Synthetic Cinnamides and Cinnamates: Antimicrobial Activity, Mechanism of Action, and In Silico Study
Abstract
1. Introduction
2. Results and Discussion
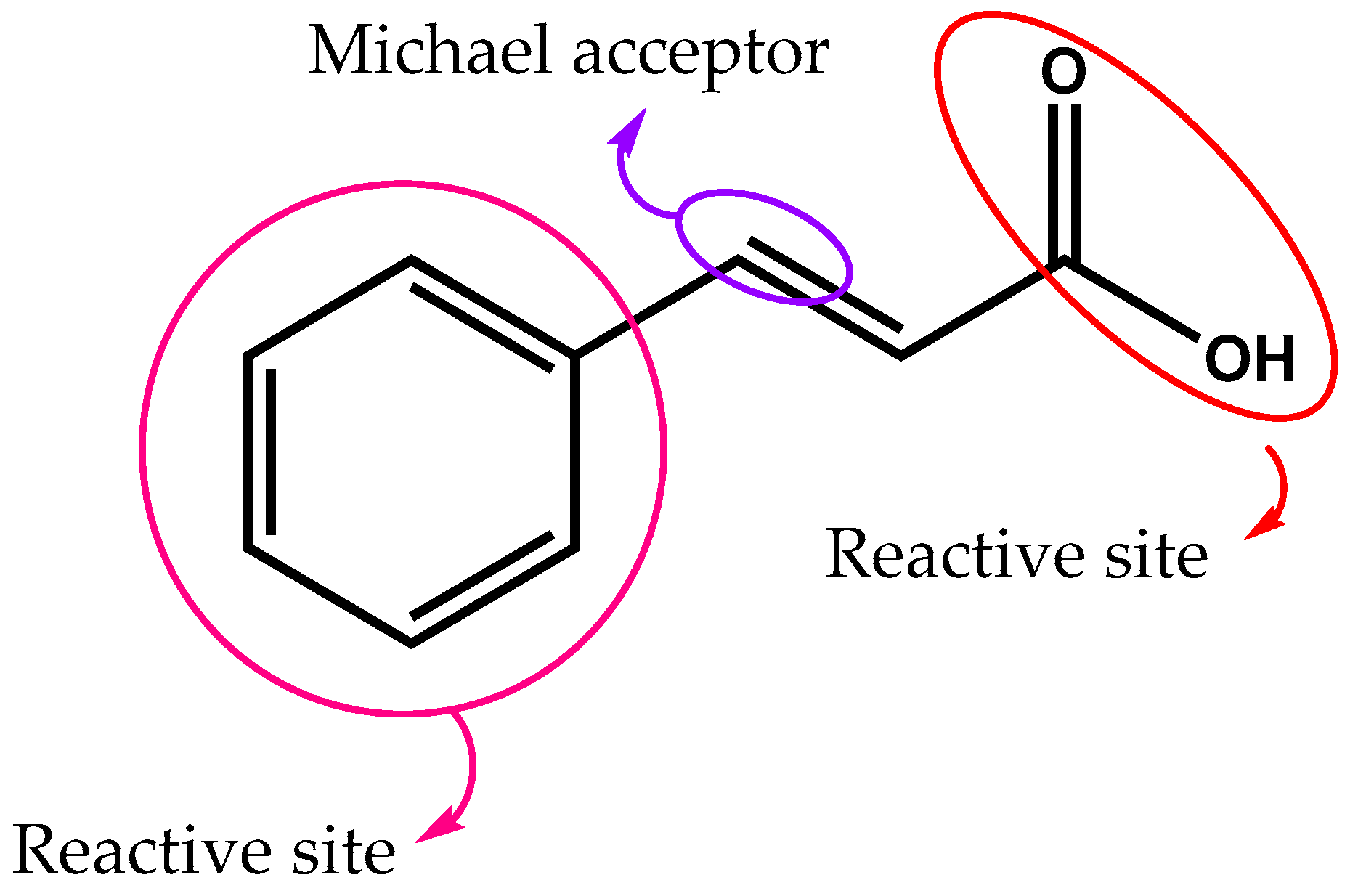
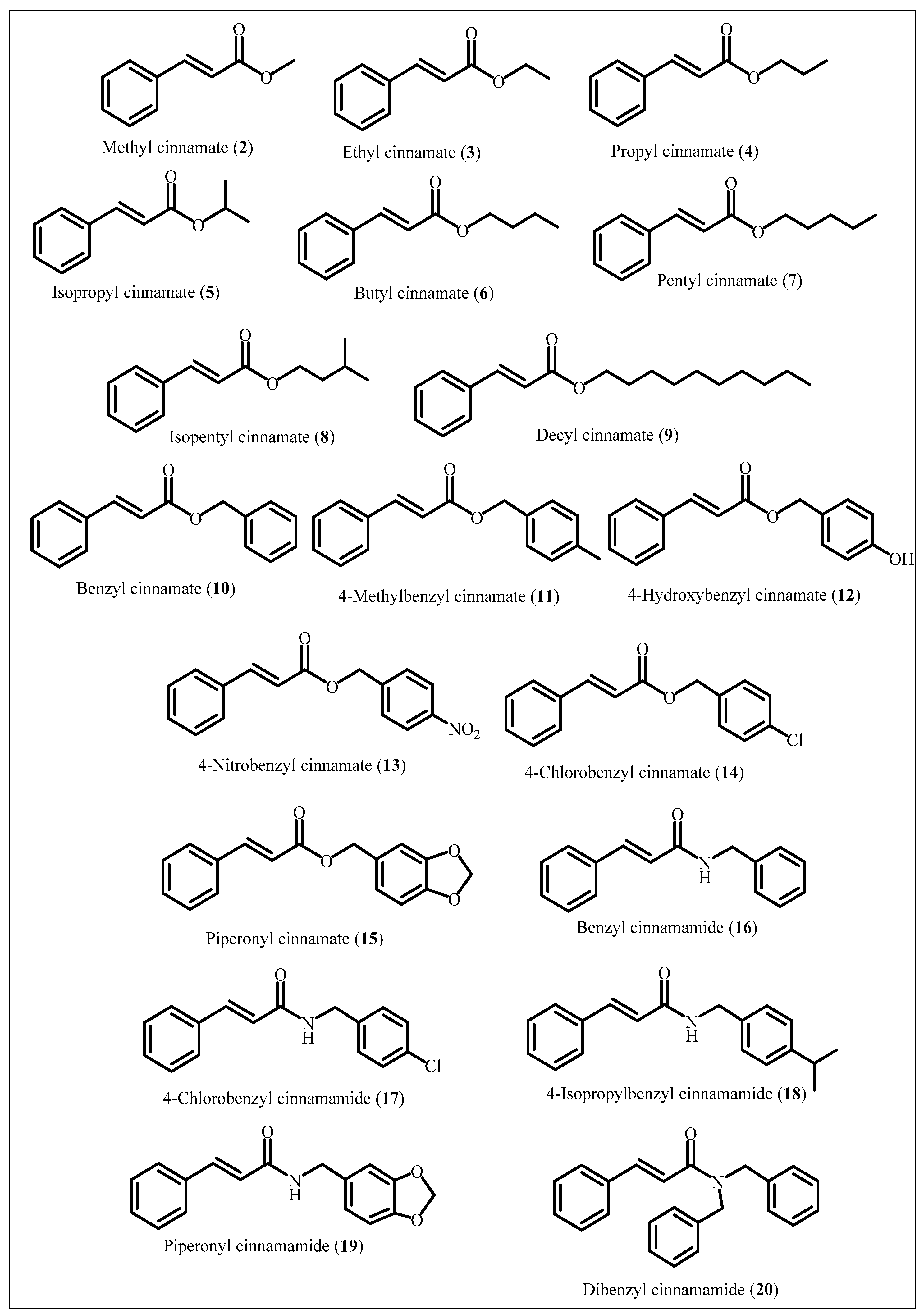
2.1. Chemistry
2.2. Evaluation of Antifungal Activity
2.2.1. Minimum Fungicidal Concentration (MFC)
2.2.2. Mechanism of Action
2.2.3. Association Tests—Checkerboard Method
2.3. Evaluation of the Antibacterial Activity of the Derivatives
2.3.1. Minimum Bactericidal Concentration (MBC)
2.3.2. Association Test Using the Checkerboard Method
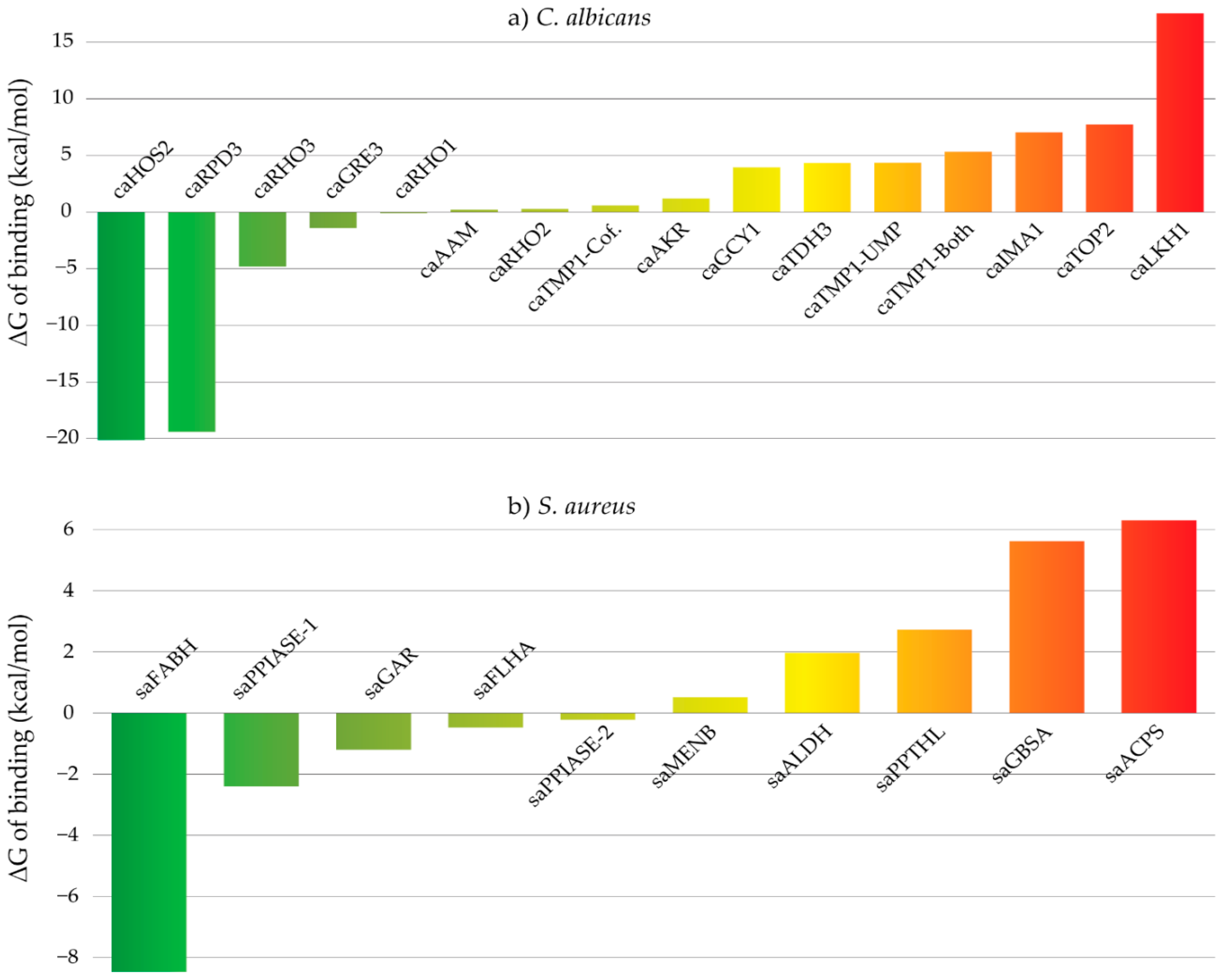
2.4. Molecular Docking
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemistry
5.1.1. Synthesis of Compounds 2–8
5.1.2. Synthesis of compounds 9 and 14
5.1.3. Synthesis of Compounds 10–13 and 15–20
5.2. Antimicrobial Tests
5.2.1. Microorganisms
5.2.2. Determination of Minimum Inhibitory Concentration (MIC)
5.2.3. Determination of Minimum Fungicidal Concentration (MFC)
5.2.4. Determination of Minimum Bactericidal Concentration (MBC)
5.2.5. Mechanism of Antifungal Action for Amides
5.2.6. Association Study Using the Checkerboard Method
5.3. Modeling Methods
5.3.1. Target Selection
5.3.2. Molecular Docking
5.3.3. Molecular Dynamics Simulations and Free Energy Calculations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- França, S.B.; Correia, P.R.d.S.; Castro, I.B.D.d.; Júnior, E.F.d.S.; Barros, M.E.d.S.B.; Lima, D.J.d.P. Synthesis, Applications and Structure-Activity Relationship (SAR) of Cinnamic Acid Derivatives: A Review. Res. Soc. Dev. 2021, 10, e28010111691. [Google Scholar] [CrossRef]
- Da Silveira E Sá, R.D.C.; Andrade, L.N.; De Oliveira, R.D.R.B.; De Sousa, D.P. A Review on Anti-Inflammatory Activity of Phenylpropanoids Found in Essential Oils. Molecules 2014, 19, 1459–1480. [Google Scholar] [CrossRef] [PubMed]
- Bhullar, K.S.; Lassalle-Claux, G.; Touaibia, M.; Vasantha Rupasinghe, H.P. Antihypertensive Effect of Caffeic Acid and Its Analogs through Dual Renin-Angiotensin-Aldosterone System Inhibition. Eur. J. Pharmacol. 2014, 730, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Ververidis, F.; Trantas, E.; Douglas, C.; Vollmer, G.; Kretzschmar, G.; Panopoulos, N. Biotechnology of Flavonoids and Other Phenylpropanoid-Derived Natural Products. Part I: Chemical Diversity, Impacts on Plant Biology and Human Health. Biotechnol. J. 2007, 2, 1214–1234. [Google Scholar] [CrossRef] [PubMed]
- Godoy, M.E.; Rotelli, A.; Pelzer, L.; Tonn, C.E. Antiinflammatory Activity of Cinnamic Acid Esters. Molecules 2000, 5, 547–548. [Google Scholar] [CrossRef]
- Bairwa, R.; Kakwani, M.; Tawari, N.R.; Lalchandani, J.; Ray, M.K.; Rajan, M.G.R.; Degani, M.S. Novel Molecular Hybrids of Cinnamic Acids and Guanylhydrazones as Potential Antitubercular Agents. Bioorg. Med. Chem. Lett. 2010, 20, 1623–1625. [Google Scholar] [CrossRef]
- Chen, Y.L.; Huang, S.T.; Sun, F.M.; Chiang, Y.L.; Chiang, C.J.; Tsai, C.M.; Weng, C.J. Transformation of Cinnamic Acid from Trans- to Cis-Form Raises a Notable Bactericidal and Synergistic Activity against Multiple-Drug Resistant Mycobacterium Tuberculosis. Eur. J. Pharm. Sci. 2011, 43, 188–194. [Google Scholar] [CrossRef]
- Guzman, J.D.; Mortazavi, P.N.; Munshi, T.; Evangelopoulos, D.; McHugh, T.D.; Gibbons, S.; Malkinson, J.; Bhakta, S. 2-Hydroxy-Substituted Cinnamic Acids and Acetanilides Are Selective Growth Inhibitors of Mycobacterium Tuberculosis. Medchemcomm 2013, 5, 47–50. [Google Scholar] [CrossRef]
- Teixeira, C.; Ventura, C.; Gomes, J.R.B.; Gomes, P.; Martins, F. Cinnamic Derivatives as Antitubercular Agents: Characterization by Quantitative Structure–Activity Relationship Studies. Molecules 2020, 25, 456. [Google Scholar] [CrossRef]
- Gravina, H.D.; Tafuri, N.F.; Silva Júnior, A.; Fietto, J.L.R.; Oliveira, T.T.; Diaz, M.A.N.; Almeida, M.R. In Vitro Assessment of the Antiviral Potential of Trans-Cinnamic Acid, Quercetin and Morin against Equid Herpesvirus 1. Res. Vet. Sci. 2011, 91, e158–e162. [Google Scholar] [CrossRef]
- Anantharaju, P.G.; Gowda, P.C.; Vimalambike, M.G.; Madhunapantula, S.V. An Overview on the Role of Dietary Phenolics for the Treatment of Cancers. Nutr. J. 2016, 15, 1–16. [Google Scholar] [CrossRef] [PubMed]
- De, P.; Baltas, M.; Bedos-Belval, F. Cinnamic Acid Derivatives as Anticancer Agents-a Review. Curr. Med. Chem. 2011, 18, 1672–1703. [Google Scholar] [CrossRef] [PubMed]
- Croft, S.L.; Sundar, S.; Fairlamb, A.H. Drug Resistance in Leishmaniasis. Clin. Microbiol. Rev. 2006, 19, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Vale, J.A.d.; Rodrigues, M.P.; Lima, Â.M.A.; Santiago, S.S.; Lima, G.D.d.A.; Almeida, A.A.; Oliveira, L.L.d.; Bressan, G.C.; Teixeira, R.R.; Machado-Neves, M. Synthesis of Cinnamic Acid Ester Derivatives with Antiproliferative and Antimetastatic Activities on Murine Melanoma Cells. Biomed. Pharmacother. 2022, 148, 112689. [Google Scholar] [CrossRef] [PubMed]
