NMR-Based Metabolomics: A New Paradigm to Unravel Defense-Related Metabolites in Insect-Resistant Cotton Variety through Different Multivariate Data Analysis Approaches
Abstract
1. Introduction
2. Results
2.1. 1H-NMR Identification of Metabolites in Cotton Varieties
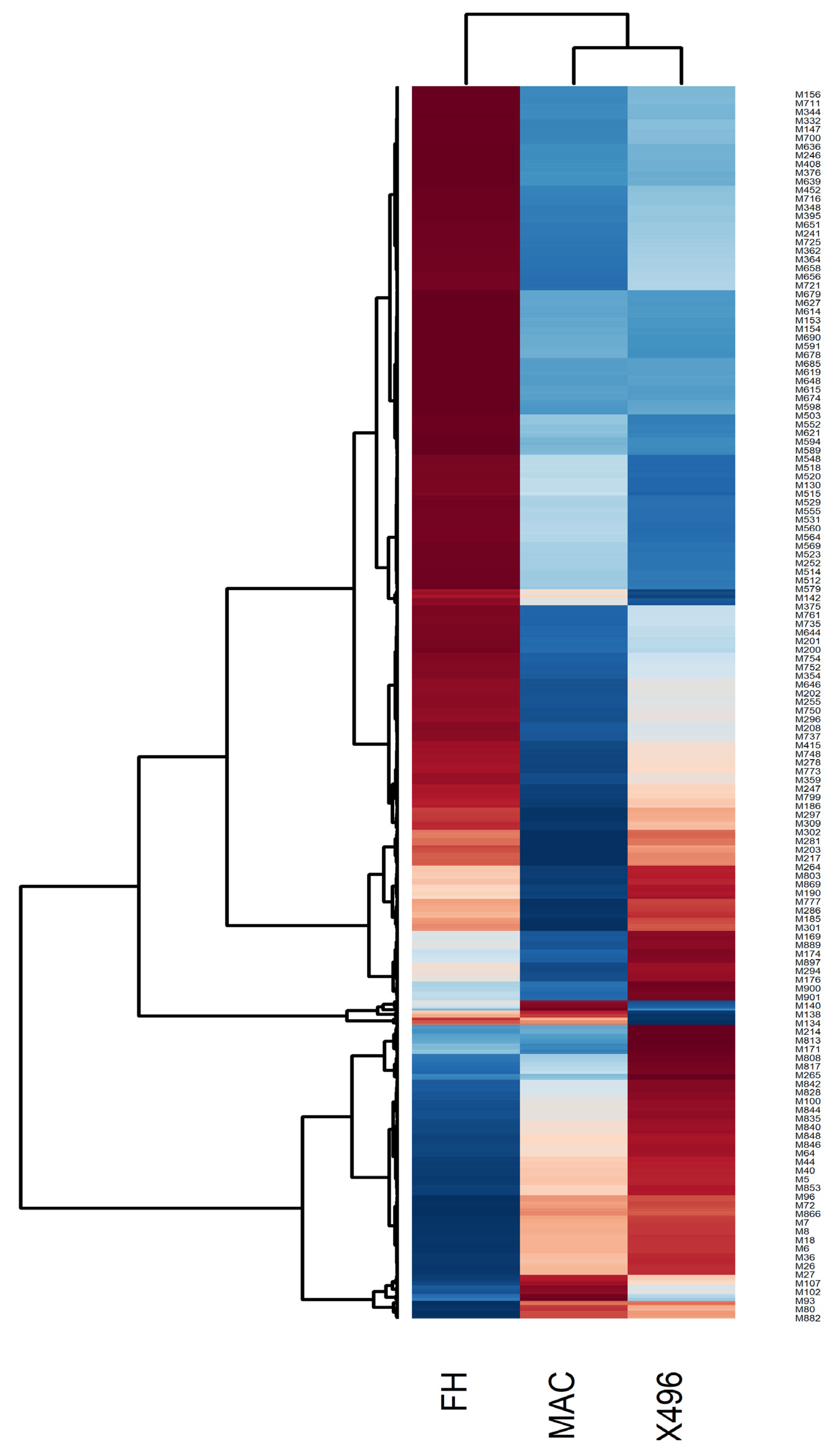
2.2. Heat map Analysis
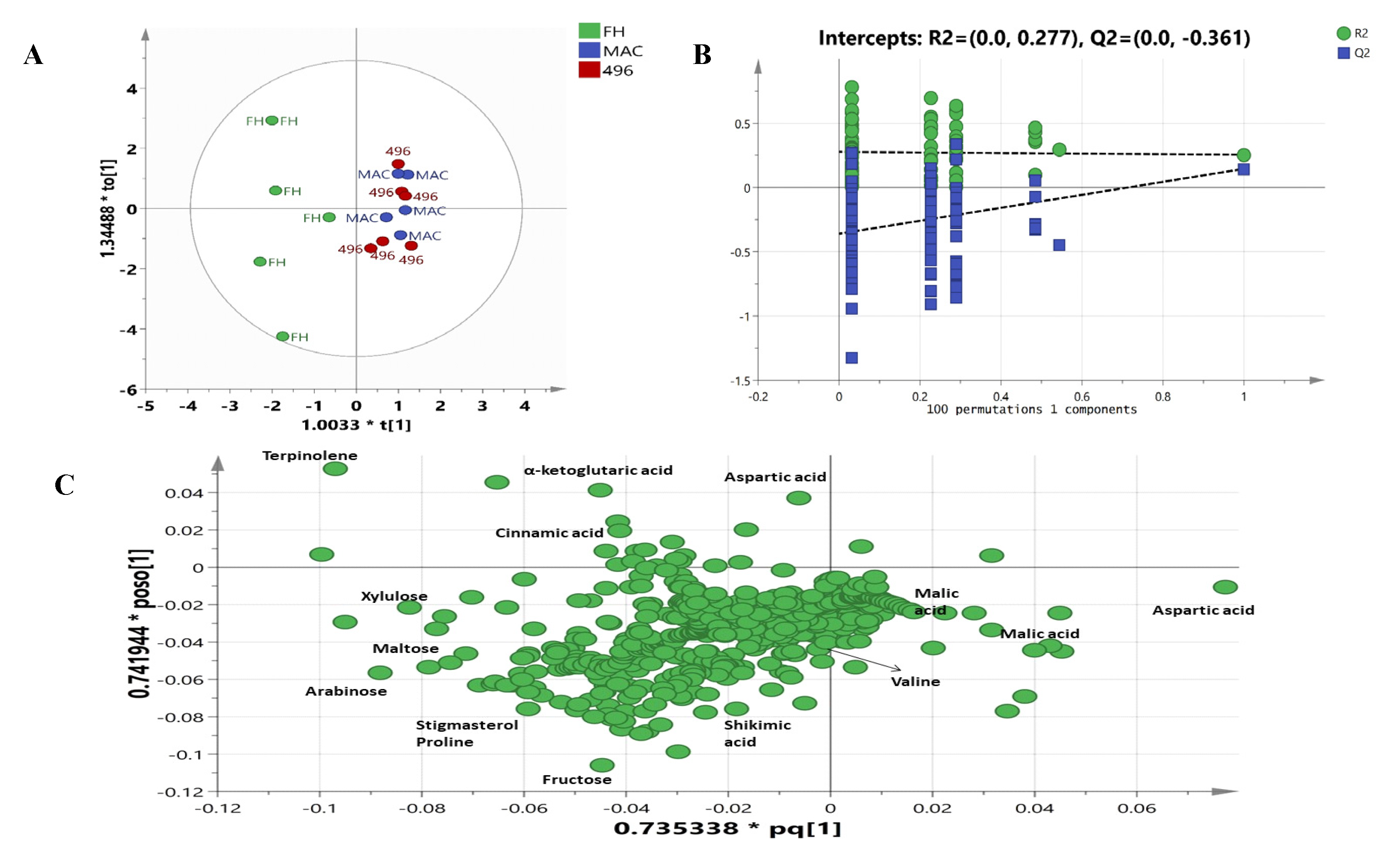
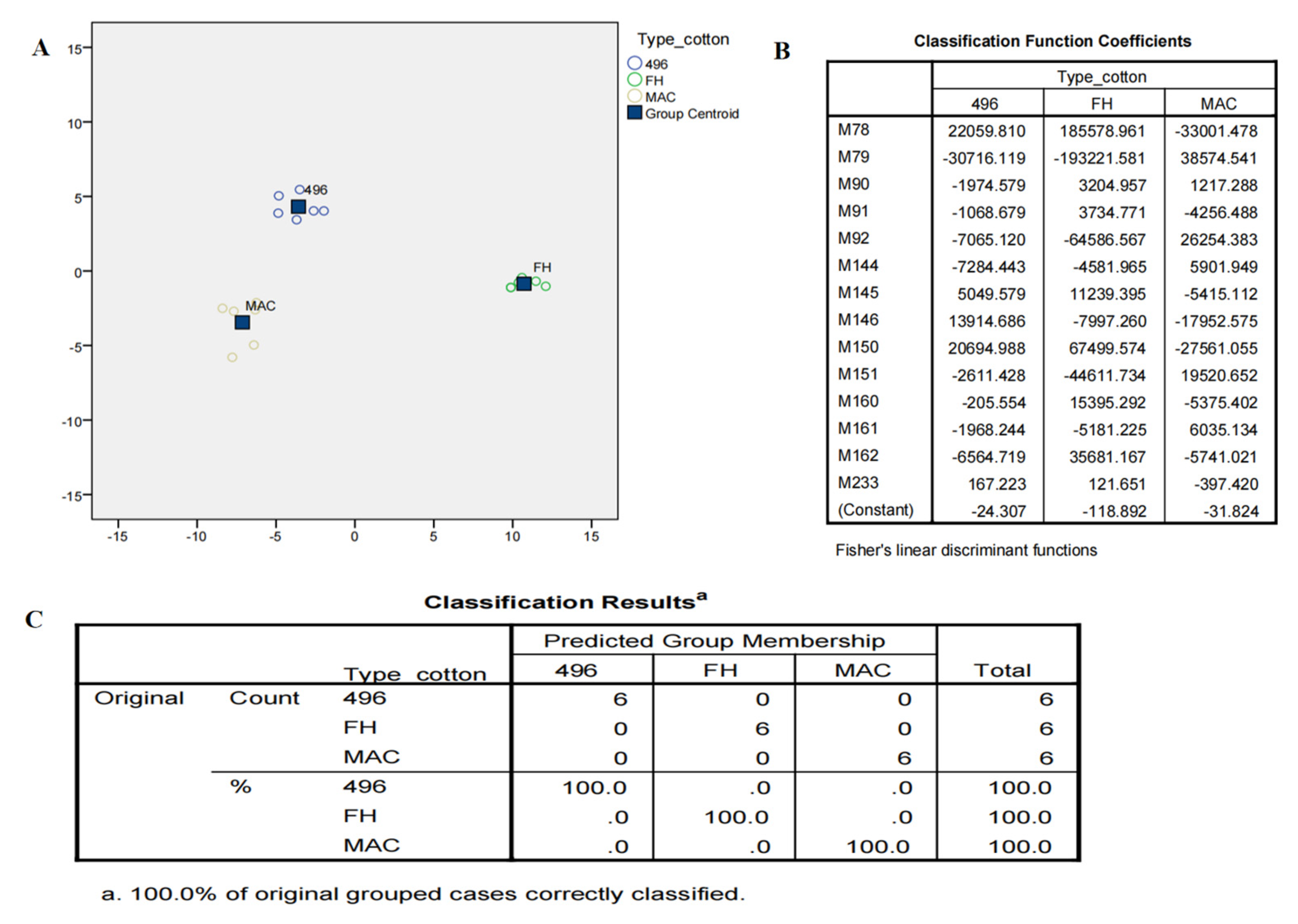
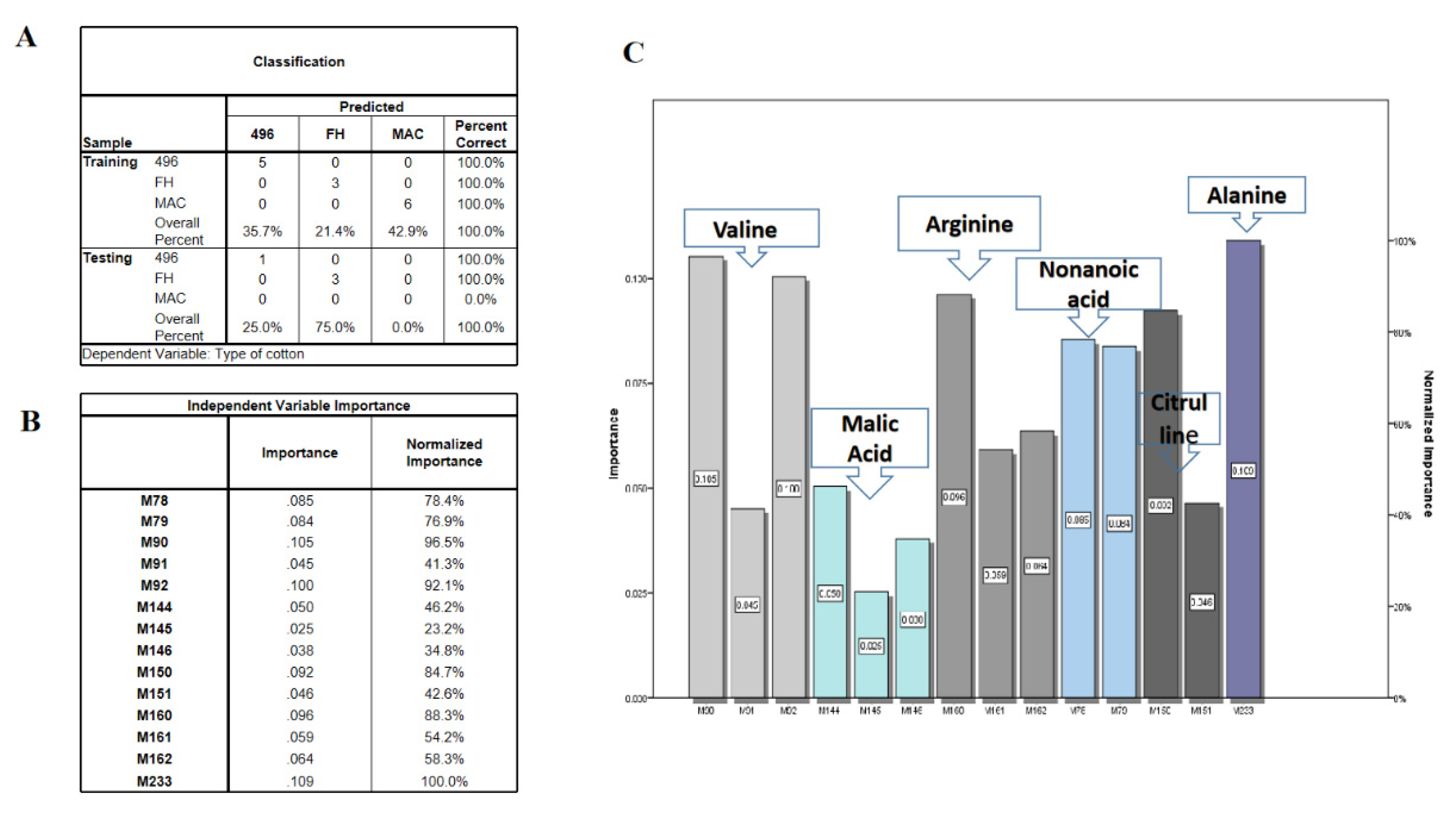
2.3. Multivariate Data Analysis
2.4. Metabolic Pathway Analysis
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Sample Collection
4.3. NMR Sample Preparation
4.4. NMR Acquisition
4.5. Data Processing
4.6. Multivariate Data Analysis (MvDA)
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Malik, C.P.; Sanadhya, D. Genetic Engineering and GM Crops. J. Plant Sci. Res. 2018, 34, 251–258. [Google Scholar] [CrossRef]
- Julia, B.S.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar]
- Nix, A.; Paull, C.C.; Colgrave, M. Flavonoid profile of the cotton plant, Gossypium hirsutum: A review. Plants 2017, 6, 43. [Google Scholar] [CrossRef]
- Ashraf, S.; Sangi, A.H.; Hassan, Z.Y.; Luqman, M. Future of cotton sector in Pakistan: A 2025 Outlook. Pak. J. Agric. Sci. 2018, 31, 145. [Google Scholar] [CrossRef]
- Nehra, V.; Saharan, B.S.; Choudhary, M. Evaluation of Brevibacillus brevis as a potential plant growth promoting rhizobacteria for cotton (Gossypium hirsutum) crop. SpringerPlus 2016, 5, 948. [Google Scholar] [CrossRef]
