Lactobacillus plantarum Metabolites Elicit Anticancer Effects by Inhibiting Autophagy-Related Responses
Abstract
1. Introduction
2. Results
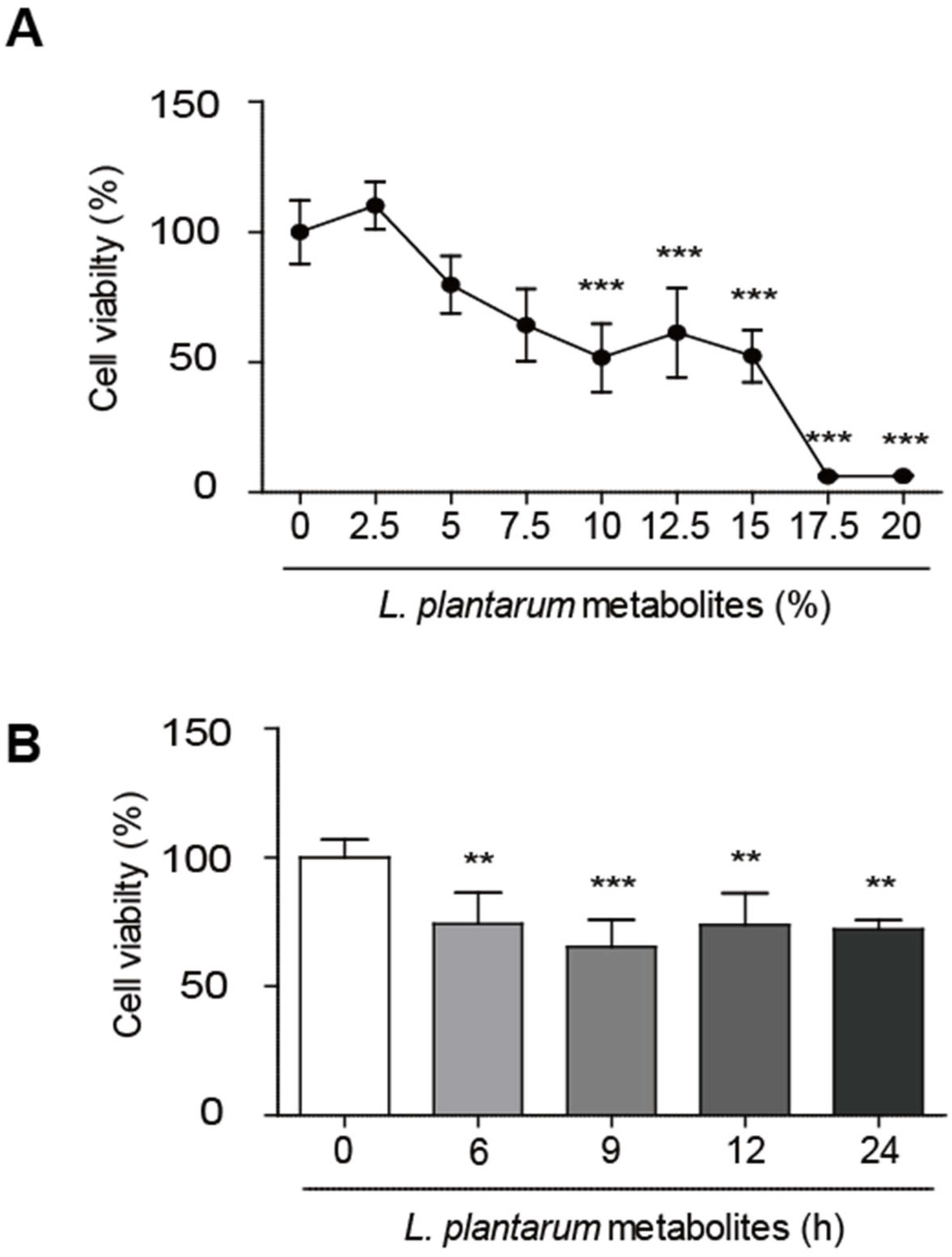
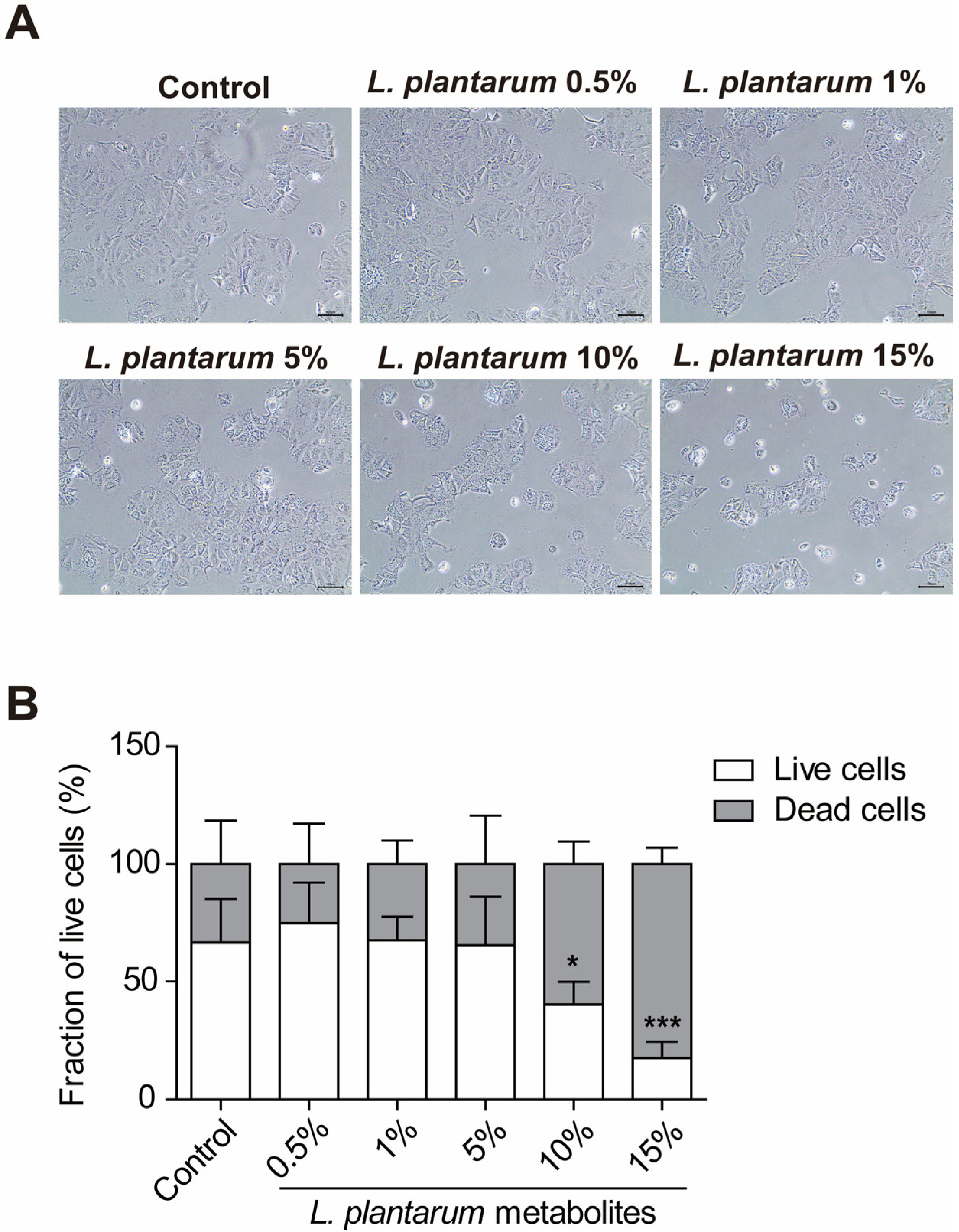
2.1. L. plantarum Metabolites Reduced the Cell Viability of Caco-2 Human Colorectal Cancer Cells
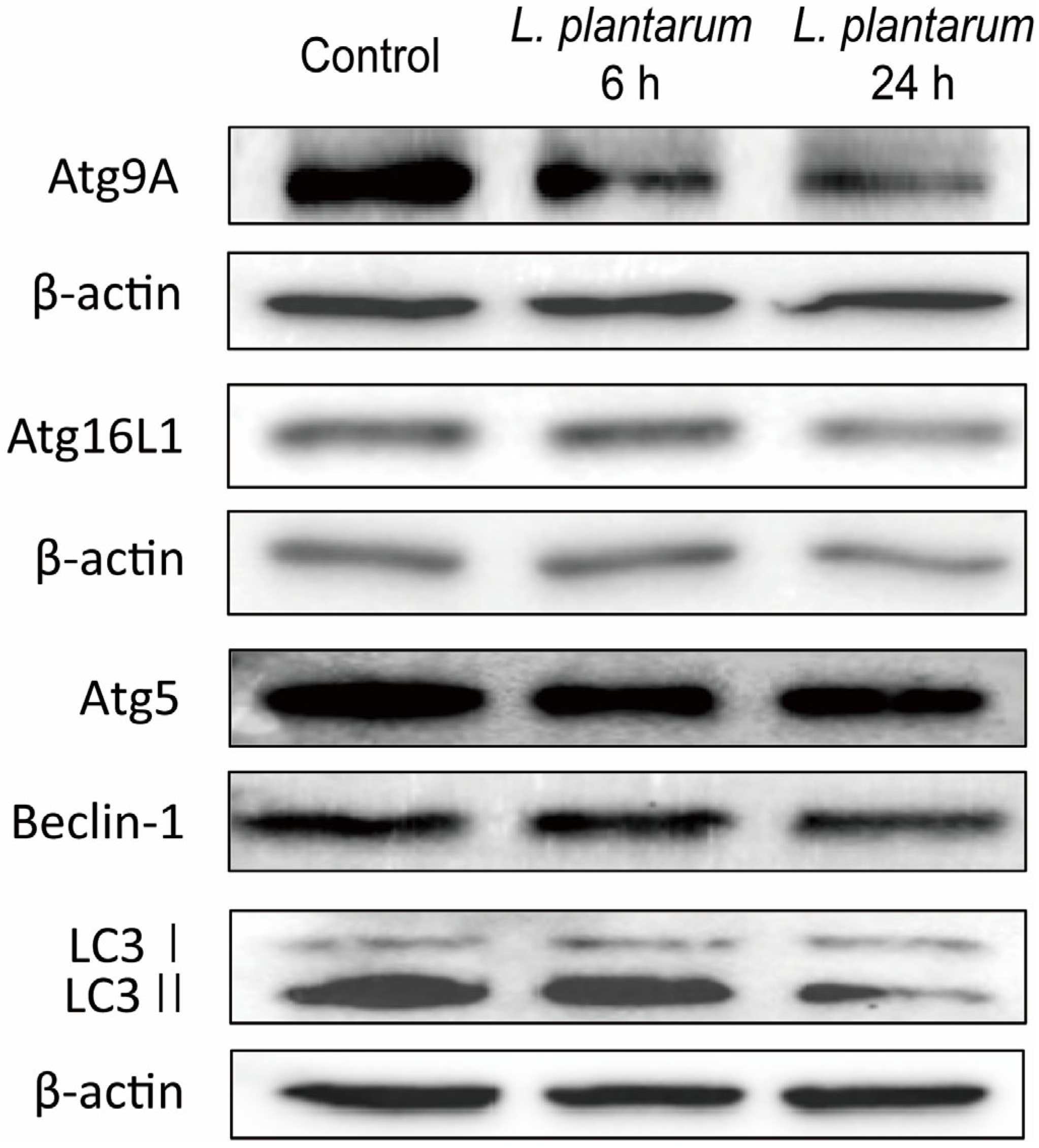
2.2. L. plantarum Metabolites Downregulated the Expression of Autophagy Markers in Caco-2 Cells
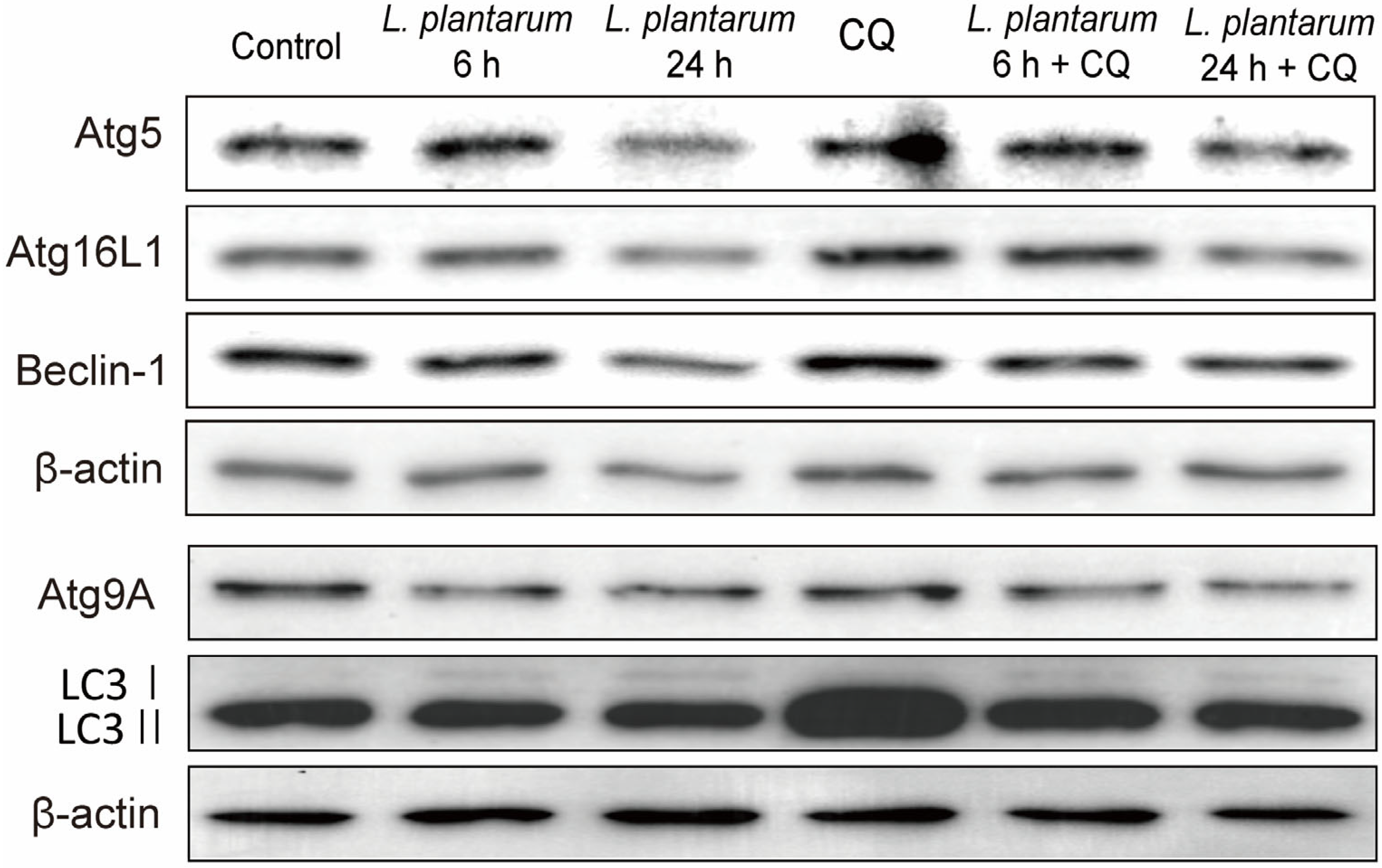
2.3. L. plantarum Metabolites Enhanced the Autophagy Inhibition Ability of CQ in Caco-2 Cells
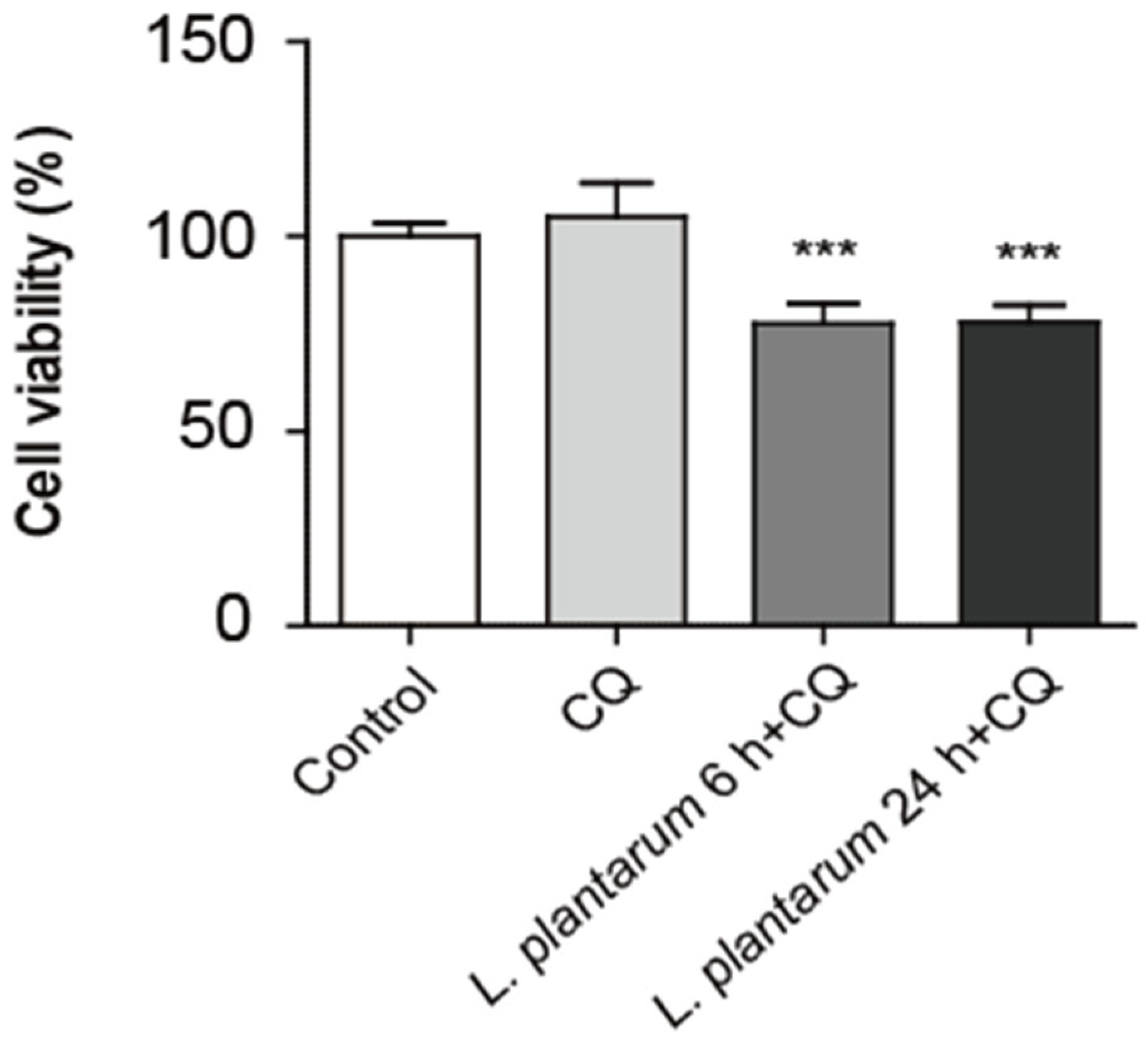
2.4. Co-Treatment with L. plantarum Metabolites and CQ Decreased Caco-2 Cell Viability More than CQ Treatment Alone
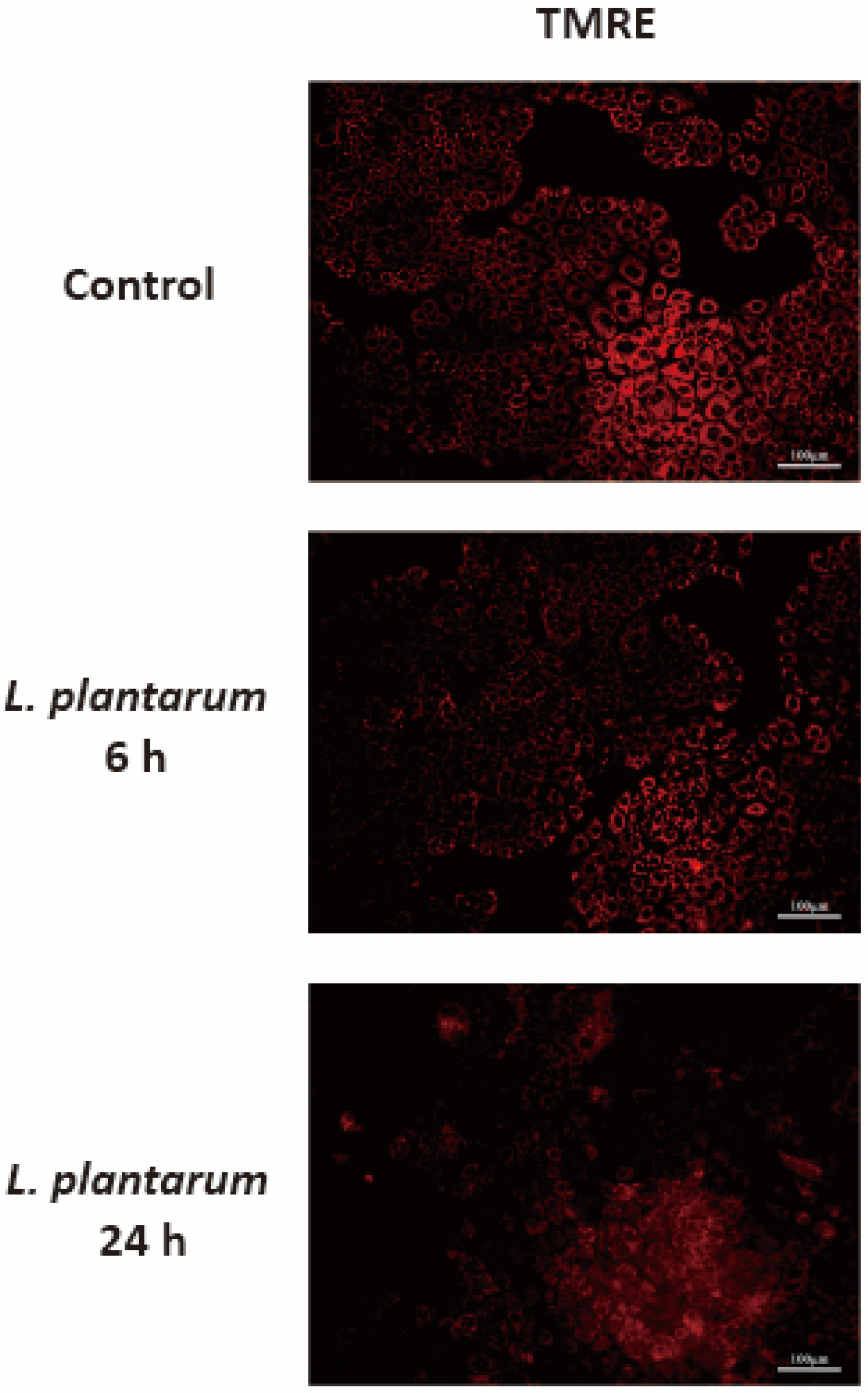
2.5. L. plantarum Metabolites Induced Mitochondrial Dysfunction in Caco-2 Cells
3. Discussion
4. Materials and Methods
4.1. Preparation of L. plantarum Metabolites
4.2. Cell Culture and Treatment
4.3. Cell Viability Assay
4.4. Western Blot Analysis
4.5. TMRE Staining
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Nogueira, L.; Mariotto, A.B.; Rowland, J.H.; Yabroff, K.R.; Alfano, C.M.; Jemal, A.; Kramer, J.L.; Siegel, R.L. Cancer treatment and survivorship statistics, 2019. CA A Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Abnet, C.C.; Neale, R.E.; Vignat, J.; Giovannucci, E.L.; McGlynn, K.A.; Bray, F. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology 2020, 159, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef]
- Hull, M.A.; Rees, C.J.; Sharp, L.; Koo, S. A risk-stratified approach to colorectal cancer prevention and diagnosis. Nature reviews. Gastroenterol. Hepatol. 2020, 17, 773–780. [Google Scholar]
- Lee, K.C.; Wu, K.L.; Yen, C.K.; Chang, S.F.; Chen, C.N.; Lu, Y.C. Inhibition of NLRP3 by Fermented Quercetin Decreases Resistin-Induced Chemoresistance to 5-Fluorouracil in Human Colorectal Cancer Cells. Pharmaceuticals 2022, 15, 798. [Google Scholar] [CrossRef]
- Rawla, P.; Barsouk, A.; Hadjinicolaou, A.V.; Barsouk, A. Immunotherapies and Targeted Therapies in the Treatment of Metastatic Colorectal Cancer. Med. Sci. 2019, 7, 83. [Google Scholar] [CrossRef]
- Her, J.Y.; Lee, Y.; Kim, S.J.; Heo, G.; Choo, J.; Kim, Y.; Howe, C.; Rhee, S.H.; Yu, H.S.; Chung, H.Y.; et al. Blockage of protease-activated receptor 2 exacerbates inflammation in high-fat environment partly through autophagy inhibition. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 320, G30–G42. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, Y.; Heo, G.; Jeong, S.; Park, S.; Yoo, J.W.; Jung, Y.; Im, E. Modulation of Intestinal Epithelial Permeability via Protease-Activated Receptor-2-Induced Autophagy. Cells 2022, 11, 878. [Google Scholar] [CrossRef]