- Pontiki, E.; Hadjipavlou-Litina, D.; Litinas, K.; Geromichalos, G. Novel Cinnamic Acid Derivatives as Antioxidant and Anticancer Agents: Design, Synthesis and Modeling Studies. Molecules 2014, 19, 9655–9674. [Google Scholar] [CrossRef]
- Wang, R.; Yang, W.; Fan, Y.; Dehaen, W.; Li, Y.; Li, H.; Wang, W.; Zheng, Q.; Huai, Q. Design and Synthesis of the Novel Oleanolic Acid-Cinnamic Acid Ester Derivatives and Glycyrrhetinic Acid-Cinnamic Acid Ester Derivatives with Cytotoxic Properties. Bioorg. Chem. 2019, 88, 102951. [Google Scholar] [CrossRef]
- Gunia-Krzyżak, A.; Słoczyńska, K.; Popiół, J.; Koczurkiewicz, P.; Marona, H.; Pękala, E. Cinnamic Acid Derivatives in Cosmetics: Current Use and Future Prospects. Int. J. Cosmet. Sci. 2018, 40, 356–366. [Google Scholar] [CrossRef]
- Martínez-Soriano, P.A.; Macías-Pérez, J.R.; María Velázquez, A.; del Carmen Camacho-Enriquez, B.; Pretelín-Castillo, G.; Ruiz-Sánchez, M.B.; Abrego-Reyes, V.H.; Villa-Treviño, S.; Angeles, E. Solvent-Free Synthesis of Carboxylic Acids and Amide Analogs of CAPE (Caffeic Acid Phenethyl Ester) under Infrared Irradiation Conditions. Green Sustain. Chem. 2015, 5, 81–91. [Google Scholar] [CrossRef]
- Imai, M.; Yokoe, H.; Tsubuki, M.; Takahashi, N. Growth Inhibition of Human Breast and Prostate Cancer Cells by Cinnamic Acid Derivatives and Their Mechanism of Action. Biol. Pharm. Bull. 2019, 42, 1134–1139. [Google Scholar] [CrossRef]
- Auger, C.; Laurent, N.; Laurent, C.; Besançon, P.; Caporiccio, B.; Teissédre, P.L.; Rouanet, J.M. Hydroxycinnamic Acids Do Not Prevent Aortic Atherosclerosis in Hypercholesterolemic Golden Syrian Hamsters. Life Sci. 2004, 74, 2365–2377. [Google Scholar] [CrossRef]
- Guzman, J.D. Natural Cinnamic Acids, Synthetic Derivatives and Hybrids with Antimicrobial Activity. Molecules 2014, 19, 19292–19349. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Han, J.M.; Kim, H.; Kim, E.; Jeong, T.S.; Lee, W.S.; Cho, K.H. Synthesis of Cinnamic Acid Derivatives and Their Inhibitory Effects on LDL-Oxidation, Acyl-CoA:Cholesterol Acyltransferase-1 and -2 Activity, and Decrease of HDL-Particle Size. Bioorg. Med. Chem. Lett. 2004, 14, 4677–4681. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.K.; Park, Y.B.; Moon, S.S.; Bok, S.H.; Kim, D.J.; Ha, T.Y.; Jeong, T.S.; Jeong, K.S.; Choi, M.S. Hypocholesterolemic and Antioxidant Properties of 3-(4-Hydroxyl)Propanoic Acid Derivatives in High-Cholesterol Fed Rats. Chem. Biol. Interact. 2007, 170, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Mnafgui, K.; Derbali, A.; Sayadi, S.; Gharsallah, N.; Elfeki, A.; Allouche, N. Anti-Obesity and Cardioprotective Effects of Cinnamic Acid in High Fat Diet- Induced Obese Rats. J. Food Sci. Technol. 2015, 52, 4369–4377. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.S.; Shin, J.C. Characterization of Antioxidant Alkaloids and Phenolic Acids from Anthocyanin-Pigmented Rice (Oryza sativa cv. Heugjinjubyeo). Food Chem. 2007, 104, 1670–1677. [Google Scholar] [CrossRef]
- Farah, A.; Monteiro, M.; Donangelo, C.M.; Lafay, S. Chlorogenic Acids from Green Coffee Extract Are Highly Bioavailable in Humans. J. Nutr. 2008, 138, 2309–2315. [Google Scholar] [CrossRef]
- Prakash, B.; Singh, P.; Mishra, P.K.; Dubey, N.K. Safety Assessment of Zanthoxylum alatum Roxb. Essential Oil, Its Antifungal, Antiaflatoxin, Antioxidant Activity and Efficacy as Antimicrobial in Preservation of Piper nigrum L. Fruits. Int. J. Food Microbiol. 2012, 153, 183–191. [Google Scholar] [CrossRef]
- Sova, M. Antioxidant and Antimicrobial Activities of Cinnamic Acid Derivatives. Mini Rev. Med. Chem. 2012, 12, 749–767. [Google Scholar] [CrossRef]
- Chao, L.K.; Hua, K.F.; Hsu, H.Y.; Cheng, S.S.; Lin, I.F.; Chen, C.J.; Chen, S.T.; Chang, S.T. Cinnamaldehyde Inhibits Pro-Inflammatory Cytokines Secretion from Monocytes/Macrophages through Suppression of Intracellular Signaling. Food Chem. Toxicol. 2008, 46, 220–231. [Google Scholar] [CrossRef]
- Hanci, D.; Altun, H.; Çetinkaya, E.A.; Muluk, N.B.; Cengiz, B.P.; Cingi, C. Cinnamaldehyde Is an Effective Anti-Inflammatory Agent for Treatment of Allergic Rhinitis in a Rat Model. Int. J. Pediatr. Otorhinolaryngol. 2016, 84, 81–87. [Google Scholar] [CrossRef]
- Muhammad, J.S.; Zaidi, S.F.; Shaharyar, S.; Refaat, A.; Usmanghani, K.; Saiki, I.; Sugiyama, T. Anti-Inflammatory Effect of Cinnamaldehyde in Helicobacter Pylori Induced Gastric Inflammation. Biol. Pharm. Bull. 2015, 38, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Tanigawa, N.; Sunghwa, F.; Ninomiya, M.; Hagiwara, M.; Matsushita, K.; Koketsu, M. Morroniside Cinnamic Acid Conjugate as an Anti-Inflammatory Agent. Bioorg. Med. Chem. Lett. 2010, 20, 4855–4857. [Google Scholar] [CrossRef] [PubMed]
- Adisakwattana, S.; Moonsan, P.; Yibchok-Anun, S. Insulin-Releasing Properties of a Series of Cinnamic Acid Derivatives In Vitro and In Vivo. J. Agric. Food Chem. 2008, 56, 7838–7844. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Martinez, E.; Bobadilla, R.; Morales-Rios, M.; Muriel, P.; Perez-Alvarez, V. Trans-3-Phenyl-2-Propenoic Acid (Cinnamic Acid) Derivatives: Structure-Activity Relationship as Hepatoprotective Agents. Med. Chem. 2007, 3, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Alvarez, V.; Bobadilla, R.A.; Muriel, P. Structure-Hepatoprotective Activity Relationship of 3,4-Dihydroxycinnamic Acid (Caffeic Acid) Derivatives. J. Appl. Toxicol. 2001, 21, 527–531. [Google Scholar] [CrossRef] [PubMed]