- Egbuta, M.A.; McIntosh, S.; Waters, D.L.E.; Vancov, T.; Liu, L. Biological importance of cotton by-products relative to chemical constituents of the cotton plant. Molecules 2017, 22, 93. [Google Scholar] [CrossRef] [PubMed]
- Tokel, D.; Genc, B.N.; Ozyigit, I.J. Economic impacts of Bt (Bacillus thuringiensis) cotton. J. Nat. Fibers 2021, 19, 4622–4639. [Google Scholar] [CrossRef]
- Bravo, A.; Likitvivatanavong, S.; Gill, S.S.; Soberón, M. Bacillus thuringiensis: A story of a successful bioinsecticide. Insect Biochem. Mol. Biol. 2011, 41, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Bilal, M.F.; Saleem, M.F.; Wahid, M.A.; Shakeel, A.; Maqbool, M. Adoption of Bt cotton: Threats and challenges. Chil. J. Agric. Res. 2012, 72, 419. [Google Scholar] [CrossRef]
- Pichersky, E.; Gang, D.R. Genetics and biochemistry of secondary metabolites in plants: An evolutionary perspective. Trends Plant Sci. 2000, 5, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Deborde, C.; Moing, A.; Roch, L.; Jacob, D.; Rolin, D.; Giraudeau, P. Plant metabolism as studied by NMR spectroscopy. Prog. Nucl. Mag. Res. Sp. 2017, 102, 61–97. [Google Scholar] [CrossRef] [PubMed]
- Bloem, E.; Haneklaus, S.; Schnug, E. Significance of Sulfur Compounds in the Protection of Plants against Pests and Diseases. J. Plant Nutr. 2005, 28, 763–784. [Google Scholar] [CrossRef]
- Mazid, M.; Khan, T.A.; Mohammad, F. Role of secondary metabolites in defense mechanisms of plants. Biol. Med. 2011, 3, 232–249. [Google Scholar]
