Using HPLC with In-Column Derivatization to Authenticate Coffee Samples
Abstract
:1. Introduction
2. Experimental Section
2.1. Chemicals and Coffee Capsules
2.2. Preparation of Samples and Reagents
2.3. Instrumentation and Chromatographic Columns
2.4. Separation Conditions
3. Results and Discussions
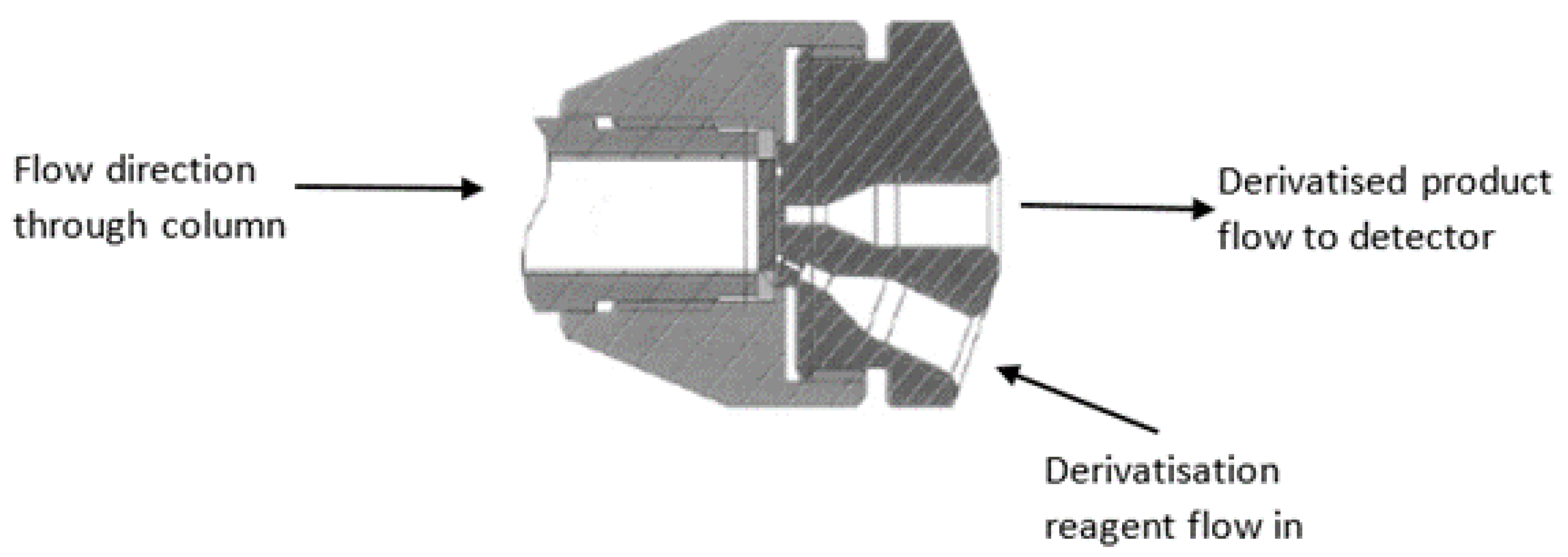
3.1. In-Column Derivatization
3.2. Coffee Samples
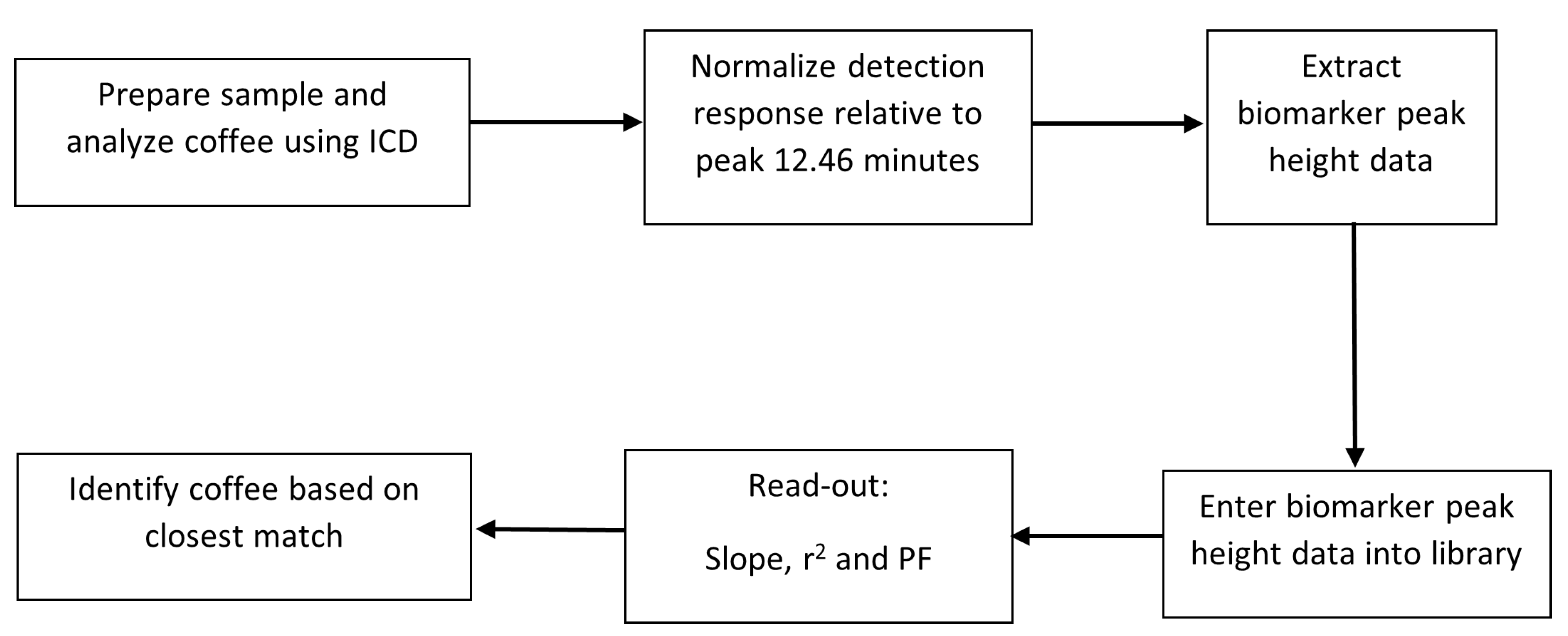
3.3. Coffee Assays, Data Treatment, and Analysis
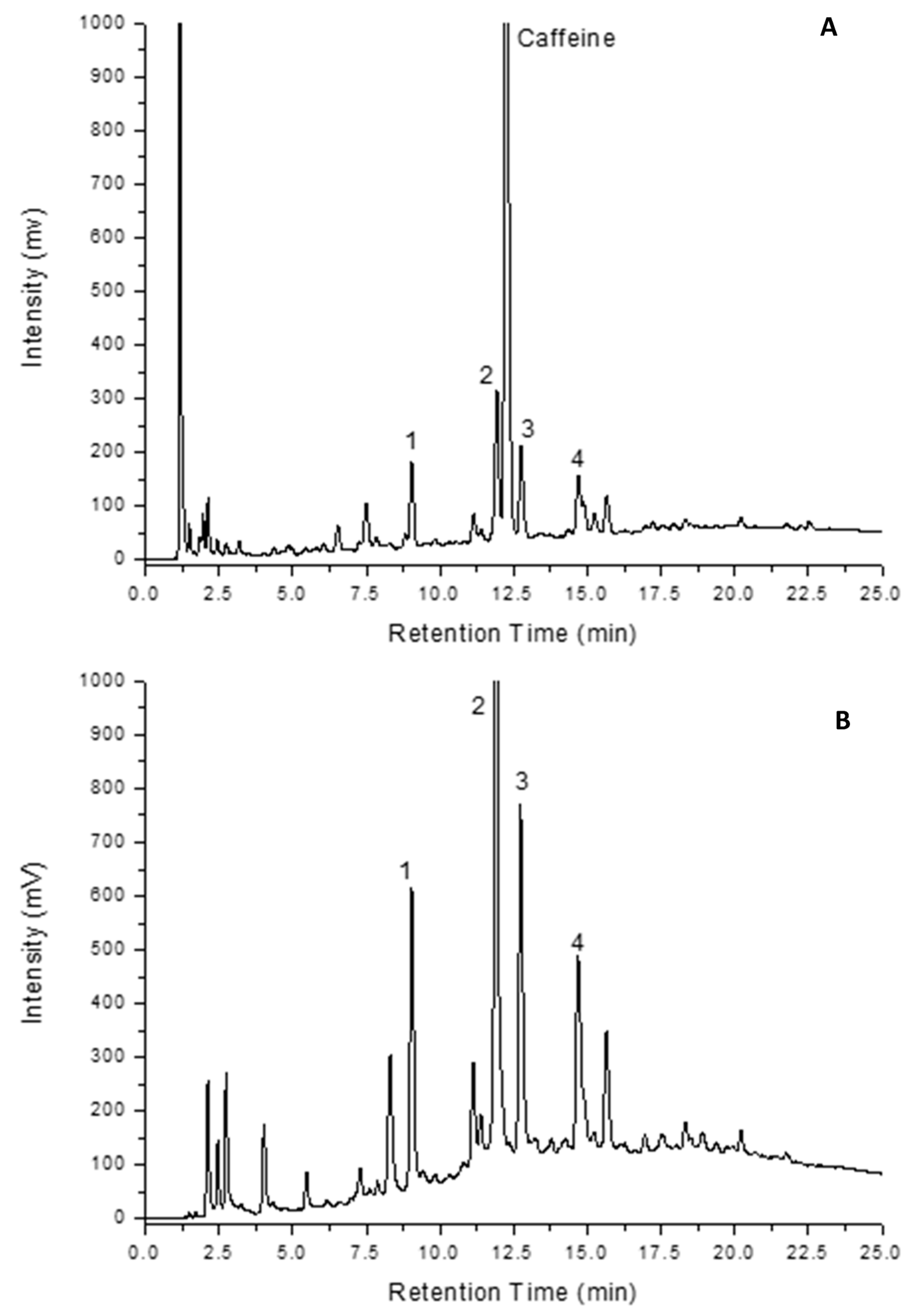
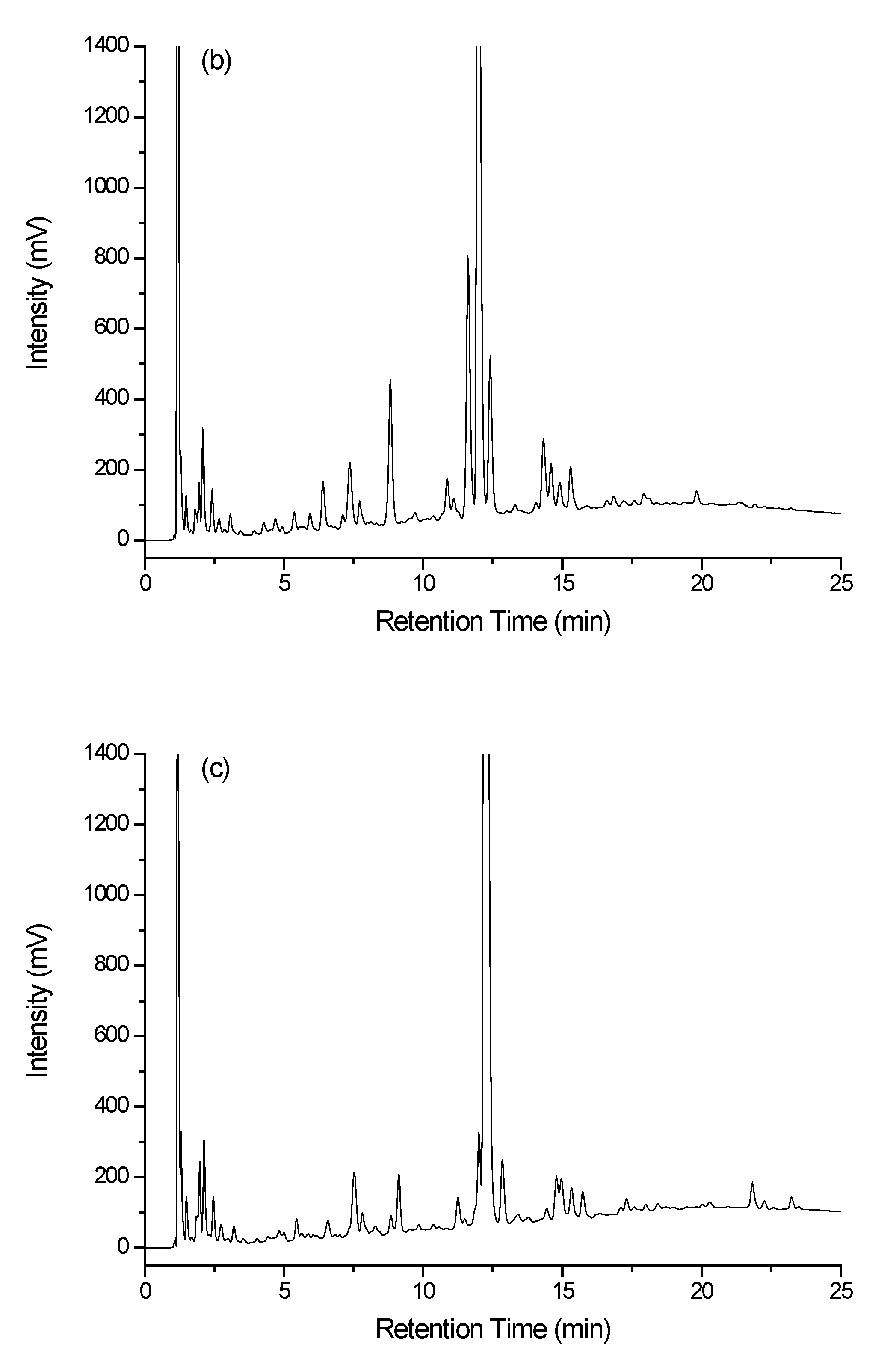
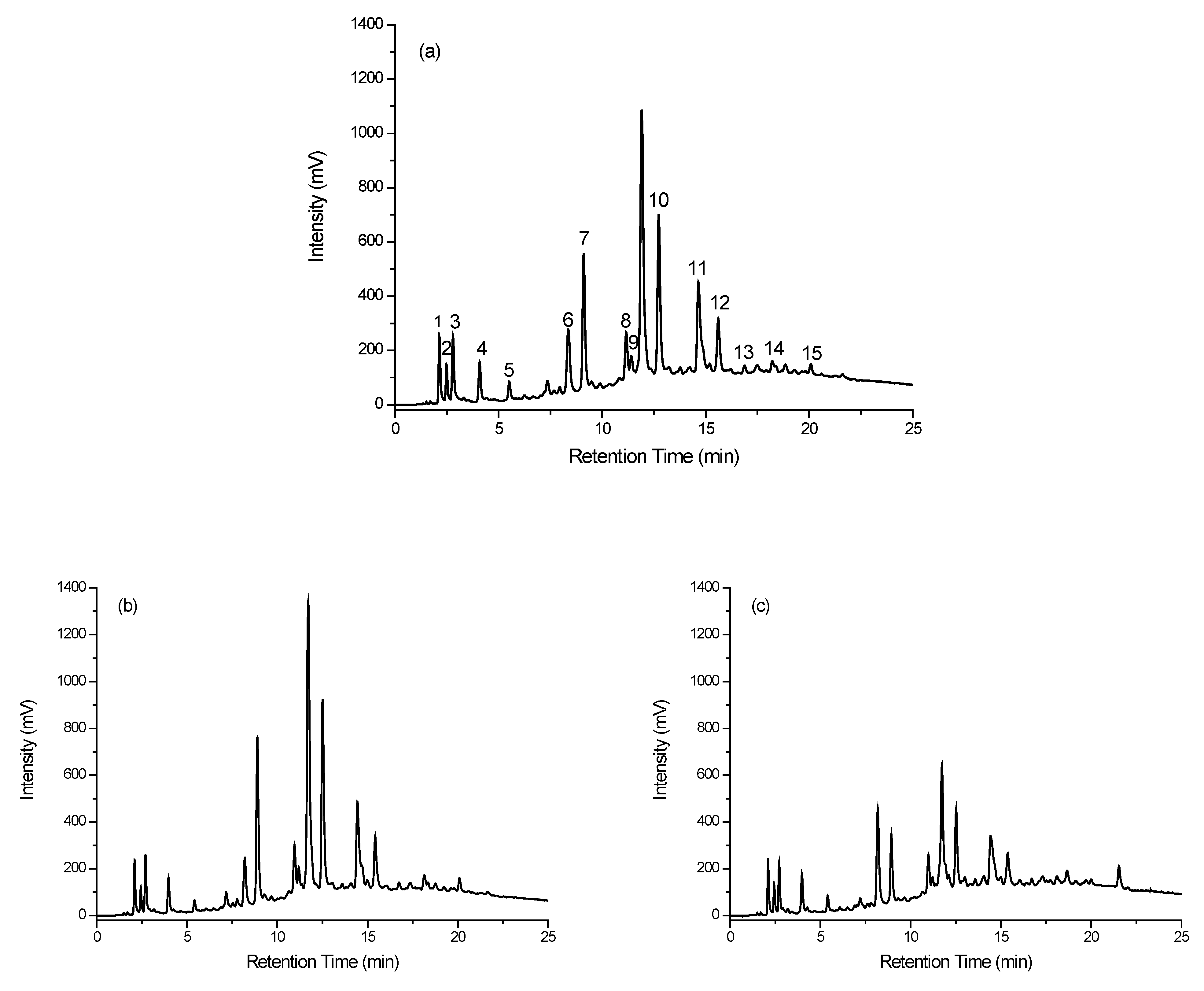
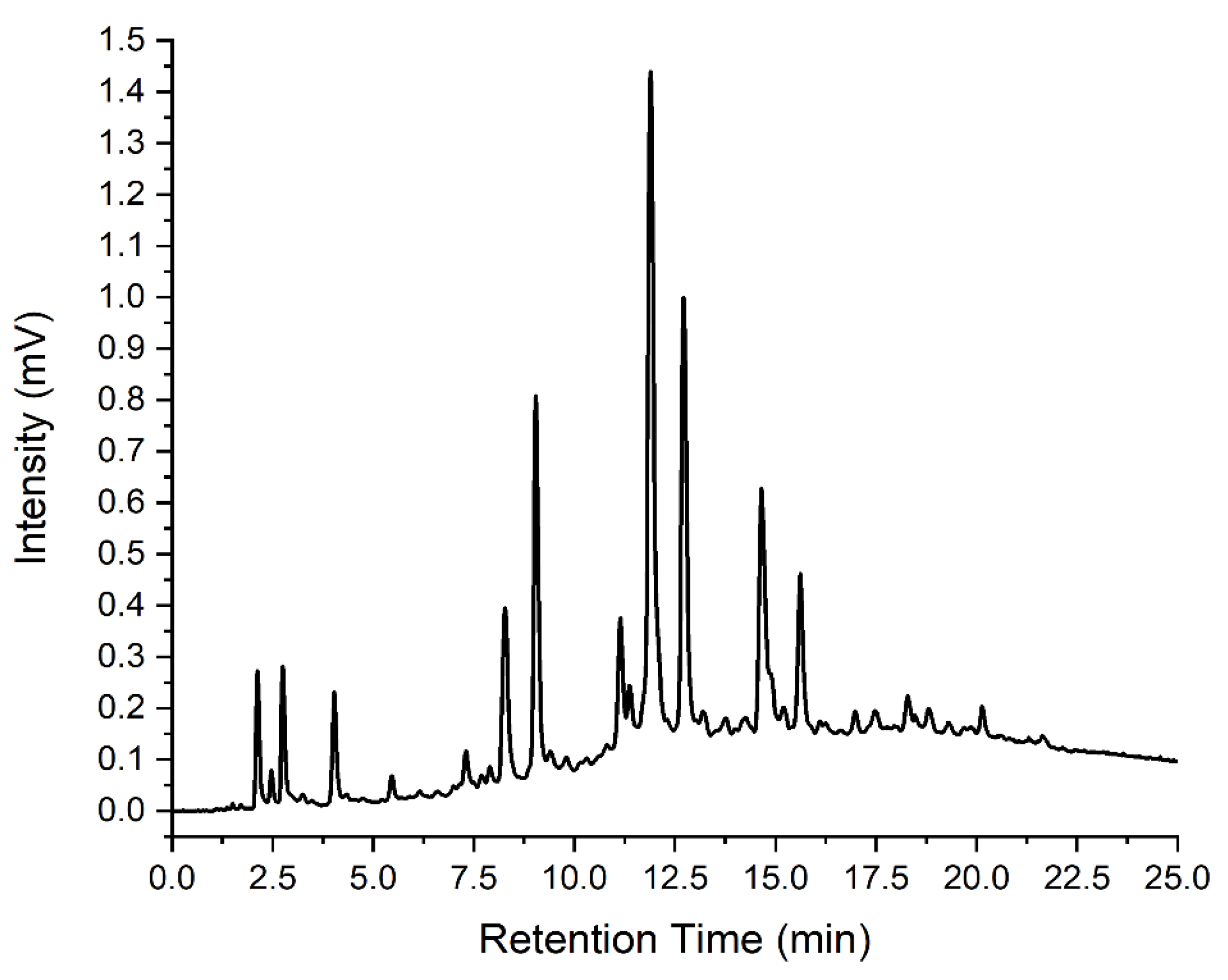
3.3.1. Separation
3.3.2. Data Treatment
3.3.3. Data Analysis
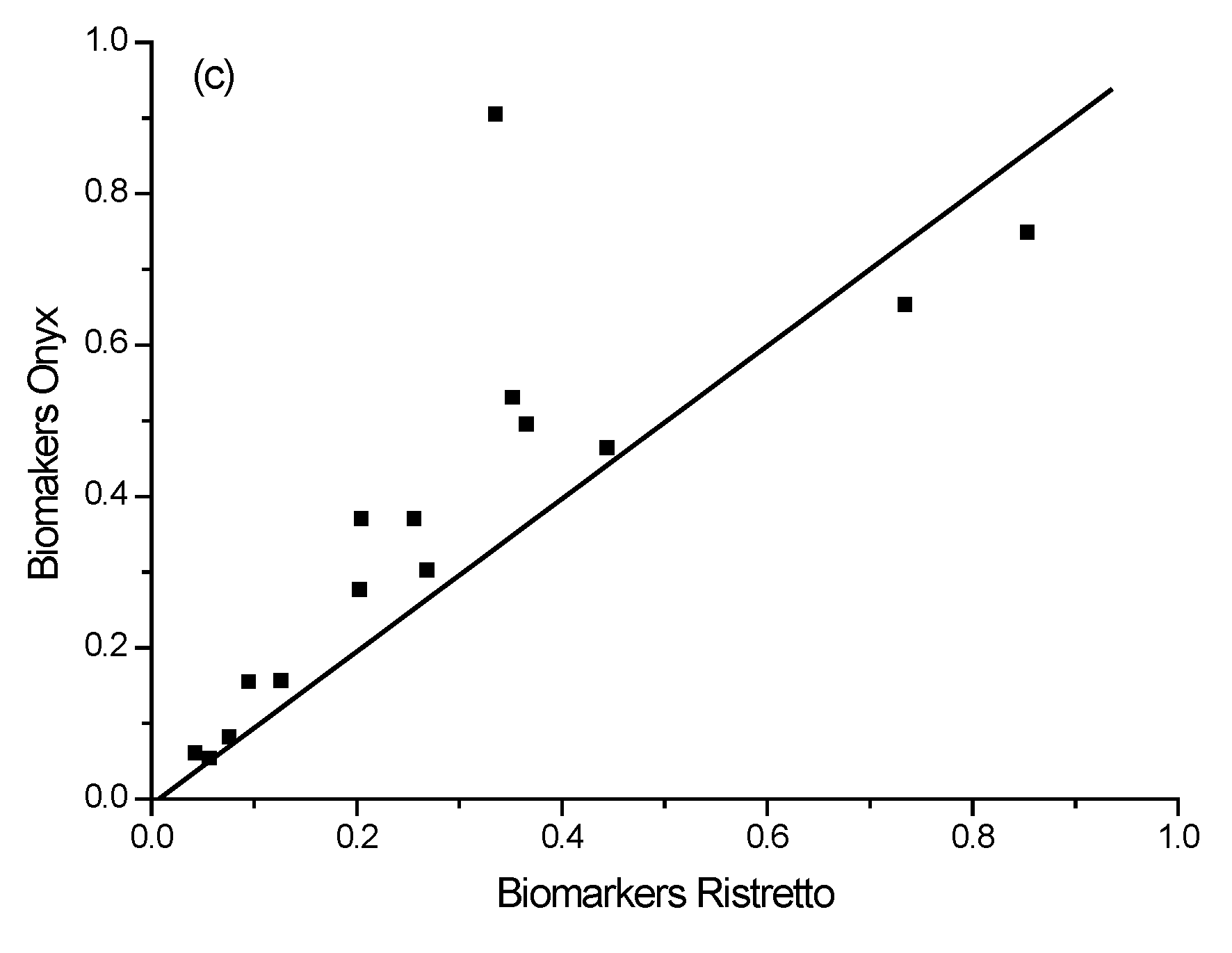
3.3.4. Testing the Library Matching
3.3.5. Unknown Sample Assignment to Library Data
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Milani, M.I.; Rossini, E.L.; Catelani, T.A.; Pezza, L.; Toci, A.T.; Pezza, H.R. Authentication of roasted and ground coffee samples containing multiple adulterants using NMR and a chemometric approach. Food Control 2020, 112, 107104. [Google Scholar] [CrossRef]
- Perez, M.; Domínguez-López, I.; López-Yerena, A.; Vallverdú Queralt, A. Current strategies to guarantee the authenticity of coffee. Crit. Rev. Food Sci. Nutr. 2021, 63, 539–554. [Google Scholar] [CrossRef] [PubMed]
- ICO. World Coffee Consumption. 2019. Available online: http://www.ico.org/prices/new-consumption-table.pdf (accessed on 29 October 2022).
- Burns, D.T.; Walker, M.J. Critical Review of Analytical and Bioanalytical Verification of the Authenticity of Coffee. J. AOAC Int. 2020, 103, 283–294. [Google Scholar] [CrossRef]
- Coffee Pods and Capsules Market|2022—27|Industry Share, Size, Growth—Mordor Intelligence. (n.d.). Available online: https://www.mordorintelligence.com/industry-reports/coffee-pods-and-capsules-market (accessed on 10 April 2022).
- Núñez, N.; Saurina, J.; Núñez, O. Authenticity Assessment and Fraud Quantitation of Coffee Adulterated with Chicory, Barley, and Flours by Untargeted HPLC-UV-FLD Fingerprinting and Chemometrics. Foods 2021, 10, 840. [Google Scholar] [CrossRef]