- Eun, H.J.; Sung, R.K.; In, K.H.; Tae, Y.H. Hypoglycemic Effects of a Phenolic Acid Fraction of Rice Bran and Ferulic Acid in C57BL/KsJ-Db/Db Mice. J. Agric. Food Chem. 2007, 55, 9800–9804. [Google Scholar]
- Sharma, P. Cinnamic Acid Derivatives: A New Chapter of Various Pharmacological Activities. J. Chem. Pharm. Res. 2011, 3, 403–423. [Google Scholar]
- Adisakwattana, S. Cinnamic Acid and Its Derivatives: Mechanisms for Prevention and Management of Diabetes and Its Complications. Nutrients 2017, 9, 163. [Google Scholar] [CrossRef]
- Wiesner, J.; Mitsch, A.; Wißner, P.; Jomaa, H.; Schlitzer, M. Structure—Activity Relationships of Novel Anti-Malarial Agents. Part 2: Cinnamic Acid Derivatives. Bioorg. Med. Chem. Lett. 2001, 11, 423–424. [Google Scholar] [CrossRef]
- Yoon, B.H.; Jung, J.W.; Lee, J.J.; Cho, Y.W.; Jang, C.G.; Jin, C.; Oh, T.H.; Ryu, J.H. Anxiolytic-like Effects of Sinapic Acid in Mice. Life Sci. 2007, 81, 234–240. [Google Scholar] [CrossRef]
- Yabe, T.; Hirahara, H.; Harada, N.; Ito, N.; Nagai, T.; Sanagi, T.; Yamada, H. Ferulic Acid Induces Neural Progenitor Cell Proliferation In Vitro and In Vivo. Neuroscience 2010, 165, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S.; Roy, A.; Jana, M.; Pahan, K. Cinnamic Acid Activates PPARα to Stimulate Lysosomal Biogenesis and Lower Amyloid Plaque Pathology in an Alzheimer’s Disease Mouse Model. Neurobiol. Dis. 2019, 124, 379–395. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.S.; Hou, J.W.; Liu, Y.; Ding, Y.; Zhang, Y.; Li, L.; Zhang, T. Design, Synthesis and Evaluation of Novel Cinnamic Acid Derivatives Bearing N-Benzyl Pyridinium Moiety as Multifunctional Cholinesterase Inhibitors for Alzheimer’s Disease. J. Enzyme Inhib. Med. Chem. 2017, 32, 776–788. [Google Scholar] [CrossRef] [PubMed]
- Szwajgier, D.; Borowiec, K.; Pustelniak, K. The Neuroprotective Effects of Phenolic Acids: Molecular Mechanism of Action. Nutrients 2017, 9, 477. [Google Scholar] [CrossRef] [PubMed]
- Szwajgier, D.; Baranowska-Wojcik, E.; Borowiec, K. Phenolic Acids Exert Anticholinesterase and Cognition-Improving Effects. Curr. Alzheimer Res. 2018, 15, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.X.; Wang, H.; Cui, H.R.; Guo, W.B.; Zhou, F.; Cai, D.S.; Xu, B.; Jia, X.H.; Huang, X.M.; Yang, Y.Q.; et al. Design, Synthesis and Biological Evaluation of Cinnamic Acid Derivatives with Synergetic Neuroprotection and Angiogenesis Effect. Eur. J. Med. Chem. 2019, 183, 111695. [Google Scholar] [CrossRef]
- Cheng, S.S.; Liu, J.Y.; Tsai, K.H.; Chen, W.J.; Chang, S.T. Chemical Composition and Mosquito Larvicidal Activity of Essential Oils from Leaves of Different Cinnamomum Osmophloeum Provenances. J. Agric. Food Chem. 2004, 52, 4395. [Google Scholar] [CrossRef]
- Dias, C.N.; Moraes, D.F.C. Essential Oils and Their Compounds as Aedes aegypti L. (Diptera: Culicidae) Larvicides: Review. Parasitol. Res. 2014, 113, 565–592. [Google Scholar] [CrossRef]
- Fujiwara, G.M.; Annies, V.; de Oliveira, C.F.; Lara, R.A.; Gabriel, M.M.; Betim, F.C.M.; Nadal, J.M.; Farago, P.V.; Dias, J.F.G.; Miguel, O.G.; et al. Evaluation of Larvicidal Activity and Ecotoxicity of Linalool, Methyl Cinnamate and Methyl Cinnamate/Linalool in Combination against Aedes Aegypti. Ecotoxicol. Environ. Saf. 2017, 139, 238–244. [Google Scholar] [CrossRef]
- Seo, S.M.; Park, H.M.; Park, I.K. Larvicidal Activity of Ajowan (Trachyspermum ammi) and Peru Balsam (Myroxylon pereira) Oils and Blends of Their Constituents against Mosquito, Aedes Aegypti, Acute Toxicity on Water Flea, Daphnia Magna, and Aqueous Residue. J. Agric. Food Chem. 2012, 60, 5909–5914. [Google Scholar] [CrossRef]
- Araújo, M.O.; Pérez-Castillo, Y.; Oliveira, L.H.G.; Nunes, F.C.; Sousa, D.P.d. Larvicidal Activity of Cinnamic Acid Derivatives: Investigating Alternative Products for Aedes aegypti L. Control. Molecules 2020, 26, 61. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.P.; Tomaz, D.C.; Ângelo de Souza, L.; Onofre, T.S.; Aquiles de Menezes, W.; Almeida-Silva, J.; Suarez-Fontes, A.M.; Rogéria de Almeida, M.; Manoel da Silva, A.; Bressan, G.C.; et al. Synthesis of Cinnamic Acid Derivatives and Leishmanicidal Activity against Leishmania Braziliensis. Eur. J. Med. Chem. 2019, 183, 111688. [Google Scholar] [CrossRef] [PubMed]
- Letizia, C.S.; Cocchiara, J.; Lalko, J.; Lapczynski, A.; Api, A.M. Fragrance Material Review on Cinnamyl Alcohol. Food Chem. Toxicol. 2005, 43, 837–866. [Google Scholar] [CrossRef] [PubMed]
- Anwar, A.; Siddiqui, R.; Shah, M.R.; Khan, N.A. Gold Nanoparticle-Conjugated Cinnamic Acid Exhibits Antiacanthamoebic and Antibacterial Properties. Antimicrob. Agents Chemother. 2018, 62, e00630-18. [Google Scholar] [CrossRef]