- Divekar, P.A.; Narayana, S.; Divekar, B.A.; Kumar, R.; Gadratagi, B.G.; Ray, A.; Singh, A.K.; Rani, V.; Singh, V.; Singh, A.K.; et al. Plant secondary metabolites as defense tools against herbivores for sustainable crop protection. Int. J. Mol. Sci. 2022, 23, 2690. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Chikayama, E.; Tsuboi, Y.; Kuromori, T.; Shinozaki, K.; Kikuchi, J.; Hirayama, T. Top-down phenomics of Arabidopsis thaliana metabolic profiling by one-and two-dimensional nuclear magnetic resonance spectroscopy and transcriptome analysis of albino mutants. J. Biol. Chem. 2007, 282, 18532–18541. [Google Scholar] [CrossRef]
- Hong, J.; Yang, L.; Zhang, D.; Shi, J. Plant metabolomics: An indispensable system biology tool for plant science. Int. J. Mol. Sci. 2016, 17, 767. [Google Scholar] [CrossRef] [PubMed]
- Hussain, R.M.F.; Kim, H.K.; Khurshid, M.; Akhtar, M.T.; Linthorst, H.J. Overexpression of AtWRKY50 is correlated with enhanced production of sinapic derivatives in Arabidopsis. Metabolomics 2018, 14, 25. [Google Scholar] [CrossRef]
- Brennan, L. NMR-based metabolomics: From sample preparation to applications in nutrition research. Prog. Nucl. Magn. Reson. Spectrosc. 2014, 83, 42–49. [Google Scholar] [CrossRef]
- Moco, S. Studying metabolism by NMR-based metabolomics. Front. Mol. Biosci. 2022, 372, 882487. [Google Scholar] [CrossRef]
- David, A.; Rostkowski, P. Analytical techniques in metabolomics. In Environmental Metabolomics; Elsevier: Amsterdam, The Netherlands, 2020; pp. 35–64. [Google Scholar]
- Antonio, J.G.M.; Brasil, J.M.; Cruz, G.C.F.; de Souza, R.N.; Tasic, L. NMR-based metabolomics strategies: Plants, animals and humans. Anal. Methods 2017, 9, 1078–1096. [Google Scholar]