- Fain, S.J.; Quiñones, M.; Álvarez-Berríos, N.L.; Parés-Ramos, I.K.; Gould, W.A. Climate change and coffee: Assessing vulnerability by modeling future climate suitability in the Caribbean island of Puerto Rico. Clim. Chang. 2018, 146, 175–186. [Google Scholar] [CrossRef]
- Toci, A.T.; Farah, A.; Pezza, H.R.; Pezza, L. Coffee Adulteration: More than Two Decades of Research. Crit. Rev. Anal. Chem. 2016, 46, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Cuadros-Rodríguez, L.; Ortega-Gavilán, F.; Martín-Torres, S.; Arroyo-Cerezo, A.; Jiménez-Carvelo, A.M. Chromatographic Fingerprinting and Food Identity/Quality: Potentials and Challenges. J. Agric. Food Chem. 2021, 69, 14428–14434. [Google Scholar] [CrossRef]
- Ellis, D.I.; Brewster, V.L.; Dunn, W.B.; Allwood, J.W.; Golovanov, A.P.; Goodacre, R. Fingerprinting food: Current technologies for the detection of food adulteration and contamination. Chem. Soc. Rev. 2012, 41, 5706–5727. [Google Scholar] [CrossRef] [PubMed]
- Górska-Horczyczak, E.; Zalewska, M.; Wierzbicka, A. Chromatographic fingerprint application possibilities in food authentication. Eur. Food Res. Technol. 2022, 248, 1163–1177. [Google Scholar] [CrossRef]
- Medina, S.; Pereira, J.A.; Silva, P.; Perestrelo, R.; Câmara, J.S. Food fingerprints–A valuable tool to monitor food authenticity and safety. Food Chem. 2019, 278, 144–162. [Google Scholar] [CrossRef]
- Xie, X.; Zhang, Y.; Yue, Z.; Wang, K.; Mai, X.; Liu, Y.; Zhu, M.; Fan, H.; Zhang, W. Multi-fingerprint profiling analysis for screening and quantification of illegal adulterated antidiabetics in a functional food using HPLC coupled to diode array detection/fluorescence detection. Microchem. J. 2019, 149, 103995. [Google Scholar] [CrossRef]
- Azadmard-Damirchi, S.; Torbati, M. Adulterations in Some Edible Oils and Fats and Their Detection Methods. J. Food Qual. Hazards Control 2015, 2, 38–44. [Google Scholar]
- González-Pereira, A.; Otero, P.; Fraga-Corral, M.; Garcia-Oliveira, P.; Carpena, M.; Prieto, M.A.; Simal-Gandara, J. State-of-the-Art of Analytical Techniques to Determine Food Fraud in Olive Oils. Foods 2021, 10, 484. [Google Scholar] [CrossRef]