- Bisogno, F.; Mascoti, L.; Sanchez, C.; Garibotto, F.; Giannini, F.; Kurina-Sanz, M.; Enriz, R. Structure-Antifungal Activity Relationship of Cinnamic Acid Derivatives. J. Agric. Food Chem. 2007, 55, 10635–10640. [Google Scholar] [CrossRef]
- Schmidt, E.; Bail, S.; Friedl, S.M.; Jirovetz, L.; Buchbauer, G.; Wanner, J.; Denkova, Z.; Slavchev, A.; Stoyanova, A.; Geissler, M. Antimicrobial Activities of Single Aroma Compounds 1. Nat. Prod. Commun. 2010, 5, 1365–1368. [Google Scholar] [CrossRef]
- Utchariyakiat, I.; Surassmo, S.; Jaturanpinyo, M.; Khuntayaporn, P.; Chomnawang, M.T. Efficacy of Cinnamon Bark Oil and Cinnamaldehyde on Anti-Multidrug Resistant Pseudomonas Aeruginosa and the Synergistic Effects in Combination with Other Antimicrobial Agents. BMC Complement. Altern. Med. 2016, 16, 158. [Google Scholar] [CrossRef]
- Zhu, J.; Zhu, H.; Kobamoto, N.; Yasuda, M.; Tawata, S. Fungitoxic and Phytotoxic Activities of Cinnamic Acid Esters and Amides. J. Pestic. Sci. 2000, 25, 263–266. [Google Scholar] [CrossRef]
- Carvalho, S.A.; Kaiser, M.; Brun, R.; Da Silva, E.F.; Fraga, C.A.M. Antiprotozoal Activity of (E)-Cinnamic N-Acylhydrazone Derivatives. Molecules 2014, 19, 20374–20381. [Google Scholar] [CrossRef]
- Chiriac, C.I.; Tanasa, F.; Onciu, M. A Novel Approach in Cinnamic Acid Synthesis: Direct Synthesis of Cinnamic Acids from Aromatic Aldehydes and Aliphatic Carboxylic Acids in the Presence of Boron Tribromide. Molecules 2005, 10, 481–487. [Google Scholar] [CrossRef]
- Hemaiswarya, S.; Doble, M. Synergistic Interaction of Phenylpropanoids with Antibiotics against Bacteria. J. Med. Microbiol. 2010, 59, 1469–1476. [Google Scholar] [CrossRef] [PubMed]
- Jităreanu, A.; Pădureanu, S.; Tătărîngă, G.; Tuchiluș, C.; Stănescu, U. Turkish Journal of Biology. Evaluation of Phytotoxic and Mutagenic Effects of Some Cinnamic Acid Derivatives Using the Triticum Test. Turk. J. Biol. 2013, 37, 748–756. [Google Scholar] [CrossRef]
- Kim, J.H.; Campbell, B.C.; Mahoney, N.E.; Chan, K.L.; Molyneux, R.J. Identification of Phenolics for Control of Aspergillus Flavus Using Saccharomyces Cerevisiae in a Model Target-Gene Bioassay. J. Agric. Food Chem. 2004, 52, 7814–7821. [Google Scholar] [CrossRef] [PubMed]
- Korošec, B.; Sova, M.; Turk, S.; Kraševec, N.; Novak, M.; Lah, L.; Stojan, J.; Podobnik, B.; Berne, S.; Zupanec, N.; et al. Antifungal Activity of Cinnamic Acid Derivatives Involves Inhibition of Benzoate 4-Hydroxylase (CYP53). J. Appl. Microbiol. 2014, 116, 955–966. [Google Scholar] [CrossRef]
- Narasimhan, B.; Belsare, D.; Pharande, D.; Mourya, V.; Dhake, A. Esters, Amides and Substituted Derivatives of Cinnamic Acid: Synthesis, Antimicrobial Activity and QSAR Investigations. Eur. J. Med. Chem. 2004, 39, 827–834. [Google Scholar] [CrossRef]
- Naz, S.; Ahmad, S.; Ajaz Rasool, S.; Asad Sayeed, S.; Siddiqi, R. Antibacterial Activity Directed Isolation of Compounds from Onosma Hispidum. Microbiol. Res. 2006, 161, 43–48. [Google Scholar] [CrossRef]
- Aminov, R.I. A Brief History of the Antibiotic Era: Lessons Learned and Challenges for the Future. Front. Microbiol. 2010, 1, 134. [Google Scholar] [CrossRef]
- Harbottle, H.; Thakur, S.; Zhao, S.; White, D.G. Genetics of Antimicrobial Resistance. Anim. Biotechnol. 2007, 17, 111–124. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global Trends in Antimicrobial Use in Food Animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef]
- Li, Y.G.; Wang, J.X.; Zhang, G.N.; Zhu, M.; You, X.F.; Hu, X.X.; Zhang, F.; Wang, Y.C. Antibacterial Activity and Structure-Activity Relationship of a Series of Newly Synthesized Pleuromutilin Derivatives. Chem. Biodivers. 2019, 16, e1800560. [Google Scholar] [CrossRef]
- Cai, R.; Miao, M.; Yue, T.; Zhang, Y.; Cui, L.; Wang, Z.; Yuan, Y. Antibacterial Activity and Mechanism of Cinnamic Acid and Chlorogenic Acid against Alicyclobacillus Acidoterrestris Vegetative Cells in Apple Juice. Int. J. Food Sci. Technol. 2019, 54, 1697–1705. [Google Scholar] [CrossRef]
- Godlewska-żyłkiewicz, B.; Świsłocka, R.; Kalinowska, M.; Golonko, A.; Świderski, G.; Arciszewska, Ż.; Nalewajko-Sieliwoniuk, E.; Naumowicz, M.; Lewandowski, W. Biologically Active Compounds of Plants: Structure-Related Antioxidant, Microbiological and Cytotoxic Activity of Selected Carboxylic Acids. Materials 2020, 13, 4454. [Google Scholar] [CrossRef]
- Lopes, S.P.; Castillo, Y.P.; Monteiro, M.L.; de Menezes, R.R.P.P.B.; Almeida, R.N.; Martins, A.M.C.; de Sousa, D.P. Trypanocidal Mechanism of Action and In Silico Studies of P-Coumaric Acid Derivatives. Int. J. Mol. Sci. 2019, 20, 5916. [Google Scholar] [CrossRef] [PubMed]
- Lopes, S.P.; Yepe, L.M.; Pérez-Castillo, Y.; Robledo, S.M.; De Sousa, D.P. Alkyl and Aryl Derivatives Based on P-Coumaric Acid Modification and Inhibitory Action against Leishmania Braziliensis and Plasmodium Falciparum. Molecules 2020, 25, 3178. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Cai, Y.; Sun, X.; Du, X.; Jiang, Z.; Ni, H.; Yang, Y.; Chen, F. Tyrosinase inhibition by p-coumaric acid ethyl ester identified from camellia pollen. Food Sci. Nutr. 2021, 9, 389–400. [Google Scholar] [CrossRef] [PubMed]