- Emwas, A.H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR spectroscopy for metabolomics research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Wang, T.; Peng, Y.; Xia, B.; Qu, L.J. Distinguishing transgenic from non-transgenic Arabidopsis plants by 1H NMR-based metabolic fingerprinting. JGG 2009, 36, 621–628. [Google Scholar] [CrossRef]
- Leiss, K.A.; Choi, Y.H.; Abdel-Farid, I.B.; Verpoorte, R.; Klinkhamer, P.G.L. NMR metabolomics of thrips (Frankliniella occidentalis) resistance in Senecio hybrids. J. Chem. Ecol. 2009, 35, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Becerra-Martinez, E.; Hernandez, Y.P.; Gloria, E.L.; Jimenez, M.G.B.; Martinez, D.H.; Vallejo, L.G.Z.; Ruano, N.V. 1H-NMR metabolomics profiling of recombinant tobacco plants holding a promoter of a sesquiterpene cyclase. Phytochem. Anal. 2020, 31, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Bisht, H.; Bhatnagar, M.; Bisht, S.; Murthy, A.K. Study on variation of polar metabolites in control and water stressed Gossypium hirsutum L. using NMR spectroscopy. Asian J. Appl. Chem. 2018, 1, 1–12. [Google Scholar] [CrossRef]
- Yang, Q.; Zhao, D.; Liu, Q. Connections between Amino Acid Metabolisms in Plants: Lysine as an Example. Front. Plant Sci. 2020, 11, 928. [Google Scholar] [CrossRef]
- Taylor, R.E.; French, A.D.; Gamble, G.R.; Himmelsbach, D.S.; Stipanovic, R.D.; Thibodeaux, D.P.; Wakelyn, P.J.; Dybowski, C. 1H and 13C solid-state NMR of Gossypium barbadense (Pima) cotton. J. Mol. Struct. 2008, 878, 177–184. [Google Scholar] [CrossRef]
- NMRFAM. Available online: http://mmcd.nmrfam.wisc.edu/ (accessed on 10 June 2020).