- Saha, S.; Panigrahi, D.P.; Patil, S.; Bhutia, S.K. Autophagy in health and disease: A comprehensive review. Biomed. Pharmacother. 2018, 104, 485–495. [Google Scholar] [CrossRef]
- Csizmadia, T.; Juhász, G. Crinophagy mechanisms and its potential role in human health and disease. Prog. Mol. Biol. Transl. Sci. 2020, 172, 239–255. [Google Scholar] [PubMed]
- Chung, K.W.; Kim, K.M.; Choi, Y.J.; An, H.J.; Lee, B.; Kim, D.H.; Lee, E.K.; Im, E.; Lee, J.; Im, D.S.; et al. The critical role played by endotoxin-induced liver autophagy in the maintenance of lipid metabolism during sepsis. Autophagy 2017, 13, 1113–1129. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.W.; Chung, H.Y. The Effects of Calorie Restriction on Autophagy: Role on Aging Intervention. Nutrients 2019, 11, 2923. [Google Scholar] [CrossRef] [PubMed]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef]
- Kim, J.; Hlaing, S.P.; Lee, J.; Saparbayeva, A.; Kim, S.; Hwang, D.S.; Lee, E.H.; Yoon, I.-S.; Yun, H.; Kim, M.-S. Exfoliated bentonite/alginate nanocomposite hydrogel enhances intestinal delivery of probiotics by resistance to gastric pH and on-demand disintegration. Carbohydr. Polym. 2021, 272, 118462. [Google Scholar] [CrossRef]
- Paolillo, R.; Carratelli, C.R.; Sorrentino, S.; Mazzola, N.; Rizzo, A. Immunomodulatory effects of Lactobacillus plantarum on human colon cancer cells. Int. Immunopharmacol. 2009, 9, 1265–1271. [Google Scholar] [CrossRef]
- Chuah, L.-O.; Foo, H.L.; Loh, T.C.; Mohammed Alitheen, N.B.; Yeap, S.K.; Abdul Mutalib, N.E.; Abdul Rahim, R.; Yusoff, K. Postbiotic metabolites produced by Lactobacillus plantarum strains exert selective cytotoxicity effects on cancer cells. BMC Complement. Altern. Med. 2019, 19, 114. [Google Scholar] [CrossRef]
- Prakoeswa, C.R.S.; Bonita, L.; Karim, A.; Herwanto, N.; Umborowati, M.A.; Setyaningrum, T.; Hidayati, A.N.; Surono, I.S. Beneficial effect of Lactobacillus plantarum IS-10506 supplementation in adults with atopic dermatitis: A randomized controlled trial. J. Dermatol. Treat. 2022, 33, 1491–1498. [Google Scholar] [CrossRef]
- Le, B.; Yang, S.H. Efficacy of Lactobacillus plantarum in prevention of inflammatory bowel disease. Toxicol. Rep. 2018, 5, 314–317. [Google Scholar] [CrossRef]