- de Morais, M.C.; Perez-Castillo, Y.; Silva, V.R.; de Souza Santos, L.; Soares, M.B.P.; Bezerra, D.P.; de Castro, R.D.; de Sousa, D.P. Cytotoxic and Antifungal Amides Derived from Ferulic Acid: Molecular Docking and Mechanism of Action. Biomed Res. Int. 2021, 2021, 3598000. [Google Scholar] [CrossRef]
- Lima, T.C.; Ferreira, A.R.; Silva, D.F.; Lima, E.O.; de Sousa, D.P. Antifungal Activity of Cinnamic Acid and Benzoic Acid Esters against Candida albicans Strains. Nat. Prod. Res. 2017, 32, 572–575. [Google Scholar] [CrossRef]
- Silva, R.H.N.; Andrade, A.C.M.; Nóbrega, D.F.; Castro, R.D.D.; Pessôa, H.L.F.; Rani, N.; De Sousa, D.P. Antimicrobial Activity of 4-Chlorocinnamic Acid Derivatives. Biomed Res. Int. 2019, 2019, 3941242. [Google Scholar] [CrossRef]
- Sardana, K.; Gupta, A.; Sadhasivam, S.; Gautam, R.K.; Khurana, A.; Saini, S.; Gupta, S.; Ghosh, S. Checkerboard Analysis To Evaluate Synergistic Combinations of Existing Antifungal Drugs and Propylene Glycol Monocaprylate in Isolates from Recalcitrant Tinea Corporis and Cruris Patients Harboring Squalene Epoxidase Gene Mutation. Antimicrob. Agents Chemother. 2021, 65, e00321-21. [Google Scholar] [CrossRef]
- Wang, J.; Morin, P.; Wang, W.; Kollman, P.A. Use of MM-PBSA in Reproducing the Binding Free Energies to HIV-1 RT of TIBO Derivatives and Predicting the Binding Mode to HIV-1 RT of Efavirenz by Docking and MM-PBSA. J. Am. Chem. Soc. 2001, 123, 5221–5230. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand-Protein Interaction Diagrams for Drug Discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Robbins, N.; O’Meara, T.R.; Cowen, L.E. Extensive Functional Redundancy in the Regulation of Candida Albicans Drug Resistance and Morphogenesis by Lysine Deacetylases Hos2, Hda1, Rpd3 and Rpd31. Mol. Microbiol. 2017, 103, 635–656. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Messer, S.A.; Georgopapadakou, N.; Martell, L.A.; Besterman, J.M.; Diekema, D.J. Activity of MGCD290, a Hos2 Histone Deacetylase Inhibitor, in Combination with Azole Antifungals against Opportunistic Fungal Pathogens. J. Clin. Microbiol. 2009, 47, 3797–3804. [Google Scholar] [CrossRef]
- Robbins, N.; Leach, M.D.; Cowen, L.E. Lysine Deacetylases Hda1 and Rpd3 Regulate Hsp90 Function Thereby Governing Fungal Drug Resistance. Cell Rep. 2012, 2, 878–888. [Google Scholar] [CrossRef]
- McCarthy, M.W.; Kontoyiannis, D.P.; Cornely, O.A.; Perfect, J.R.; Walsh, T.J. Novel Agents and Drug Targets to Meet the Challenges of Resistant Fungi. J. Infect. Dis. 2017, 216 (Suppl. 3), S474–S483. [Google Scholar] [CrossRef]
- Su, S.; Li, X.; Yang, X.; Li, Y.; Chen, X.; Sun, S.; Jia, S. Histone Acetylation/Deacetylation in Candida Albicans and Their Potential as Antifungal Targets. Future Microbiol. 2020, 15, 1075–1090. [Google Scholar] [CrossRef]
- Perez, M.; Castillo, Y. Bacterial Beta-Ketoacyl-Acyl Carrier Protein Synthase III (FabH): An Attractive Target for the Design of New Broad-Spectrum Antimicrobial Agents. Mini Rev. Med. Chem. 2008, 8, 36–45. [Google Scholar] [CrossRef]
- Pishchany, G.; Mevers, E.; Ndousse-Fetter, S.; Horvath, D.J.; Paludo, C.R.; Silva-Junior, E.A.; Koren, S.; Skaar, E.P.; Clardy, J.; Kolter, R. Amycomicin Is a Potent and Specific Antibiotic Discovered with a Targeted Interaction Screen. Proc. Natl. Acad. Sci. USA 2018, 115, 10124–10129. [Google Scholar] [CrossRef]
- Wang, J.; Ye, X.; Yang, X.; Cai, Y.; Wang, S.; Tang, J.; Sachdeva, M.; Qian, Y.; Hu, W.; Leeds, J.A.; et al. Discovery of Novel Antibiotics as Covalent Inhibitors of Fatty Acid Synthesis. ACS Chem. Biol. 2020, 15, 1826–1834. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kodali, S.; Sang, H.L.; Galgoci, A.; Painter, R.; Dorso, K.; Racine, F.; Motyl, M.; Hernandez, L.; Tinney, E.; et al. Discovery of Platencin, a Dual FabF and FabH Inhibitor with in Vivo Antibiotic Properties. Proc. Natl. Acad. Sci. USA 2007, 104, 7612–7616. [Google Scholar] [CrossRef]
- He, X.; Reynolds, K.A. Purification, Characterization, and Identification of Novel Inhibitors of the Beta-Ketoacyl-Acyl Carrier Protein Synthase III (FabH) from Staphylococcus Aureus. Antimicrob. Agents Chemother. 2002, 46, 1310–1318. [Google Scholar] [CrossRef] [PubMed]
- Dimmock, J.R.; Murthi Kandepu, N.; Hetherington, M.; Wilson Quail, J.; Pugazhenthi, U.; Sudom, A.M.; Chamankhah, M.; Rose, P.; Pass, E.; Allen, T.M.; et al. Cytotoxic Activities of Mannich Bases of Chalcones and Related Compounds. J. Med. Chem. 1998, 41, 1014–1026. [Google Scholar] [CrossRef] [PubMed]
- Uno, J.; Shigematsu, M.L.; Arai, T. Primary Site of Action of Ketoconazole on Candida Albicans. Antimicrob. Agents Chemother. 1982, 21, 912–918. [Google Scholar] [CrossRef]
- Pushkareva, V.I.; Slezina, M.P.; Korostyleva, T.V.; Shcherbakova, L.A.; Istomina, E.A.; Ermolaeva, S.A.; Ogarkova, O.A.; Odintsova, T.I. Antimicrobial Activity of Wild Plant Seed Extracts against Human Bacterial and Plant Fungal Pathogens. Am. J. Plant Sci. 2017, 8, 1572–1592. [Google Scholar] [CrossRef]
