Experimental Validation of MHC Class I and II Peptide-Based Potential Vaccine Candidates for Human Papilloma Virus Using Sprague-Dawly Models
Abstract
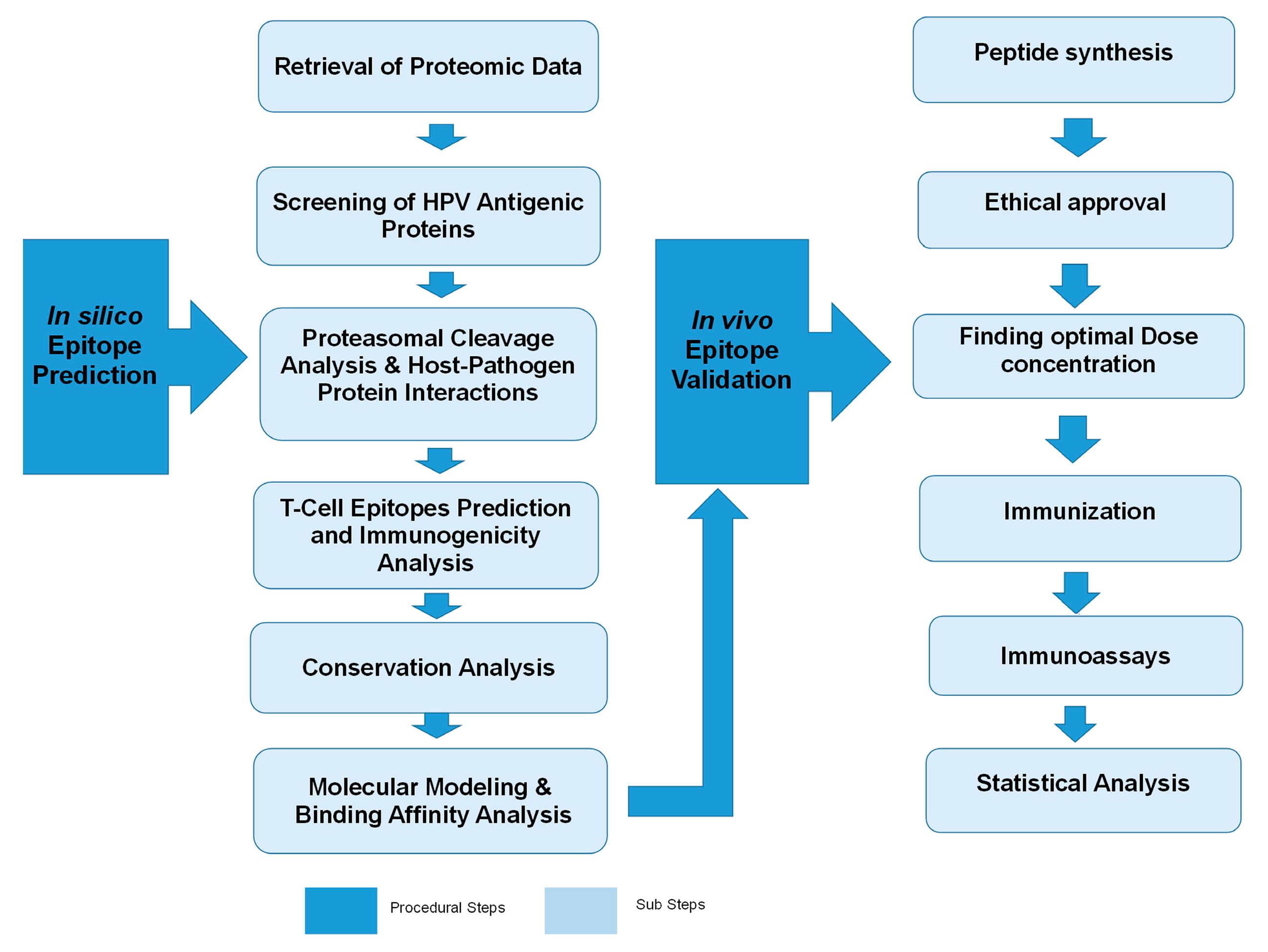
1. Introduction
2. Results
2.1. Prediction of T-Cell Epitopes
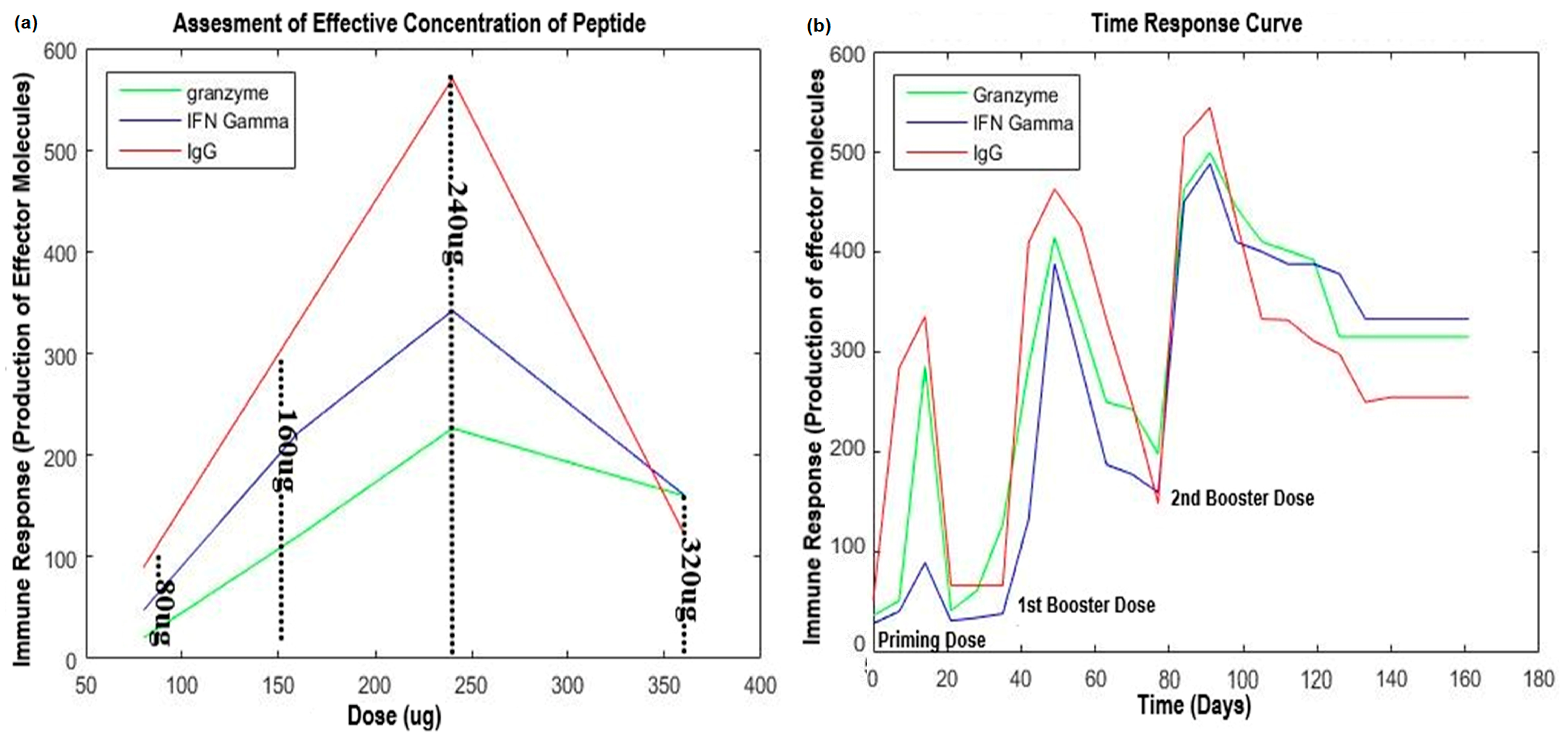
2.2. Validation of Potential Vaccine Candidates
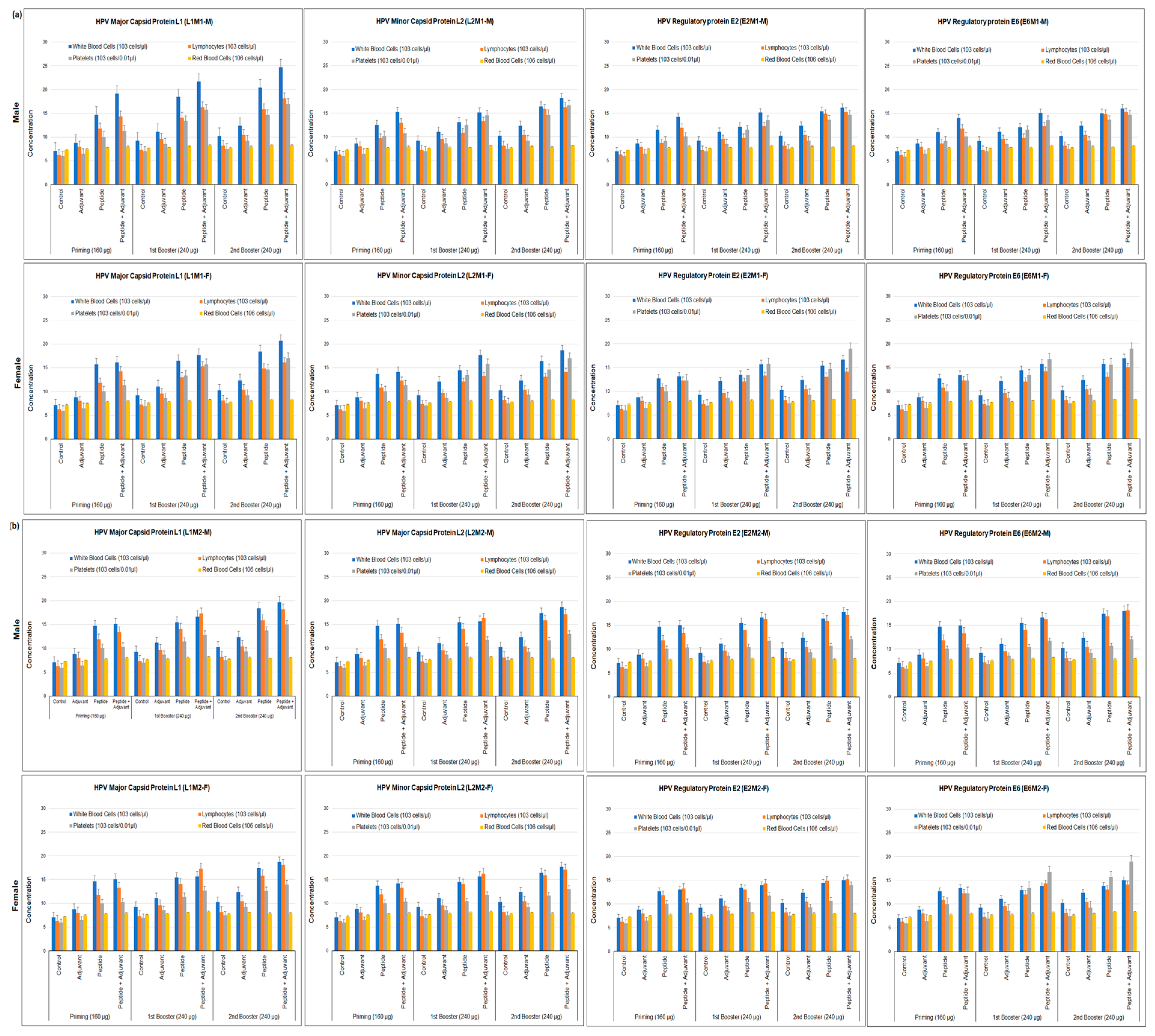
2.3. Hematological Assays
2.4. IgG ELISA Assay
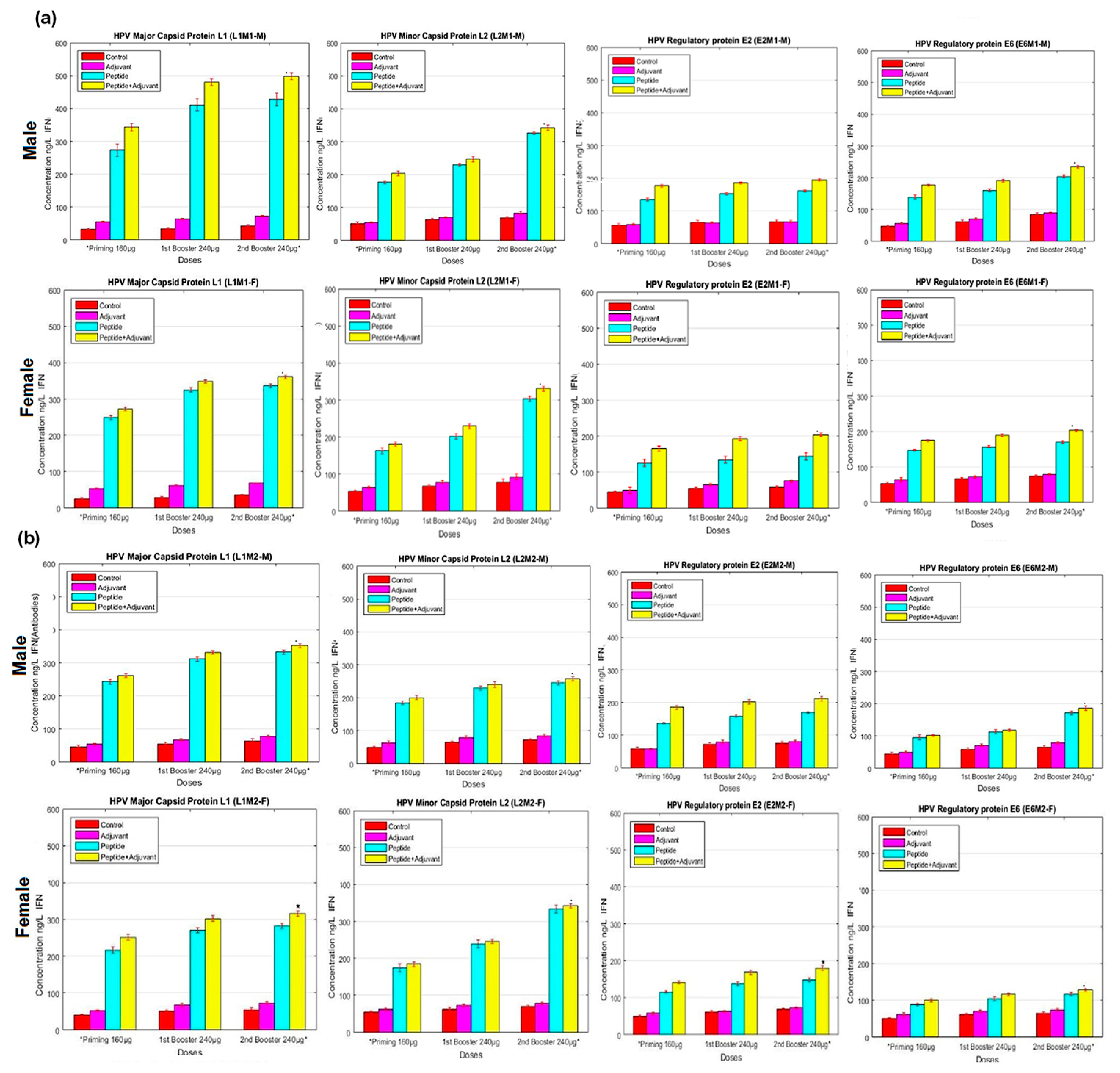
2.5. IFN-γ ELISA Assay
2.6. Granzyme B Assay
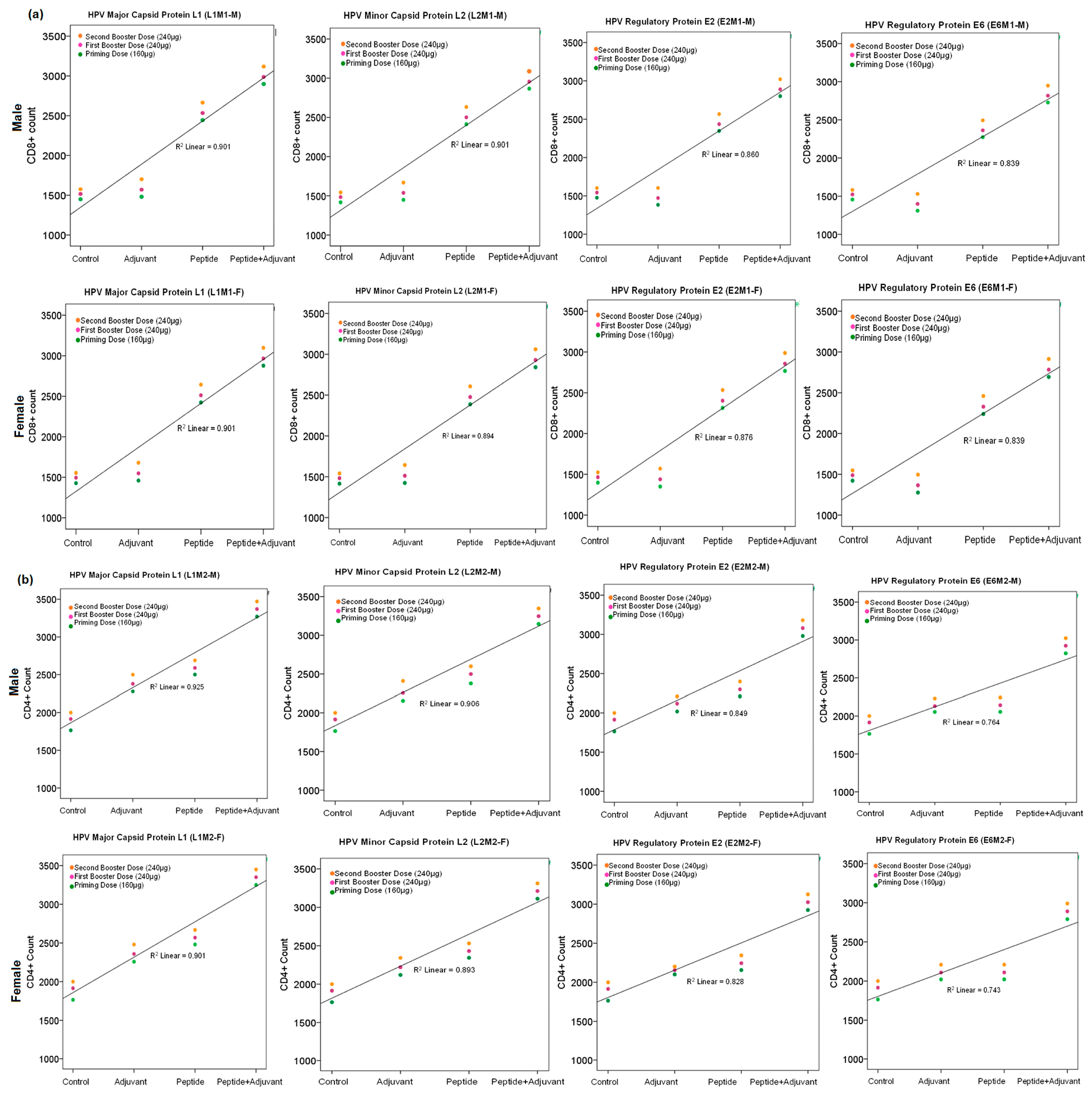
2.7. CD4+ and CD8+ Cell Count
3. Discussion
4. Materials and Methods
4.1. Ethical Approval
4.2. Retrieval of Viral Proteomic Data and Prediction of MHC Class I and II Binding Epitopes