- Valdez, J.C.; Peral, M.C.; Rachid, M.; Santana, M.; Perdigon, G. Interference of Lactobacillus plantarum with Pseudomonas aeruginosa in vitro and in infected burns: The potential use of probiotics in wound treatment. Clin. Microbiol. Infect. 2005, 11, 472–479. [Google Scholar] [CrossRef]
- Liu, J.; Gu, Z.; Song, F.; Zhang, H.; Zhao, J.; Chen, W. Lactobacillus plantarum ZS2058 and Lactobacillus rhamnosus GG use different mechanisms to prevent Salmonella infection in vivo. Front. Microbiol. 2019, 10, 299. [Google Scholar] [CrossRef] [PubMed]
- Pourramezan, Z.; Oloomi, M.; Kasra-Kermanshahi, R. Antioxidant and Anticancer Activities of Lactobacillus Hilgardii Strain AG12a. Int. J. Prev. Med. 2020, 11, 132. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.Y.; Kang, H.R.; Cho, S.K. Changes Over the Fermentation Period in Phenolic Compounds and Antioxidant and Anticancer Activities of Blueberries Fermented by Lactobacillus plantarum. J. Food Sci. 2019, 84, 2347–2356. [Google Scholar] [CrossRef]
- Riaz Rajoka, M.S.; Zhao, H.; Lu, Y.; Lian, Z.; Li, N.; Hussain, N.; Shao, D.; Jin, M.; Li, Q.; Shi, J. Anticancer potential against cervix cancer (HeLa) cell line of probiotic Lactobacillus casei and Lactobacillus paracasei strains isolated from human breast milk. Food Funct. 2018, 9, 2705–2715. [Google Scholar] [CrossRef] [PubMed]
- Sheng, S.; Fu, Y.; Pan, N.; Zhang, H.; Xiu, L.; Liang, Y.; Liu, Y.; Liu, B.; Ma, C.; Du, R.; et al. Novel exopolysaccharide derived from probiotic Lactobacillus pantheris TCP102 strain with immune-enhancing and anticancer activities. Front. Microbiol. 2022, 13, 1015270. [Google Scholar] [CrossRef]
- Kamaluddin, W.N.F.W.; Rismayuddin, N.A.R.; Ismail, A.F.; Aidid, E.M.; Othman, N.; Mohamad, N.A.H.; Arzmi, M.H. Probiotic inhibits oral carcinogenesis: A systematic review and meta-analysis. Arch. Oral Biol. 2020, 118, 104855. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, F.; Jiang, L.; Liu, R.; Zhang, L.; Lei, X.; Li, J.; Jiang, J.; Guo, H.; Fang, B.; et al. Lactobacillus salivarius REN inhibits rat oral cancer induced by 4-nitroquioline 1-oxide. Cancer Prev. Res. 2013, 6, 686–694. [Google Scholar] [CrossRef]
- Yan, S.; Qiao, L.; Dou, X.; Song, X.; Chen, Y.; Zhang, B.; Xu, C. Biogenic selenium nanoparticles by Lactobacillus casei ATCC 393 alleviate the intestinal permeability, mitochondrial dysfunction and mitophagy induced by oxidative stress. Food Funct. 2021, 12, 7068–7080. [Google Scholar] [CrossRef]