- HMDB. Available online: https://hmdb.ca/ (accessed on 31 November 2019).
- Boonchaisri, S.; Rochfort, S.; Stevenson, T.; Dias, D.A. Recent developments in metabolomics-based research in understanding transgenic grass metabolism. Metabolomics 2019, 15, 47. [Google Scholar] [CrossRef] [PubMed]
- Bastien, C.; Pluskal, T.; Aubry, S.; Weng, J.K. Contribution of untargeted metabolomics for future assessment of biotech crops. Trends Plant Sci. 2018, 23, 1047–1056. [Google Scholar]
- Levandi, T.; Leon, C.; Kaljurand, M.; Garcia-Cañas, V.; Cifuentes, A. Capillary electrophoresis time-of-flight mass spectrometry for comparative metabolomics of transgenic versus conventional maize. Anal. Chem. 2008, 80, 6329–6335. [Google Scholar] [CrossRef]
- Ning, K.; Ding, C.; Zhu, W.; Dong, Y.; Shen, Y.; Su, X. Comparative metabolomic analysis of the cambium tissue of non-transgenic and multi-gene transgenic poplar (Populus × euramericana ‘Guariento’). Front. Plant Sci. 2018, 9, 1201. [Google Scholar] [CrossRef] [PubMed]
- Schenck, C.A.; Maeda, H.A. Tyrosine biosynthesis, metabolism, and catabolism in plants. Phytochemistry 2018, 149, 82–102. [Google Scholar] [CrossRef]
- Zemanová, V.; Pavlík, M.; Pavlíková, D.; Tlustoš, P. The significance of methionine, histidine and tryptophan in plant responses and adaptation to cadmium stress. Plant Soil Environ. 2014, 60, 426–432. [Google Scholar] [CrossRef]
- Mikkelsen, M.D.; Halkier, B.A. Metabolic engineering of valine-and isoleucine-derived glucosinolates in Arabidopsis expressing CYP79D2 from cassava. Plant Physiol. 2003, 131, 773–779. [Google Scholar] [CrossRef]
- Tambel, L.I.M.; Zhou, M.; Chen, Y.; Zhang, X.; Chen, Y.; Chen, D. Amino acids application enhances flowers insecticidal protein content in Bt cotton. J. Cotton Sci. 2019, 2, 7. [Google Scholar] [CrossRef]
- Ramos-Ruiz, R.; Martinez, F.; Knauf-Beiter, G. The effects of GABA in plants. Cogent Food Agric. 2019, 5, 1670553. [Google Scholar] [CrossRef]
- Shelp, B.J.; Aghdam, M.S.; Flaherty, E.J. γ-Aminobutyrate (GABA) regulated plant defense: Mechanisms and opportunities. Plants 2021, 10, 1939. [Google Scholar] [CrossRef]
- Scholz, S.S.; Reichelt, M.; Mekonnen, D.W.; Ludewig, F.; Mithöfer, A. Insect herbivory-elicited GABA accumulation in plants is a wound-induced, direct, systemic, and jasmonate-independent defense response. Front. Plant Sci. 2015, 6, 1128. [Google Scholar] [CrossRef]
- Bown, A.W.; Hall, D.E.; MacGregor, K.B. Insect footsteps on leaves stimulate the accumulation of 4-aminobutyrate and can be visualized through increased chlorophyll fluorescence and superoxide production. Plant Physiol. 2002, 129, 1430–1434. [Google Scholar] [CrossRef]
- Trouvelot, S.; Héloir, M.C.; Poinssot, B.; Gauthier, A.; Paris, F.; Guillier, C.; Combier, M.; Trdá, L.; Daire, X.; Adrian, M. Carbohydrates in plant immunity and plant protection: Roles and potential application as foliar sprays. Front. Plant Sci. 2014, 5, 592. [Google Scholar] [CrossRef]
- Krasavina, M.S.; Burmistrova, N.A.; Raldugina, G.N. The role of carbohydrates in plant resistance to abiotic stresses. In Emerging Technologies and Management of Crop Stress Tolerance; Academic Press: Cambridge, MA, USA, 2014; pp. 229–270. [Google Scholar]