- Pinheiro, L.S.; Filho, A.A.d.O.; Guerra, F.Q.S.; de Menezes, C.P.; dos Santos, S.G.; de Sousa, J.P.; Dantas, T.B.; Lima, E.d.O. Antifungal Activity of the Essential Oil Isolated from Laurus Nobilis L. against Cryptococcus Neoformans Strains. J. Appl. Pharm. Sci. 2017, 7, 115–118. [Google Scholar]
- Perez-Castillo, Y.; Montes, R.C.; da Silva, C.R.; Neto, J.B.d.A.; Dias, C.d.S.; Duarte, A.B.S.; Júnior, H.V.N.; de Sousa, D.P. Antifungal Activity of N-(4-Halobenzyl)Amides against Candida Spp. and Molecular Modeling Studies. Int. J. Mol. Sci. 2022, 23, 419. [Google Scholar] [CrossRef]
- Keiser, M.J.; Roth, B.L.; Armbruster, B.N.; Ernsberger, P.; Irwin, J.J.; Shoichet, B.K. Relating Protein Pharmacology by Ligand Chemistry. Nat. Biotechnol. 2007, 25, 197–206. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A Better Web Interface. Nucleic Acids Res. 2008, 36 (Suppl. 2), W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, P.C.D.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer Generation with OMEGA: Algorithm and Validation Using High Quality Structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 2010, 50, 572–584. [Google Scholar] [CrossRef] [PubMed]
- OpenEye Scientific Software. QUACPAC. Santa Fe, NM: OpenEye Scientific Software. Available online: http://www.eyesopen.com (accessed on 26 May 2022).
- Bienert, S.; Waterhouse, A.; De Beer, T.A.P.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-New Features and Functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef]
- Perez-Castillo, Y.; Lima, T.C.; Ferreira, A.R.; Silva, C.R.; Campos, R.S.; Neto, J.B.A.; Magalhães, H.I.F.; Cavalcanti, B.C.; Júnior, H.V.N.; De Sousa, D.P. Bioactivity and Molecular Docking Studies of Derivatives from Cinnamic and Benzoic Acids. Biomed Res. Int. 2020, 2020, 6345429. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and Validation of a Genetic Algorithm for Flexible Docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Case, I.Y.B.-S.D.A.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E.; Cruzeiro, V.W.D.; Darden, T.A.; Duke, D.G.R.E.; Gilson, M.K.; Gohlke, H.; Goetz, A.W.; et al. AMBER; University of California: San Francisco, CA, USA, 2018. [Google Scholar]
- Pang, Y.-P. Novel Zinc Protein Molecular Dynamics Simulations: Steps Toward Antiangiogenesis for Cancer Treatment. Mol. Model. Annu. 1999, 5, 196–202. [Google Scholar] [CrossRef]
- Machado, M.R.; Pantano, S. Split the Charge Difference in Two! A Rule of Thumb for Adding Proper Amounts of Ions in MD Simulations. J. Chem. Theory Comput. 2020, 16, 1367–1372. [Google Scholar] [CrossRef]
- Iranpoor, N.; Firouzabadi, H.; Riazi, A.; Pedrood, K. Regioselective hydrocarbonylation of phenylacetylene to α,β-unsaturated esters and thioesters with Fe(CO)5 and Mo(CO)6. J. Organomet. Chem. 2016, 822, 67–73. [Google Scholar] [CrossRef]
- Lutjen, A.B.; Quirk, M.A.; Barbera, A.M.; Kolonko, E.M. Synthesis of (E)-cinnamyl ester derivatives via a greener Steglich esterification. Bioorg. Med. Chem. 2018, 26, 5291–5298. [Google Scholar] [CrossRef]
- Jakovetić, S.M.; Jugović, B.Z.; Gvozdenović, M.M.; Bezbradica, D.I.; Antov, M.G.; Mijin, D.Ž.; Knežević-Jugović, Z.D. Synthesis of Aliphatic Esters of Cinnamic Acid as Potential Lipophilic Antioxidants Catalyzed by Lipase B from Candida antarctica. Appl. Biochem. Biotechnol. 2013, 170, 1560–1573. [Google Scholar] [CrossRef]
- Sova, M.; Perdih, A.; Kotnik, M.; Kristan, K.; Rižner, T.L.; Solmajer, T.; Gobec, S. Flavonoids and cinnamic acid esters as inhibitors of fungal 17β-hydroxysteroid dehydrogenase: A synthesis, QSAR and modelling study. Bioorg. Med. Chem. 2006, 14, 7404–7418. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Ouchi, H.; Takahata, H. Carboxamidation of carboxylic acids with 1-tert-butoxy-2-tert-butoxycarbonyl-1,2-dihydroisoquinoline (BBDI) without bases. Tetrahedron 2008, 64, 11129–11135. [Google Scholar] [CrossRef]
- Allen, C.L.; Chhatwal, A.R.; Williams, J.M.J. Direct amide formation from unactivated carboxylic acids and amines. Chem. Commun. 2011, 48, 666–668. [Google Scholar] [CrossRef] [PubMed]
- Barajas, J.G.H.; Méndez, L.Y.V.; Kouznetsov, V.V.; Stashenko, E.E. Efficient synthesis of new N-benzyl- or N-(2-furylmethyl)cinnamamides promoted by the ‘green’ catalyst boric acid, and their spectral analysis. Synthesis 2008, 3, 0377–0382. [Google Scholar] [CrossRef]
- Khaldoun, K.; Safer, A.; Saidi-Besbes, S.; Carboni, B.; Le Guevel, R.; Carreaux, F. An Efficient Solvent-Free Microwave-Assisted Synthesis of Cinnamamides by Amidation Reaction Using Phenylboronic Acid/Lewis Base Co-catalytic System. Synth. J. Synth. Org. Chem. 2019, 51, 3891–3900. [Google Scholar] [CrossRef]
- Yasui, Y.; Tsuchida, S.; Miyabe, H.; Takemoto, Y. One-pot amidation of olefins through Pd-catalyzed coupling of alkylboranes and carbamoyl chlorides. J. Org. Chem. 2007, 72, 5898–5900. [Google Scholar] [CrossRef]


| Compounds | C. albicans ATCC-76485 | C. tropicalis ATCC-13803 | C. glabrata ATCC-90030 | A. flavus LM-171 | P. citrinum ATCC-4001 |