- Danezis, G.P.; Tsagkaris, A.S.; Camin, F.; Brusic, V.; Georgiou, C.A. Food authentication: Techniques, trends & emerging approaches. Trends Anal. Chem. 2016, 85, 123–132. [Google Scholar]
- van de Merbel, N.C. Advances in Liquid Chromatography–Tandem Mass Spectrometry (LC–MS–MS)-Based Quantitation of Biopharmaceuticals in Biological Samples. LCGC Eur. 2015, 1, 38–42. [Google Scholar]
- Cserháti, T.; Forgács, E.; Deyl, Z.; Miksik, I. Chromatography in authenticity and traceability tests of vegetable oils and dairy products: A review. Biomed. Chromatogr. BMC 2005, 19, 183–190. [Google Scholar] [CrossRef]
- Bikrani, S.; Jiménez-Carvelo, A.M.; Nechar, M.; Bagur-González, M.G.; Souhail, B.; Cuadros-Rodríguez, L. Authentication of the Geographical Origin of Margarines and Fat-Spread Products from Liquid Chromatographic UV-Absorption Fingerprints and Chemometrics. Foods 2019, 8, 588. [Google Scholar] [CrossRef]
- Núñez, N.; Collado, X.; Martínez, C.; Saurina, J.; Núñez, O. Authentication of the Origin, Variety and Roasting Degree of Coffee Samples by Non-Targeted HPLC-UV Fingerprinting and Chemometrics. Application to the Detection and Quantitation of Adulterated Coffee Samples. Foods 2020, 9, 378. [Google Scholar] [CrossRef]
- Celik, S.E.; Ozyürek, M.; Güçlü, K.; Apak, R. Determination of antioxidants by a novel on-line HPLC-cupric reducing antioxidant capacity (CUPRAC) assay with post-column detection. Anal. Chim. Acta 2010, 674, 79–88. [Google Scholar] [CrossRef]
- Perrett, D. Liquid Chromatography of Amino Acids and their Derivatives. In Chemistry and Biochemistry of the Amino Acids; Barrett, G.C., Ed.; Springer: Dordrecht, The Netherlands.
- Radjai, M.K.; Hatch, R.T. Fast determination of free amino acids by ion-pair high-performance liquid chromatography using on-line post-column derivatization. J. Chromatogr. A 1980, 196, 319–322. [Google Scholar] [CrossRef]