- Rautengarten, C.; Birdseye, D.; Pattathil, S.; McFarlane, H.E.; Saez-Aguayo, S.; Orellana, A.; Persson, S.; Hahn, M.G.; Scheller, H.V.; Heazlewood, J.L.; et al. The elaborate route for UDP-arabinose delivery into the Golgi of plants. Proc. Natl. Acad. Sci. USA 2017, 114, 4261–4266. [Google Scholar] [CrossRef] [PubMed]
- Zayed, O.M. The Role of Plant Cell Wall Arabinose in Salt Stress Sensing and Adaptation. Ph.D. Thesis, Purdue University Graduate School, West Lafayette, IN, USA, 2019. [Google Scholar]
- Araujo, W.L.; Nesi, A.N.; Fernie, A.R. Fumarate: Multiple functions of a simple metabolite. Phytochemistry 2011, 72, 838–843. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, L.; Chang, Y.; Lu, X.; Zhu, Z.; Xu, G. Alteration of leaf metabolism in Bt-transgenic rice (Oryza sativa L.) and its wild type under insecticide stress. J. Proteome Res. 2012, 11, 4351–4360. [Google Scholar] [CrossRef] [PubMed]
- Fahnenstich, H.; Saigo, M.; Andreo, C.; Drincovich, M.F.; Flügge, U.; Maurino, V.G. Malate and Fumarate Emerge as Key Players in Primary Metabolism: Arabidopsis thaliana Overexpressing C 4-NADP-ME Offer a Way to Manipulate the Levels of Malate and to Analyse the Physiological Consequences. In Photosynthesis: Energy from the Sun; Springer: Dordrecht, The Netherlands, 2008; pp. 971–975. [Google Scholar]
- Dumont, S.; Rivoal, J. Consequences of oxidative stress on plant glycolytic and respiratory metabolism. Front. Plant Sci. 2019, 10, 166. [Google Scholar] [CrossRef] [PubMed]
- Sies, H.; Berndt, C.; Jones, D.P. Oxidative stress. Annu. Rev. Biochem. 2017, 86, 715–748. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Shi, L.X.; Yang, C.W.; Yan, C.R.; Zhong, X.L.; Liu, Q.; Xia, X.; Li, H.R. Comparison of ionomic and metabolites response under alkali stress in old and young leaves of cotton (Gossypium hirsutum L.) seedlings. Front. Plant Sci. 2016, 7, 1785. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A.; Achnine, L.; Kota, P.; Liu, C.J.; Reddy, M.S.; Wang, L.J. The phenylpropanoid pathway and plant defence—A genomics perspective. Mol. Plant Pathol. 2002, 3, 371–390. [Google Scholar] [CrossRef] [PubMed]
- Farré-Armengol, G.; Filella, I.; Llusià, J.; Peñuelas, J. β-Ocimene, a key floral and foliar volatile involved in multiple interactions between plants and other organisms. Molecules 2017, 22, 1148. [Google Scholar] [CrossRef]
- Navia-Giné, W.G.; Yuan, J.S.; Mauromoustakos, A.; Murphy, J.B.; Chen, F.; Korth, K.L. Medicago truncatula (E)-β-ocimene synthase is induced by insect herbivory with corresponding increases in emission of volatile ocimene. Plant Physiol. Biochem. 2009, 47, 416–425. [Google Scholar] [CrossRef]
- Kang, Z.W.; Liu, F.H.; Zhang, Z.F.; Tian, H.G.; Liu, T.X. Volatile β-ocimene can regulate developmental performance of peach aphid Myzus persicae through activation of defense responses in Chinese cabbage Brassica pekinensis. Front. Plant Sci. 2018, 9, 708. [Google Scholar] [CrossRef]