|---|---|---|---|---|---|
| 1 | + | + | + | + | + |
| 2 | 128/789.19 | 128/789.19 | 128/789.19 | 256/1578.16 | 256/1578.16 |
| 3 | 128/726.36 | 128/726.36 | 128/726.36 | 128/726.36 | 128/726.36 |
| 4 | 128/672.83 | 128/672.83 | 128/672.83 | 128/672.83 | 128/672.83 |
| 5 | 128/672.83 | 128/672.83 | 128/672.83 | + | + |
| 6 | 128/626.62 | 128/626.62 | 128/626.62 | 128/626.62 | 128/626.62 |
| 7 | + | + | + | 512/2345.39 | 512/2345.39 |
| 8 | + | + | + | + | + |
| 9 | 256/1101.92 | 256/1101.92 | 256/1101.92 | 256/1101.92 | 256/1101.92 |
| 10 | 256/1075.63 | 256/1075.63 | 256/1075.63 | 512/2151.26 | 512/2151.26 |
| 11 | + | + | + | + | + |
| 12 | + | + | + | + | + |
| 13 | + | + | + | + | + |
| 14 | + | + | + | + | + |
| 15 | + | + | + | + | + |
| 16 | + | + | + | + | + |
| 17 | + | + | + | 512/2021.31 | 512/2021.31 |
| 18 | 512/1832.62 | 512/1832.62 | 256/916.31 | 256/916.31 | 256/916.31 |
| 19 | + | + | + | + | + |
| 20 | + | + | + | + | + |
| Nystatin | - | - | - | - | - |
| Control medium | - | - | - | - | - |
| Fungal growth control | + | + | + | + | + |
| Compound | C. albicans (ATCC-76485) | C. tropicalis (ATCC-13803) | C. glabrata (ATCC-90030) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MIC (µM) | MFC (µM) | MFC/ MIC | MIC (µM) | MFC (µM) | MFC/ MIC | MIC (µM) | MFC (µM) | MFC/ MIC | |
| 3 | 726.36 | 1452.72 | 2 | 726.36 | 1452.72 | 2 | 726.36 | 1452.72 | 2 |
| 4 | 672.83 | 672.83 | 1 | 672.83 | 672.83 | 1 | 672.83 | 672.83 | 1 |
| 6 | 626.62 | 626.62 | 1 | 626.62 | 626.62 | 1 | 626.62 | 626.62 | 1 |
| A. flavus (LM-171) | P. citrinum (ATCC-4001) | ||||||||
| MIC (µM) | MFC (µM) | MFC/ MIC | MIC (µM) | MFC (µM) | MFC/ MIC | ||||
| 3 | 726.36 | 726.36 | 1 | 726.36 | 726.36 | 1 | |||
| 4 | 672.83 | 1345.66 | 2 | 672.83 | 1345.66 | 2 | |||
| 6 | 626.62 | 626.62 | 1 | 626.62 | 626.62 | 1 | |||
| C. albicans ATCC-76485 | ||||
|---|---|---|---|---|
| Compounds | with Ergosterol | without Ergosterol | with Sorbitol | without Sorbitol |
| 3 | 672.83 µM | 2691.32 µM | 672.83 µM | 5382.64 µM |
| 6 | 626.62 µM | 2506.48 µM | 626.62 µM | 5012.96 µM |
| Caspofungin | - | - | 0.5 µg/mL | 1.0 µg/mL |
| Nystatin | 8.05 µM | 129.0 µM | - | - |
| P. citrinum ATCC-4001 | ||||
|---|---|---|---|---|
| Compounds | with Ergosterol | without Ergosterol | with Sorbitol | without Sorbitol |
| 4 | 672.83 µM | 2691.32 µM | 672.83 µM | 2691.32 µM |
| 6 | 626.62 µM | 5012.96 µM | 626.62 µM | 5012.96 µM |
| Caspofungin | - | - | 0.5 µg/mL | 1.0 µg/mL |
| Nystatin | 8.05 µM | 129.0 µM | - | - |
| Micro-Organism | Compound | MIC (µM) | FIC | FICI | Result | |
|---|---|---|---|---|---|---|
| Alone | Combined | |||||
| C. albicans | 6 | 626.62 | 313.31 | 0.5 | 0.9 | Additive |
| Nystatin | 8.0 | 3.2 | 0.4 | |||
| Compounds | S. aureus ATCC-35903 | S. epidermidis ATCC-12228 | P. aeruginosa ATCC-25853 |
|---|---|---|---|
| 1 | 128/841.05 | 512/3364.21 | 512/3364.21 |
| 2 | 128/789.19 | 128/789.19 | 128/789.19 |
| 3 | 128/726.36 | 128/726.36 | 128/726.36 |
| 4 | 128/672.83 | 128/672.83 | 128/672.83 |
| 5 | 128/672.83 | 128/672.83 | 128/672.83 |
| 6 | 128/626.62 | 128/626.62 | 128/626.62 |
| 7 | + | 128/586.34 | 128/586.34 |
| 8 | + | + | + |
| 9 | 128/550.96 | 128/550.96 | 128/550.96 |
| 10 | 128/537.81 | 128/537.81 | 256/1075.63 |
| 11 | + | + | + |
| 12 | + | + | + |
| 13 | + | + | + |
| 14 | + | + | + |
| 15 | + | + | + |
| 16 | + | + | + |
| 17 | + | + | + |
| 18 | 128/458.15 | 128/458.15 | 128/458.15 |
| 19 | + | + | + |
| 20 | + | + | + |
| Amoxicillin | - | - | - |
| Control medium | - | - | - |
| Bacterial growth control | + | + | + |
| Compound | S. aureus ATCC-35903 | S. epidermidis ATCC-12228 | P. aeruginosa ATCC-25853 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MIC (µM) | MFC (µM) | MFC/ MIC | MIC (µM) | MFC (µM) | MFC/ MIC | MIC (µM) | MFC (µM) | MFC/ MIC | |
| 6 | 626.62 | 1253.24 | 2 | 626.62 | 1253.24 | 2 | 626.62 | 1253.24 | 2 |
| 9 | 550.96 | 1101.92 | 2 | 550.96 | 1101.92 | 2 | 550.96 | 550.96 | 1 |
| 18 | 458.15 | 458.15 | 1 | 458.15 | 458.15 | 1 | 458.15 | 458.15 | 1 |
| Micro-Organism | Compound | MIC (µg/mL) | FIC | FICI | Result | |
|---|---|---|---|---|---|---|
| Alone | Combined | |||||
| S. aureus | 18 | 128.0 | 64.0 | 0.5 | 0.53 | Additive |
| Amoxicillin | 0.015 | 0.0005 | 0.033 | |||
| UniProt Accession | ID (a) | Compound | Description (b) | Source (c) |
|---|---|---|---|---|
| C. albicans | ||||
| A0A1D8PLH1 | caTIM10 | 6 | Mitochondrial import inner membrane translocase subunit TIM10 | Homology |
| A0A1D8PHZ5 | caRHO2 | 6 | Rho family GTPase | Homology |
| P0CY33 | caCDC42 | 6 | Cell division control protein 42 homolog | Homology |
| O42825 | caRHO1 | 6 | GTP-binding protein RHO1 | Homology |
| A0A1D8PH96 | caRHO3 | 6 | Rho family GTPase | Homology |
| Q5ADT3 | caAKR | 6 | Aldo/keto reductase | Homology |
| Q5ADT4 | caGCY1 | 6 | Glycerol 2-dehydrogenase | Homology |
| P12461 | caTMP1 | 6 | Thymidylate synthase | Homology |
| Q5ADM7 | caTDH3 | 6 | Glyceraldehyde-3-phosphate dehydrogenase | PDB |