4.3. Conservation Analysis
4.4. Synthesis of Peptides
4.5. Dose Optimization and Animal Immunization
4.6. Hematological Assays
4.7. IgG ELISA Assay
4.8. IFN-γ ELISA Assay
4.9. Granzyme B Assay
4.10. CD4+ and CD8+ Cell Count Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arbyn, M.; Weiderpass, E.; Bruni, L.; de Sanjosé, S.; Saraiya, M.; Ferlay, J.; Bray, F. Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. Lancet Glob. Health 2020, 8, e191–e203. [Google Scholar] [CrossRef]
- Wheeler, C.M.; Castellsagué, X.; Garland, S.M.; Szarewski, A.; Paavonen, J.; Naud, P.; Salmerón, J.; Chow, S.N.; Apter, D.; Kitchener, H.; et al. Cross-protective efficacy of HPV-16/18 AS04-adjuvanted vaccine against cervical infection and precancer caused by non-vaccine oncogenic HPV types: 4-year end-of-study analysis of the randomised, double-blind PATRICIA trial. Lancet Oncol. 2012, 13, 100–110. [Google Scholar] [CrossRef]
- Dilley, S.; Miller, K.M.; Huh, W.K. Human papillomavirus vaccination: Ongoing challenges and future directions. Gynecol. Oncol. 2020, 156, 498–502. [Google Scholar] [CrossRef]
- Souayah, N.; Michas-Martin, P.A.; Nasar, A.; Krivitskaya, N.; Yacoub, H.A.; Khan, H.; Qureshi, A.I. Guillain–Barré syndrome after Gardasil vaccination: Data from vaccine adverse event reporting system 2006–2009. Vaccine 2011, 29, 886–889. [Google Scholar] [CrossRef]