- Park, S.H.; Scheffler, J.; Scheffler, B.; Cantrell, C.L.; Pauli, C.S. Chemical defense responses of upland cotton, Gossypium hirsutum L. to physical wounding. Plant Direc. 2019, 3, e00141. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, R.; Narasimhan, K.; Swarup, S. Metabolomics and its role in understanding cellular responses in plants. Plant Cell Rep. 2005, 24, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Afendi, F.M.; Altaf-Ul-Amin, M.; Takahashi, H.; Nakamura, K.; Kanaya, S. Metabolomics of medicinal plants: The importance of multivariate analysis of analytical chemistry data. Curr. Comput. Aided Drug Des. 2010, 6, 179–196. [Google Scholar] [CrossRef]
- Møller, S.F.; von Frese, J.; Bro, R. Robust methods for multivariate data analysis. J. Chemom. 2005, 19, 549–563. [Google Scholar] [CrossRef]
- Choi, H.K.; Choi, Y.H.; Verberne, M.; Lefeber, A.W.M.; Erkelens, C.; Verpoorte, R. Metabolic fingerprinting of wild type and transgenic tobacco plants by 1H NMR and multivariate analysis technique. Phytochemistry 2004, 65, 857–864. [Google Scholar] [CrossRef]
- Johnson, H.E.; Lloyd, A.J.; Mur, L.A.J.; Smith, A.R.; Causton, D.R. The application of MANOVA to analyse Arabidopsis thaliana metabolomic data from factorially designed experiments. Metabolomics 2007, 3, 517–530. [Google Scholar] [CrossRef]
- Tharwat, A.; Gaber, T.; Ibrahim, A.; Hassanien, A.E. Linear discriminant analysis: A detailed tutorial. AI Commun. 2017, 30, 169–190. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments, Philosophical Transactions of the Royal Society A: Mathematical. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar]
- Balakrishnama, S.; Ganapathiraju, A.; Picone, J. Linear discriminant analysis for signal processing problems. In Proceedings of the IEEE Southeastcon’99, Technology on the Brink of 2000 (Cat. No. 99CH36300), Lexington, KY, USA, 25–28 March 1999; pp. 78–81. [Google Scholar]
- Alves, M.V.S.; Maciel, L.I.L.; Ramalho, R.R.F.; Lima, L.A.S.; Vaz, B.G.; Morais, C.L.M.; Passos, J.O.S.; Pegado, R.; Lima, K.M.G. Multivariate classification techniques and mass spectrometry as a tool in the screening of patients with fibromyalgia. Sci. Rep. 2021, 11, 22625. [Google Scholar] [CrossRef]
- Mahadevan, S.; Shah, S.L.; Marrie, T.J.; Slupsky, C.M. Analysis of metabolomic data using supporting vector machines. Anal. Chem. 2008, 80, 7562–7570. [Google Scholar] [CrossRef]
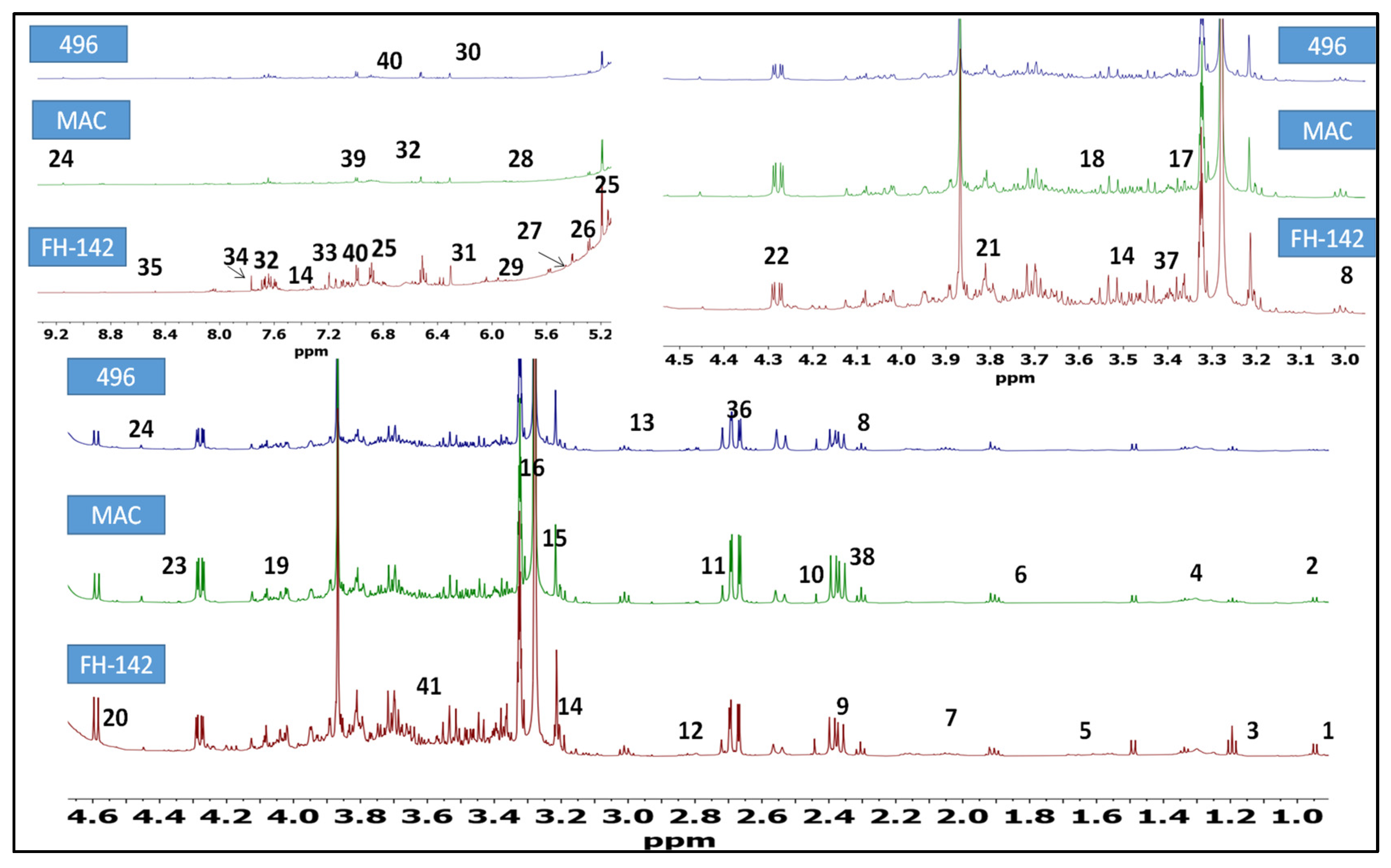


| Sr. No. | Metabolites | Chemical Shift (ppm) | Multiplicity | Assignment Methods |
|---|---|---|---|---|
| 1 | Nonanoic acid | 0.82 | Doublet | JRES/1D-HNMR |
| 2 | Valine | 0.95 | Doublet | JRES/1D-HNMR |
| 3 | Alanine | 1.48 | Doublet | JRES/1D-HNMR |
| 4 | Citrulline | 1.56 | Multiplet | JRES/1D-HNMR |
| 5 | Arginine/myristic acid | 1.68 | Multiplet | JRES/1D-HNMR |
| 6 | Limonene | 1.91 | Multiplet | JRES/1D-HNMR |
| 7 | Linoleic acid | 2.06 | Multiplet | JRES/1D-HNMR |
| 8 | γ-aminobutyric acid | 2.30 3.01 | triplet triplet | JRES/1D-HNMR |
| 9 | Malic acid | 2.39 | doublet of doublet | JRES/1D-HNMR |
| 10 | Succinic acid | 2.45 | Singlet | JRES/1D-HNMR |
| 11 | Terpinolene | 2.71 | Singlet | JRES/1D-HNMR |
| 12 | di-allylic methylene | 2.80 | Multiplet | JRES/1D-HNMR |
| 13 | Asparagine | 2.94 | Multiplet | JRES/1D-HNMR |
| 14 | Tryptophan | 3.15 | singlet | JRES/1D-HNMR |
| 3.50 | doublet | |||
| 7.53 | doublet | |||
| 15 | Choline | 3.20 | Singlet | JRES/1D-HNMR |