- Jones, A.; Pravadali-Cekic, S.; Dennis, G.R.; Shalliker, R.A. Post column derivatisation analyses review. Is post-column derivatisation incompatible with modern HPLC columns? Anal. Chim. Acta 2015, 889, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.; Pravadali-Cekic, S.; Hua, S.; Kocic, D.; Camenzuli, M.; Dennis, G.; Shalliker, A. Post Column Derivatization Using Reaction Flow High Performance Liquid Chromatography Columns. J. Vis. Exp. 2016, 110, 53462. [Google Scholar]


| Brand/Flavor | Type of Bean | Flavor Intensity | Description | Mass of Coffee Actual/g |
|---|---|---|---|---|
| L’Or Ristretto | Unknown, made in France with coffee from multiple origins | 11 | Notes of ginger and cardamom. | 5.586 |
| L’Or Organic Ristretto | Unknown | 11 | Notes of ginger and cardamom. | 5.798 |
| L’Or Profondo | Unknown | 8 | Intense and spicy aromas, roasted almonds, and licorice. | 5.906 |
| L’Or Colombia | Unknown | Smooth and sweet blend, notes of citrus fruit. | 5.464 | |
| L’Or Papua New Guinea | Unknown, single-origin blend from Papua New Guinea | 7 | Vibrant, fruity taste notes and deeper roasted notes. | 5.473 |
| L’Or Sontuoso | Unknown | 8 | Notes of crème brulée, nutty marzipan, and sweet cream. | 5.552 |
| L’Or Arabica Catuai | Brazilian coffee, grown inside a yellow coffee cherry. Single varietal | 7 | Zesty, sweet, and roasty. | 5.516 |
| L’Or Guatemala | Unknown | 7 | Mild-roasted flavor with light and zesty notes. | 5.627 |
| L’Or Onyx 12 | Unknown | 12 | Dark roasted, with spicy notes. | 5.952 |
| L’Or Or Absolu | Unknown | 9 | Woody notes and reserved sweetness. Intricate balance of delicate sweet caramel and complementing spice. | 5.414 |
| L’Or Ultimo | No Information | 13 | Caramel fruitiness with deeply roasted notes. | 5.741 |
| Starbucks House | Blend of fine Latin American beans 100% Arabica | 8 | Balanced tastes of nuts and cocoa. | 5.905 |
| Starbucks Roast | Blend of Latin American and Asia/Pacific coffees | 11 | Rich and caramelly sweetness. | 5.736 |
| Starbucks Colombia | Single origin 100% Arabica | 7 | Round body, juicy taste, and signature nutty finish. | 5.870 |
| Woolworths Intense | Unknown | 12 | Rich and full-bodied dark roast. | 5.687 |