- Zhang, W.; Zhu, Y.H.; Yang, G.Y.; Liu, X.; Xia, B.; Hu, X.; Su, J.H.; Wang, J.F. Lactobacillus rhamnosus GG Affects Microbiota and Suppresses Autophagy in the Intestines of Pigs Challenged with Salmonella Infantis. Front. Microbiol. 2018, 8, 2705. [Google Scholar] [CrossRef]
- Behzadi, E.; Hosseini, H.M.; Fooladi, A.A.I. The inhibitory impacts of Lactobacillus rhamnosus GG-derived extracellular vesicles on the growth of hepatic cancer cells. Microb. Pathog. 2017, 110, 1–6. [Google Scholar] [CrossRef]
- Zhu, T.; Mao, J.; Zhong, Y.; Huang, C.; Deng, Z.; Cui, Y.; Liu, J.; Wang, H.L. reuteri ZJ617 inhibits inflammatory and autophagy signaling pathways in gut-liver axis in piglet induced by lipopolysaccharide. J. Anim. Sci. Biotechnol. 2021, 12, 110. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Seok, H.; Ha, E.M. GABA-producing Lactobacillus plantarum inhibits metastatic properties and induces apoptosis of 5-FU-resistant colorectal cancer cells via GABAB receptor signaling. J. Microbiol. 2021, 59, 202–216. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Liu, W.; Song, Y.; Tuo, Y.; Mu, G.; Ma, F. The Effects of Lactobacillus plantarum-12 Crude Exopolysaccharides on the Cell Proliferation and Apoptosis of Human Colon Cancer (HT-29) Cells. Probiotics Antimicrob. Proteins 2021, 13, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Hiraishi, N.; Kanmura, S.; Oda, K.; Arima, S.; Kumagai, K.; Mawatari, S.; Tanoue, S.; Sasaki, F.; Hashimoto, S.; Ido, A. Extract of Lactobacillus plantarum strain 06CC2 induces JNK/p38 MAPK pathway-mediated apoptosis through endoplasmic reticulum stress in Caco2 colorectal cancer cells. Biochem. Biophys. Rep. 2019, 20, 100691. [Google Scholar] [CrossRef] [PubMed]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 and Autophagy. Methods Mol. Biol. 2008, 445, 77–88. [Google Scholar]
- Mehrpour, M.; Esclatine, A.; Beau, I.; Codogno, P. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010, 20, 748–762. [Google Scholar] [CrossRef]
- Maeda, S.; Yamamoto, H.; Kinch, L.N.; Garza, C.M.; Takahashi, S.; Otomo, C.; Grishin, N.V.; Forli, S.; Mizushima, N.; Otomo, T. Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat. Struct. Mol. Biol. 2020, 27, 1194–1201. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Q.J.; Li, X.; Yan, Y.; Backer, J.M.; Chait, B.T.; Heintz, N.; Yue, Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009, 11, 468–476. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Chen, Z.; Lu, W.; Garcia-Prieto, C.; Huang, P. The Warburg effect and its cancer therapeutic implications. J. Bioenerg. Biomembr. 2007, 39, 267–274. [Google Scholar] [CrossRef]