| A0A1D8PNK3 | caGRE3 | 6 | D-xylose reductase | Homology |
| Q5AHE2 | caUGA11 | 6 | 4-aminobutyrate aminotransferase | Homology |
| A0A1D8PJ17 | caHOS2 | 6 | Histone deacetylase | Homology |
| A0A1D8PH55 | caUGA1 | 6 | 4-aminobutyrate aminotransferase | Homology |
| A0A1D8PSA6 | caRPD3 | 6 | Histone deacetylase | Homology |
| A0A1D8PUB9 | caIMA1 | 6 | Oligo-1,6-glucosidase IMA1 | Homology |
| A0A1D8PP39 | caAAM | 6 | Aamy domain-containing protein | Homology |
| Q59NB8 | caLKH1 | 6 | Leucine aminopeptidase 2 | Homology |
| A0A1D8PMM1 | caTOP2 | 6 | DNA topoisomerase 2 | Homology |
| S. aureus | ||||
| Q5HED0 | saACPS | 18 | 4′-phosphopantetheinyl transferase AcpS | PDB |
| Q5HH38 | saMENB | 18 | 1,4-dihydroxy-2-naphthoyl-CoA synthase | PDB |
| Q7A6I1 | saPPIASE-1 | 18 | Peptidyl-prolyl cis-trans isomerase (PPIase) | Homology |
| A0A1Q8DEF0 | saPPIASE-2 | 18 | Peptidyl-prolyl cis-trans isomerase (PPIase) | Homology |
| T1YB03 | saGAR | 18 | 2,5-diketo-D-gluconic acid reductase | Homology |
| A0A8B1CIU3 | saALDH | 18 | Aldo/keto reductase | Homology |
| A7X0K2 | saFABH | 18 | 3-oxoacyl-[acyl-carrier-protein] synthase III | PDB |
| A0A5C0VWB1 | saFLHA | 18 | Alcohol dehydrogenase | Homology |
| Q9L4P8 | saGBSA | 18 | Betaine aldehyde dehydrogenase | PDB |
| A0A1Q8DC81 | saPPTHL | 18 | Alpha,alpha-phosphotrehalase | Homology |
| Target | Compound | PLP (a) | Z-PLP (b) | GS (c) | Z-GS (d) | CS (e) | Z-CS (f) | ASP (g) | Z-ASP (h) | Aggregated Z-Score |
|---|---|---|---|---|---|---|---|---|---|---|
| C. albicans | ||||||||||
| caRHO2 | 6 | 71.91 | 3.01 | 27.36 | 0.73 | 24.28 | 2.49 | 19.65 | 1.27 | 1.87 |
| caRHO1 | 6 | 71.48 | 2.58 | 30.14 | 0.84 | 20.49 | 1.33 | 19.76 | 1.13 | 1.47 |
| caRHO3 | 6 | 51.88 | 1.4 | 23.06 | 1.95 | 17.56 | 0.91 | 14.27 | 1.12 | 1.35 |
| caAKR | 6 | 47.04 | 1.22 | 12.46 | 1.03 | 21.49 | 1.65 | 29.27 | 1.82 | 1.43 |
| caGCY1 | 6 | 42.72 | 0.18 | 8.85 | 1.26 | 16.08 | 1.31 | 23.79 | 1.33 | 1.02 |
| caTMP1-Both (i) | 6 | 44.4 | 1.63 | 26.88 | 1.03 | 14.27 | −0.73 | 22.16 | 2.39 | 1.08 |
| caTMP1-UMP (j) | 6 | 51.26 | 2.06 | 20.96 | 0.32 | 11.16 | 0.65 | 17.58 | 0.37 | 0.85 |
| caTMP1-Cof. (k) | 6 | 60.42 | 1.53 | 27.43 | 1.19 | 23.54 | 1.14 | 33.79 | 3.49 | 1.84 |
| caTDH3 | 6 | 33.35 | 1.32 | 0.06 | −0.1 | 10.49 | 2.38 | 10.67 | 1.42 | 1.26 |
| caGRE3 | 6 | 44.93 | 1.38 | 19.45 | 1.24 | 13.61 | 1.87 | 19.18 | 1.44 | 1.48 |
| caUGA11 | 6 | 55.78 | 1.77 | 15.07 | 0.77 | 19.41 | 1.13 | 25.73 | 1.99 | 1.42 |
| caHOS2 | 6 | 66.22 | 1.77 | 26.47 | 0.76 | 25.33 | 2.06 | 29.1 | 1.88 | 1.62 |
| caUGA1 | 6 | 52.47 | 2.31 | 17.77 | 1.13 | 19.68 | 1.78 | 17.4 | −0.06 | 1.29 |
| caRPD3 | 6 | 64.83 | 4.04 | 43.1 | 2.61 | 21.1 | 2.2 | 26.76 | 1.84 | 2.67 |
| caIMA1 | 6 | 64.55 | 3.22 | 22.94 | 0.32 | 26.58 | 2.64 | 30.08 | 1.49 | 1.92 |
| caAAM | 6 | 40.33 | 1.7 | −21.83 | −0.15 | 11.56 | 2 | 18.72 | 2.01 | 1.39 |
| caLKH1 | 6 | 57.37 | 1.77 | 19.55 | 0.29 | 24.73 | 1.69 | 26.95 | 1.3 | 1.26 |
| caTOP2 | 6 | 40.32 | 1.31 | 8.27 | 1.7 | 12 | 2.02 | −1.69 | −0.29 | 1.18 |
| S. aureus | ||||||||||
| saACPS | 18 | 69.08 | 2.87 | 39.36 | 1.29 | 21.73 | 1.23 | 19.88 | 0.46 | 1.46 |
| saMENB | 18 | 70.68 | 2.5 | 26.92 | −0.98 | 29.31 | 2.31 | 26.4 | 0.68 | 1.13 |
| saPPIASE-1 | 18 | 64.35 | 2.69 | 19.57 | 0.51 | 24.81 | 1.88 | 20.69 | −0.21 | 1.22 |
| saPPIASE-2 | 18 | 55.7 | 1.69 | 15.71 | 0.58 | 24.29 | 2.21 | 31.95 | 3 | 1.87 |
| saGAR | 18 | 62.95 | 1.58 | 10.82 | 0.51 | 26.03 | 1.5 | 32.66 | 1.29 | 1.22 |
| saALDH | 18 | 54.21 | 1.98 | 6.7 | 0.6 | 22.61 | 2.1 | 24.56 | 2.28 | 1.74 |
| saFABH | 18 | 74.62 | 2.31 | 14.42 | 0.92 | 34.2 | 1.61 | 35.22 | 2.32 | 1.79 |
| saFLHA | 18 | 49.49 | 1.37 | −48.13 | 0.78 | 11.41 | 1.47 | 12.48 | 1.59 | 1.3 |
| saGBSA | 18 | 38.58 | 1.64 | −93.97 | 0.26 | 8.62 | 1.47 | 10.47 | 1.57 | 1.23 |
| saPPTHL | 18 | 55.63 | 0.76 | 29.66 | 1.27 | 23.05 | 1.8 | 24.44 | 0.23 | 1.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Morais, M.C.; de Oliveira Lima, E.; Perez-Castillo, Y.; de Sousa, D.P. Synthetic Cinnamides and Cinnamates: Antimicrobial Activity, Mechanism of Action, and In Silico Study. Molecules 2023, 28, 1918. https://doi.org/10.3390/molecules28041918
de Morais MC, de Oliveira Lima E, Perez-Castillo Y, de Sousa DP. Synthetic Cinnamides and Cinnamates: Antimicrobial Activity, Mechanism of Action, and In Silico Study. Molecules. 2023; 28(4):1918. https://doi.org/10.3390/molecules28041918
Chicago/Turabian Stylede Morais, Mayara Castro, Edeltrudes de Oliveira Lima, Yunierkis Perez-Castillo, and Damião Pergentino de Sousa. 2023. "Synthetic Cinnamides and Cinnamates: Antimicrobial Activity, Mechanism of Action, and In Silico Study" Molecules 28, no. 4: 1918. https://doi.org/10.3390/molecules28041918
APA Stylede Morais, M. C., de Oliveira Lima, E., Perez-Castillo, Y., & de Sousa, D. P. (2023). Synthetic Cinnamides and Cinnamates: Antimicrobial Activity, Mechanism of Action, and In Silico Study. Molecules, 28(4), 1918. https://doi.org/10.3390/molecules28041918