| Woolworths Strong | Unknown | 10 | Full-bodied dark roast. | 5.766 |
| Woolworths Decaf | Arabica | 6 | Sweet, well-balanced espresso. | 5.709 |
| Vittoria Ristretto | 100% Arabica blend | 13 | Sweet, nutty, and butterscotch. | 5.131 |
| Vittoria Espresso | 100% Arabica blend from Central and South America | A dark-roasted coffee blend featuring a rich, full and flavorsome taste. | 5.174 | |
| Starbucks Sumatra | Single-origin from Indonesia 100% Arabica | 10 | Spicy and full-bodied, with a herbal taste. | 5.797 |
| Starbucks Decaf Espresso Roast | Blend from Asia and Latin America 100% Arabica | 11 | Rich and caramelly. | 5.900 |
| Starbucks Café Verona | 100% Arabica | 10 | Roasty sweet with dark cocoa notes. | 5.780 |
| Moccona Long Black | Unknown | 8 | Fruity notes with coffee bitters and chocolate sweetness. | 5.468 |
| Daley St Med Roast | Colombian and Kenyan Arabica Beans | 8 | Notes of chocolate, licorice, and toffee. | 5.252 |
| Daley St Intense Roast | Colombian Arabica and Vietnamese Robusta Beans | 12 | Notes of treacle, malt, and prune. | 5.307 |
| Vittoria Black Valley | 100% Arabica from Brazil, Colombia, and Honduras | 15 | Syrupy and full-flavored. Smokey. | 5.245 |
| Vittoria Inca Peru | 100% Arabica from Brazil, Costa Rica, and the Cajamarca, Chirinos, and Cuzco regions of Peru | 15 | Maple syrup, roasted peanuts, and blossoms. | 5.192 |
| Moccona Barista Reserve Long Black | 100% ground coffee | 8 | Fruity notes, coffee bitters, and chocolatey sweetness. | 5.436 |
| Ari coffee, Nan (local Thailand brand) | 100% Arabica from Nan, Thailand | N/A | Medium—dark roasted | 5.300 |
| Ari coffee, Chum Phon (local Thailand brand) | 100% Arabica from Chum Phon, Thailand | N/A | Medium—dark roasted | 5.133 |
| Ari coffee, Doi Pang Khon (local Thailand brand) | 100% Arabica from Chiang Rai, Thailand | N/A | Medium—dark roasted | 5.690 |
| Ari coffee, Doi Chang (local Thailand brand) | 100% Arabica from Chiang Mai, Thailand | N/A | Medium—dark roasted | 4.780 |
| Indicator | Retention Time (Minutes) |
|---|---|
| 1 | 2.08 |
| 2 | 2.41 |
| 3 | 2.67 |
| 4 | 3.95 |
| 5 | 5.37 |
| 6 | 8.16 |
| 7 | 8.85 |