- Alfarouk, K.O.; Verduzco, D.; Rauch, C.; Muddathir, A.K.; Adil, H.H.; Elhassan, G.O.; Ibrahim, M.E.; Orozco, J.D.P.; Cardone, R.A.; Reshkin, S.J.; et al. Glycolysis, tumor metabolism, cancer growth and dissemination. A new pH-based etiopathogenic perspective and therapeutic approach to an old cancer question. Oncoscience 2014, 1, 777–802. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Outschoorn, U.E.; Lin, Z.; Ko, Y.H.; Goldberg, A.F.; Flomenberg, N.; Wang, C.; Pavlides, S.; Pestell, R.G.; Howell, A.; Sotgia, F.; et al. Understanding the metabolic basis of drug resistance: Therapeutic induction of the Warburg effect kills cancer cells. Cell Cycle 2011, 10, 2521–2528. [Google Scholar] [CrossRef]
- Xu, R.H.; Pelicano, H.; Zhou, Y.; Carew, J.S.; Feng, L.; Bhalla, K.N.; Keating, M.J.; Huang, P. Inhibition of glycolysis in cancer cells: A novel strategy to overcome drug resistance associated with mitochondrial respiratory defect and hypoxia. Cancer Res. 2005, 65, 613–621. [Google Scholar] [CrossRef]
- Cairns, R.A.; Harris, I.S.; Mak, T.W. Regulation of cancer cell metabolism. Nature reviews. Cancer 2011, 11, 85–95. [Google Scholar]
- Zhao, Y.; Butler, E.B.; Tan, M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013, 4, e532. [Google Scholar] [CrossRef]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef]
- Montagnani, F.; Chiriatti, A.; Turrisi, G.; Francini, G.; Fiorentini, G. A systematic review of FOLFOXIRI chemotherapy for the first-line treatment of metastatic colorectal cancer: Improved efficacy at the cost of increased toxicity. Color. Dis. Off. J. Assoc. Coloproctol. G. B. Irel. 2011, 13, 846–852. [Google Scholar] [CrossRef]
- Papanastasopoulos, P.; Stebbing, J. Molecular basis of 5-fluorouracil-related toxicity: Lessons from clinical practice. Anticancer. Res. 2014, 34, 1531–1535. [Google Scholar]
- An, J.; Ha, E.M. Combination Therapy of Lactobacillus plantarum Supernatant and 5-Fluouracil Increases Chemosensitivity in Colorectal Cancer Cells. J. Microbiol. Biotechnol. 2016, 26, 1490–1503. [Google Scholar] [CrossRef] [PubMed]