| 16 | Scyloinositol | 3.21 | Singlet | JRES/1D-HNMR |
| 17 | Proline | 3.40 | triplet of doublet | JRES/1D-HNMR |
| 18 | Glycine | 3.54 | Singlet | JRES/1D-HNMR |
| 19 | Shikimic acid | 4.01 | Multiplet | JRES/1D-HNMR |
| 20 | Arabinose | 3.82 4.58 | doublet of doublet doublet | JRES/1D-HNMR |
| 21 | Fructose | 3.85 | Doublet | JRES/1D-HNMR |
| 22 | Xylulose | 4.28 | doublet of doublet | JRES/1D-HNMR |
| 23 | Tartaric acid | 4.35 | Singlet | JRES/1D-HNMR |
| 24 | Trigonelline | 4.45 | singlet | JRES/1D-HNMR |
| 9.14 | singlet | |||
| 25 | E-β-ocimene | 5.12 | singlet | JRES/1D-HNMR |
| 6.79 | doublet of doublet | |||
| 26 | Stigmasterol | 5.15 | doublet of doublet | JRES/1D-HNMR |
| 27 | Maltose | 5.20 | Doublet | JRES/1D-HNMR |
| 28 | Sucrose | 5.45 | Doublet | JRES/1D-HNMR |
| 29 | Uridine | 5.95 | Doublet | JRES/1D-HNMR |
| 30 | Maleic acid | 6.05 | Singlet | JRES/1D-HNMR |
| 31 | Fumarate | 6.50 | Singlet | JRES/1D-HNMR |
| 32 | Cinnamic acid | 6.52 | doublet | JRES/1D-HNMR |
| 7.60 | multiplet | |||
| 33 | Tyrosine | 7.20 | Doublet | JRES/1D-HNMR |
| 34 | Dibutyl phthalate | 7.77 | Singlet | JRES/1D-HNMR |
| 35 | Formate | 8.47 | Singlet | JRES/1D-HNMR |
| 36 | Aspartic acid | 2.68 | Doublet of doublet | JRES/1D-HNMR |
| 37 | Aconitic acid | 3.43 | Doublet | JRES/1D-HNMR |
| 38 | α-Ketoglutaric acid | 2.43 | triplet | JRES/1D-HNMR |
| 39 | Chlorogenic acid | 7.12 | Doublet of doublet | JRES/1D-HNMR |
| 40 | Ferulic acid | 7.07 6.91 | Doublet of doublet Doublet | JRES/1D-HNMR |
| 41 | Quinic acid | 3.56 | Doublet of doublet | JRES/1D-HNMR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shami, A.A.; Akhtar, M.T.; Mumtaz, M.W.; Mukhtar, H.; Tahir, A.; Shahzad-ul-Hussan, S.; Chaudhary, S.U.; Muneer, B.; Iftikhar, H.; Neophytou, M. NMR-Based Metabolomics: A New Paradigm to Unravel Defense-Related Metabolites in Insect-Resistant Cotton Variety through Different Multivariate Data Analysis Approaches. Molecules 2023, 28, 1763. https://doi.org/10.3390/molecules28041763
Shami AA, Akhtar MT, Mumtaz MW, Mukhtar H, Tahir A, Shahzad-ul-Hussan S, Chaudhary SU, Muneer B, Iftikhar H, Neophytou M. NMR-Based Metabolomics: A New Paradigm to Unravel Defense-Related Metabolites in Insect-Resistant Cotton Variety through Different Multivariate Data Analysis Approaches. Molecules. 2023; 28(4):1763. https://doi.org/10.3390/molecules28041763
Chicago/Turabian StyleShami, Anam Amin, Muhammad Tayyab Akhtar, Muhammad Waseem Mumtaz, Hamid Mukhtar, Amna Tahir, Syed Shahzad-ul-Hussan, Safee Ullah Chaudhary, Bushra Muneer, Hafsa Iftikhar, and Marios Neophytou. 2023. "NMR-Based Metabolomics: A New Paradigm to Unravel Defense-Related Metabolites in Insect-Resistant Cotton Variety through Different Multivariate Data Analysis Approaches" Molecules 28, no. 4: 1763. https://doi.org/10.3390/molecules28041763
APA StyleShami, A. A., Akhtar, M. T., Mumtaz, M. W., Mukhtar, H., Tahir, A., Shahzad-ul-Hussan, S., Chaudhary, S. U., Muneer, B., Iftikhar, H., & Neophytou, M. (2023). NMR-Based Metabolomics: A New Paradigm to Unravel Defense-Related Metabolites in Insect-Resistant Cotton Variety through Different Multivariate Data Analysis Approaches. Molecules, 28(4), 1763. https://doi.org/10.3390/molecules28041763