| 8 | 10.88 |
| 9 | 11.12 |
| 10 | 12.46 |
| 11 | 14.44 |
| 12 | 15.45 |
| 13 | 16.76 |
| 14 | 18.19 |
| 15 | 20.12 |
| (A) | |||||||||||
| Ristretto | Organic R | Profondo | Columbia | Papua | Ultimo | Sontuoso | Arabica C | Or Ab-solu | Gua-temala | Onyx 12 | |
| Ristretto | 1.0000 | ||||||||||
| Organic R | 0.9983 | 1.0000 | |||||||||
| Profondo | 0.9492 | 0.9500 | 1.0000 | ||||||||
| Columbia | 0.9687 | 0.9700 | 1.0162 | 1.0000 | |||||||
| Papua | 0.9579 | 0.9615 | 1.0046 | 0.9864 | 1.0000 | ||||||
| Ultimo | 0.9848 | 0.9759 | 0.9882 | 0.9786 | 0.9794 | 1.0000 | |||||
| Sontuoso | 0.9532 | 0.9560 | 1.0008 | 0.9813 | 0.9943 | 0.8957 | 1.0000 | ||||
| Arabica C | 0.9178 | 0.9203 | 0.9748 | 0.9529 | 0.9643 | 0.8544 | 0.9708 | 1.0000 | |||
| Or Absolu | 0.9862 | 0.9831 | 1.0095 | 0.9848 | 1.0000 | 0.9511 | 1.0062 | 1.0223 | 1.0000 | ||
| Guatemala | 0.8802 | 0.8863 | 0.9408 | 0.9225 | 0.9344 | 0.8095 | 0.9388 | 0.9677 | 0.8619 | 1.0000 | |
| Onyx 12 | 0.7388 | 0.7202 | 0.6881 | 0.6782 | 0.6756 | 0.8086 | 0.6779 | 0.6658 | 0.7738 | 0.6333 | 1.0000 |
| (B) | |||||||||||
| Ristretto | SB House | SB Roast | SB Col | W Int | W Strong | Decaf | Ari, Nan | Ari, Chum Phon | Ari, Doi Pang Khon | ||
| Ristretto | 1.0000 | ||||||||||
| SB House | 0.9225 | 1.0000 | |||||||||
| SB Roast | 0.4429 | 0.6247 | 1.0000 | ||||||||
| SB Col | 0.9568 | 1.0111 | 1.1295 | 1.0000 | |||||||
| W Int | 0.9648 | 0.8635 | 0.6641 | 0.8665 | 1.0000 | ||||||
| W Strong | 0.9263 | 0.8093 | 0.5765 | 0.8141 | 0.9688 | 1.0000 | |||||
| Decaf | 0.8320 | 0.6838 | 0.4044 | 0.6974 | 0.8749 | 0.9076 | 1.0000 | ||||
| Ari, Nan | 0.0641 | 0.1764 | 0.3856 | 0.1520 | 0.0419 | 0.0246 | −0.0192 | 1.0000 | |||
| Ari, Chum Phon | 0.6792 | 0.7542 | 0.8381 | 0.7164 | 0.7122 | 0.7123 | 0.7041 | 1.5349 | 1.0000 | ||
| Ari, Doi Pang Khon | 0.7335 | 0.5933 | 0.3451 | 0.6082 | 0.7765 | 0.7907 | 0.8814 | 0.0158 | 0.7014 | 1.0000 | |
| (A) | |||||||||||
| Ristretto | Organic R | Profondo | Columbia | Papua | Ultimo | Sontuoso | Arabica C | Or Absolu | Guatemala | Onyx 12 | |
| Ristretto | 1.0000 | ||||||||||
| Organic R | 0.9971 | 1.0000 | |||||||||