- Longley, D.B.; Johnston, P.G. Molecular mechanisms of drug resistance. J. Pathol. 2005, 205, 275–292. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; An, J.; Ha, E.M. Lactobacillus plantarum-derived metabolites sensitize the tumor-suppressive effects of butyrate by regulating the functional expression of SMCT1 in 5-FU-resistant colorectal cancer cells. J. Microbiol. 2022, 60, 100–117. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Ha, E.M. Extracellular vesicles derived from Lactobacillus plantarum restore chemosensitivity through the PDK2-mediated glucose metabolic pathway in 5-FU-resistant colorectal cancer cells. J. Microbiol. 2022, 60, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Hammond, W.A.; Swaika, A.; Mody, K. Pharmacologic resistance in colorectal cancer: A review. Ther. Adv. Med. Oncol. 2016, 8, 57–84. [Google Scholar] [CrossRef]
- Kaliyamoorthy, V.; Jacop, J.P.; Thirugnanasambantham, K.; Ibrahim, H.I.M.; Kandhasamy, S. The synergic impact of lignin and Lactobacillus plantarum on DSS-induced colitis model via regulating CD44 and miR 199a alliance. World J. Microbiol. Biotechnol. 2022, 38, 233. [Google Scholar] [CrossRef]
- Park, M.; Park, E.J.; Kim, S.H.; Lee, H.J. Lactobacillus plantarum ATG-K2 and ATG-K6 Ameliorates High-Fat with High-Fructose Induced Intestinal Inflammation. Int. J. Mol. Sci. 2021, 22, 4444. [Google Scholar] [CrossRef]
- Wu, Y.; Li, A.; Liu, H.; Zhang, Z.; Zhang, C.; Ma, C.; Zhang, L.; Zhang, J. Lactobacillus plantarum HNU082 alleviates dextran sulfate sodium-induced ulcerative colitis in mice through regulating gut microbiome. Food Funct. 2022, 13, 10171–10185. [Google Scholar] [CrossRef]
- Hasannejad-Bibalan, M.; Mojtahedi, A.; Eshaghi, M.; Rohani, M.; Pourshafie, M.R.; Talebi, M. The effect of selected Lactobacillus strains on dextran sulfate sodium-induced mouse colitis model. Acta Microbiol. Et Immunol. Hung. 2020, 67, 138–142. [Google Scholar] [CrossRef]
- Yu, P.; Ke, C.; Guo, J.; Zhang, X.; Li, B. Lactobacillus plantarum L15 Alleviates Colitis by Inhibiting LPS-Mediated NF-κB Activation and Ameliorates DSS-Induced Gut Microbiota Dysbiosis. Front. Immunol. 2020, 11, 575173. [Google Scholar] [CrossRef]
- Prantera, C.; Scribano, M.L.; Falasco, G.; Andreoli, A.; Luzi, C. Ineffectiveness of probiotics in preventing recurrence after curative resection for Crohn’s disease: A randomised controlled trial with Lactobacillus GG. Gut 2002, 51, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Rogler, G. Chronic ulcerative colitis and colorectal cancer. Cancer Lett. 2014, 345, 235–241. [Google Scholar] [CrossRef]
