.A.; Pinto, M.V.; Fernandes, A.; Companon, I.; Corzana, F.; Gasser, G.; Gois, P.M. Efficient amino-sulfhydryl stapling on peptides and proteins using bifunctional NHS-activated acrylamides. Angew. Chem. Int. Ed. 2021, 60, 10850–10857. [Google Scholar] [CrossRef]
- Djaló, M.; Silva, M.J.; Faustino, H.; Pinto, S.N.; Mendonça, R.; Gois, P.M. Multivalent NHS-activated acrylates for orthogonal site-selective functionalisation of peptides at cysteine residues. Chem. Commun. 2022, 58, 7928–7931. [Google Scholar] [CrossRef]
- Li, X.; Zhang, L.; Hall, S.E.; Tam, J.P. A new ligation method for N-terminal tryptophan-containing peptides using the Pictet–Spengler reaction. Tetrahedron Lett. 2000, 41, 4069–4073. [Google Scholar] [CrossRef]
- Geoghegan, K.F.; Stroh, J.G. Site-directed conjugation of nonpeptide groups to peptides and proteins via periodate oxidation of a 2-amino alcohol. Application to modification at N-terminal serine. Bioconjug. Chem. 1992, 3, 138–146. [Google Scholar] [CrossRef]
- De Rosa, L.; Di Stasi, R.; Longhitano, L.; D’Andrea, L.D. Labeling of VEGFR1D2 through oxime ligation. Bioorg. Chem. 2019, 91, 103160. [Google Scholar] [CrossRef]
- Zhang, L.; Tam, J.P. Thiazolidine formation as a general and site-specific conjugation method for synthetic peptides and proteins. Anal. Biochem. 1996, 233, 87–93. [Google Scholar] [CrossRef]
- Maza, J.C.; Ramsey, A.V.; Mehare, M.; Krska, S.W.; Parish, C.A.; Francis, M.B. Secondary modification of oxidatively-modified proline N-termini for the construction of complex bioconjugates. Org. Biomol. Chem. 2020, 18, 1881–1885. [Google Scholar] [CrossRef]
- Purushottam, L.; Adusumalli, S.R.; Singh, U.; Unnikrishnan, V.; Rawale, D.G.; Gujrati, M.; Mishra, R.K.; Rai, V. Single-site glycine-specific labeling of proteins. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sahu, T.; Kumar, M.; Sajeev, T.; Joshi, M.; Mishra, R.K.; Rai, V. Residue-specific N-terminal glycine to aldehyde transformation renders analytically pure single-site labeled proteins. Chem. Commun. 2022. [Google Scholar] [CrossRef] [PubMed]
- Martos-Maldonado, M.C.; Hjuler, C.T.; Sørensen, K.K.; Thygesen, M.B.; Rasmussen, J.E.; Villadsen, K.; Midtgaard, S.R.; Kol, S.; Schoffelen, S.; Jensen, K.J. Selective N-terminal acylation of peptides and proteins with a Gly-His tag sequence. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Jensen, K.J.; Thygesen, M.B.; Sørensen, K.K.; Wu, S.; Treiberg, T.; Schoffelen, S. Selective acylation of proteins at Gly and Lys in His Tags. ChemBioChem 2022, 23, e202200359. [Google Scholar] [CrossRef] [PubMed]
- Brune, K.D.; Liekniņa, I.; Sutov, G.; Morris, A.R.; Jovicevic, D.; Kalniņš, G.; Kazāks, A.; Kluga, R.; Kastaljana, S.; Zajakina, A. N-terminal modification of Gly-His-tagged proteins with azidogluconolactone. ChemBioChem 2021, 22, 3199–3207. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tantipanjaporn, A.; Wong, M.-K. Development and Recent Advances in Lysine and N-Terminal Bioconjugation for Peptides and Proteins. Molecules 2023, 28, 1083. https://doi.org/10.3390/molecules28031083
Tantipanjaporn A, Wong M-K. Development and Recent Advances in Lysine and N-Terminal Bioconjugation for Peptides and Proteins. Molecules. 2023; 28(3):1083. https://doi.org/10.3390/molecules28031083
Chicago/Turabian StyleTantipanjaporn, Ajcharapan, and Man-Kin Wong. 2023. "Development and Recent Advances in Lysine and N-Terminal Bioconjugation for Peptides and Proteins" Molecules 28, no. 3: 1083. https://doi.org/10.3390/molecules28031083
APA StyleTantipanjaporn, A., & Wong, M.-K. (2023). Development and Recent Advances in Lysine and N-Terminal Bioconjugation for Peptides and Proteins. Molecules, 28(3), 1083. https://doi.org/10.3390/molecules28031083