| Profondo | 0.9757 | 0.9767 | 1.0000 | ||||||||
| Columbia | 0.9792 | 0.9814 | 0.9951 | 1.0000 | |||||||
| Papua | 0.9788 | 0.9858 | 0.9942 | 0.9946 | 1.0000 | ||||||
| Ultimo | 0.9509 | 0.9333 | 0.8841 | 0.8997 | 0.8816 | 1.0000 | |||||
| Sontuoso | 0.9780 | 0.9832 | 0.9956 | 0.9932 | 0.9976 | 0.8807 | 1.0000 | ||||
| Arabica C | 0.9554 | 0.9602 | 0.9954 | 0.9871 | 0.9888 | 0.8445 | 0.9932 | 1.0000 | |||
| Or Absolu | 0.9795 | 0.9730 | 0.9479 | 0.9361 | 0.9441 | 0.9294 | 0.9474 | 0.9280 | 1.0000 | ||
| Guatemala | 0.9302 | 0.9427 | 0.9813 | 0.9791 | 0.9826 | 0.8025 | 0.9832 | 0.9913 | 0.8854 | 1.0000 | |
| Onyx 12 | 0.6312 | 0.5994 | 0.5056 | 0.5097 | 0.4948 | 0.7712 | 0.4936 | 0.4518 | 0.6874 | 0.3863 | 1.0000 |
| (B) | |||||||||||
| Ristretto | SB House | SB Roast | SB Col | W Int | W Strong | Decaf | Ari, Nan | Ari, Chum Phon | Ari, Doi Pang Khon | ||
| Ristretto | 1.0000 | ||||||||||
| SB House | 0.8569 | 1.0000 | |||||||||
| SB Roast | 0.3653 | 0.7219 | 1.0000 | ||||||||
| SB Col | 0.8941 | 0.9915 | 0.6690 | 1.0000 | |||||||
| W Int | 0.9597 | 0.7635 | 0.2442 | 0.7927 | 1.0000 | ||||||
| W Strong | 0.9357 | 0.7093 | 0.1946 | 0.7402 | 0.9929 | 1.0000 | |||||
| Decaf | 0.8971 | 0.6017 | 0.1138 | 0.6455 | 0.9621 | 0.9787 | 1.0000 | ||||
| Ari, Nan | 0.0363 | 0.2726 | 0.7044 | 0.2088 | 0.0150 | 0.0049 | 0.0025 | 1.0000 | |||
| Ari, Chum Phon | 0.6286 | 0.7696 | 0.5140 | 0.7161 | 0.6703 | 0.6339 | 0.5213 | 0.3638 | 1.0000 | ||
| Ari, Doi Pang Khon | 0.7284 | 0.4734 | 0.0866 | 0.5129 | 0.7918 | 0.7762 | 0.8117 | 0.0000 | 0.4889 | 1.0000 | |
| (A) | |
|---|---|
| Coffee | Power Function |
| Ristretto | 2.9370 |
| Organic Ristretto | 2.7358 |
| Profondo | 1.8529 |
| Columbia | 1.8601 |
| Papua | 1.8332 |
| Ultimo | 4.5438 |
| Sontuoso | 1.8265 |
| Arabica Catuai | 1.5687 |
| Or Absolu | 4.0843 |
| Guatemala | 1.2538 |
| Onyx 12 | −7.0712 |
| (B) | |
| Coffee | Power Function |
| Starbucks House | 114.0513 |
| Starbucks Roast | −3.0854 |
| Woolworths Intense | 2.3636 |
| Woolworths Strong | 1.9145 |
| Woolworths Decaf | 1.0272 |
| Vittoria Ristretto | 1.0670 |