- Venegas, D.P.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Corrigendum: Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 1486. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Meng, L.; Zhang, C.; Zhang, F.; Fang, Y. Corrigendum to “Extracellular vesicles of Lacticaseibacillus paracasei PC-H1 induce colorectal cancer cells apoptosis via PDK1/AKT/Bcl-2 signaling pathway”. Microbiol. Res. 2022, 259, 126955. [Google Scholar] [CrossRef]
- Ganapathy-Kanniappan, S.; Geschwind, J.F. Tumor glycolysis as a target for cancer therapy: Progress and prospects. Mol. Cancer 2013, 12, 1–11. [Google Scholar] [CrossRef]
- Kawaguchi, M.; Aoki, S.; Hirao, T.; Morita, M.; Ito, K. Autophagy is an important metabolic pathway to determine leukemia cell survival following suppression of the glycolytic pathway. Biochem. Biophys. Res. Commun. 2016, 474, 188–192. [Google Scholar] [CrossRef]
- Chu, Y.; Chang, Y.; Lu, W.; Sheng, X.; Wang, S.; Xu, H.; Ma, J. Regulation of Autophagy by Glycolysis in Cancer. Cancer Manag. Res. 2020, 12, 13259. [Google Scholar] [CrossRef]
- Xu, C.; Qiao, L.; Ma, L.; Guo, Y.; Dou, X.; Yan, S.; Zhang, B.; Roman, A. Biogenic selenium nanoparticles synthesized by Lactobacillus casei ATCC 393 alleviate intestinal epithelial barrier dysfunction caused by oxidative stress via Nrf2 signaling-mediated mitochondrial pathway. Int. J. Nanomed. 2019, 14, 4491–4502. [Google Scholar] [CrossRef]
- Levy, J.M.M.; Towers, C.G.; Thorburn, A. Targeting autophagy in cancer. Nature reviews. Cancer 2017, 17, 528–542. [Google Scholar]
- Levy, J.M.M.; Zahedi, S.; Griesinger, A.M.; Morin, A.; Davies, K.D.; Aisner, D.L.; Kleinschmidt-DeMasters, B.K.; Fitzwalter, B.E.; Goodall, M.L.; Thorburn, J.; et al. Autophagy inhibition overcomes multiple mechanisms of resistance to BRAF inhibition in brain tumors. eLife 2017, 6, e19671. [Google Scholar] [CrossRef] [PubMed]
- Goussetis, D.J.; Gounaris, E.; Wu, E.J.; Vakana, E.; Sharma, B.; Bogyo, M.; Altman, J.K.; Platanias, L.C. Autophagic degradation of the BCR-ABL oncoprotein and generation of antileukemic responses by arsenic trioxide. Blood 2012, 120, 3555–3562. [Google Scholar] [CrossRef] [PubMed]
- Kocaturk, N.M.; Akkoc, Y.; Kig, C.; Bayraktar, O.; Gozuacik, D.; Kutlu, O. Autophagy as a molecular target for cancer treatment. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2019, 134, 116–137. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Hui, K.; Hu, C.; Wen, Y.; Yang, S.; Zhu, P.; Wang, L.; Xia, Y.; Qiao, Y.; Sun, W.; et al. Autophagy inhibition potentiates the anti-angiogenic property of multikinase inhibitor anlotinib through JAK2/STAT3/VEGFA signaling in non-small cell lung cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 71. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Tozzi, F.; Chen, J.; Fan, F.; Xia, L.; Wang, J.; Gao, G.; Zhang, A.; Xia, X.; Brasher, H.; et al. Intracellular ATP levels are a pivotal determinant of chemoresistance in colon cancer cells. Cancer Res. 2012, 72, 304–314. [Google Scholar] [CrossRef]
- Bhattacharya, B.; Low, S.H.; Soh, C.; Mustapa, N.K.; Beloueche-Babari, M.; Koh, K.X.; Loh, J.; Soong, R. Increased drug resistance is associated with reduced glucose levels and an enhanced glycolysis phenotype. Br. J. Pharmacol. 2014, 171, 3255–3267. [Google Scholar] [CrossRef]
- Pellegrini, P.; Strambi, A.; Zipoli, C.; Hägg-Olofsson, M.; Buoncervello, M.; Linder, S.; De Milito, A. Acidic extracellular pH neutralizes the autophagy-inhibiting activity of chloroquine: Implications for cancer therapies. Autophagy 2014, 10, 562–571. [Google Scholar] [CrossRef]
- Thévenod, F.; Lee, W.K. Live and Let Die: Roles of Autophagy in Cadmium Nephrotoxicity. Toxics 2015, 3, 130–151. [Google Scholar] [CrossRef]
- Qu, X.; Sheng, J.; Shen, L.; Su, J.; Xu, Y.; Xie, Q.; Wu, Y.; Zhang, X.; Sun, L. Autophagy inhibitor chloroquine increases sensitivity to cisplatin in QBC939 cholangiocarcinoma cells by mitochondrial ROS. PLoS ONE 2017, 12, e0173712. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, S.; Kim, Y.; Park, S.; Lee, D.; Lee, J.; Hlaing, S.P.; Yoo, J.-W.; Rhee, S.H.; Im, E. Lactobacillus plantarum Metabolites Elicit Anticancer Effects by Inhibiting Autophagy-Related Responses. Molecules 2023, 28, 1890. https://doi.org/10.3390/molecules28041890
Jeong S, Kim Y, Park S, Lee D, Lee J, Hlaing SP, Yoo J-W, Rhee SH, Im E. Lactobacillus plantarum Metabolites Elicit Anticancer Effects by Inhibiting Autophagy-Related Responses. Molecules. 2023; 28(4):1890. https://doi.org/10.3390/molecules28041890
Chicago/Turabian StyleJeong, Sihyun, Yuju Kim, Soyeong Park, Doyeon Lee, Juho Lee, Shwe Phyu Hlaing, Jin-Wook Yoo, Sang Hoon Rhee, and Eunok Im. 2023. "Lactobacillus plantarum Metabolites Elicit Anticancer Effects by Inhibiting Autophagy-Related Responses" Molecules 28, no. 4: 1890. https://doi.org/10.3390/molecules28041890
APA StyleJeong, S., Kim, Y., Park, S., Lee, D., Lee, J., Hlaing, S. P., Yoo, J.-W., Rhee, S. H., & Im, E. (2023). Lactobacillus plantarum Metabolites Elicit Anticancer Effects by Inhibiting Autophagy-Related Responses. Molecules, 28(4), 1890. https://doi.org/10.3390/molecules28041890