| Vittoria Espresso | 1.5741 |
| Sumatra | −3.0136 |
| Decaf Espresso Roast | −3.1050 |
| Café Verona | −9.2563 |
| Long Black | 1.8319 |
| Daley St Med Roast | 1.1996 |
| Daley St inter Roast | 2.2443 |
| Black Valley | 1.1064 |
| Inca Peru | 1.1441 |
| Starbucks Columbia | 14.3692 |
| Ari, Nan | −1.1064 |
| Ari, Chum Phon | −10.5532 |
| Ari, Doi Pang Khon | 1.3555 |
| Coffee | |||||
|---|---|---|---|---|---|
| Metric | Unknown 1 | Unknown 2 | Unknown 3 | Unknown 4 | Unknown 5 |
| Highest match | Unk1: Starbucks Columbia | Unk2: Guatemala | Unk3: Vittoria Espresso | Unk4: Profondo | Unk5: Or Absolu |
| Slope | 0.9765 | 0.9999 | 0.9782 | 0.9903 | 0.9711 |
| R2 | 0.9748 | 0.9887 | 0.9956 | 0.9914 | 0.9879 |
| P.F. | 14.3692 | 1.2538 | 1.5741 | 1.8529 | 4.0843 |
| Second Highest Match | No Second Match | Unk2: Inca Peru | Unk3: Arabica Catuai | Unk4: Long Black | No Second Match |
| Slope | 1.0043 | 1.0022 | 1.0007 | ||
| R2 | 0.9958 | 0.9831 | 0.9899 | ||
| P.F. | 1.1441 | 1.5687 | 1.8319 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manwaring, C.W.; Cravino, J.A.; Patel, M.; Stathakis, J.G.H.; Soliven, A.; Suktham, T.; Shalliker, R.A. Using HPLC with In-Column Derivatization to Authenticate Coffee Samples. Molecules 2023, 28, 1651. https://doi.org/10.3390/molecules28041651
Manwaring CW, Cravino JA, Patel M, Stathakis JGH, Soliven A, Suktham T, Shalliker RA. Using HPLC with In-Column Derivatization to Authenticate Coffee Samples. Molecules. 2023; 28(4):1651. https://doi.org/10.3390/molecules28041651
Chicago/Turabian StyleManwaring, Corey W., Jake A. Cravino, Margi Patel, Jonathan G. H. Stathakis, Arianne Soliven, Thirada Suktham, and Ross Andrew Shalliker. 2023. "Using HPLC with In-Column Derivatization to Authenticate Coffee Samples" Molecules 28, no. 4: 1651. https://doi.org/10.3390/molecules28041651
APA StyleManwaring, C. W., Cravino, J. A., Patel, M., Stathakis, J. G. H., Soliven, A., Suktham, T., & Shalliker, R. A. (2023). Using HPLC with In-Column Derivatization to Authenticate Coffee Samples. Molecules, 28(4), 1651. https://doi.org/10.3390/molecules28041651