- Li, W.; Qi, Y.; Cui, X.; Huo, Q.; Zhu, L.; Zhang, A.; Tan, M.; Hong, Q.; Yang, Y.; Zhang, H.; et al. Characteristic of HPV integration in the genome and transcriptome of cervical cancer tissues. Biomed. Res. Int. 2018, 2018, 6242173. [Google Scholar] [CrossRef]
- Castro, T.P.P.G.; Bussoloti Filho, I. Prevalence of human papillomavirus (HPV) in oral cavity and oropharynx. Braz. J. Otorhinolaryngol. 2006, 72, 272–281. [Google Scholar] [CrossRef]
- de Araujo Souza, P.S.; Sichero, L.; Maciag, P.C. HPV variants and HLA polymorphisms: The role of variability on the risk of cervical cancer. Fut. Med. 2009, 5, 359–370. [Google Scholar] [CrossRef]
- Hafkamp, H.C.; Manni, J.J.; Speel, E.J. Role of human papillomavirus in the development of head and neck squamous cell carcinomas. Acta Oto-Laryngol. 2004, 124, 520–526. [Google Scholar] [CrossRef]
- Groot, A.S.D.; Moise, L.; McMurry, J.A.; Martin, W. Epitope-based immunome-derived vaccines: A strategy for improved design and safety. Clin. Appl. Immunol. 2009, 2, 39–69. [Google Scholar]
- Flower, D.R.; Phadwal, K.; Macdonald, I.K.; Coveney, P.V.; Davies, M.N.; Wan, S. T-cell epitope prediction and immune complex simulation using molecular dynamics: State of the art and persisting challenges. Immunome Res. 2010, 6, 1–18. [Google Scholar] [CrossRef]
- Moxon, R.; Reche, P.A.; Rappuoli, R. Reverse vaccinology. Front. Media SA 2019, 10, 2776. [Google Scholar]
- Ong, E.; Wang, H.; Wong, M.U.; Seetharaman, M.; Valdez, N.; He, Y. Vaxign-ML: Supervised machine learning reverse vaccinology model for improved prediction of bacterial protective antigens. Bioinformatics 2020, 36, 3185–3191. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Yen, Y.-T.; Singh, S.; Kao, C.-L.; Wu-Hsieh, B.A. SARS-CoV regulates immune function-related gene expression in human monocytic cells. Viral. Immun. 2012, 25, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Ismail, M.; Sajid, Z.; Ali, A.; Wu, X.; Muhammad, S.A.; Shaikh, R.S. Prediction of Prophylactic Peptide Vaccine Candidates for Human Papillomavirus (HPV): Immunoinformatics and Reverse Vaccinology Approaches. Curr. Proteom. 2021, 18, 178–192. [Google Scholar] [CrossRef]
- Muhammad, S.A.; Zafar, S.; Rizvi, S.Z.; Imran, I.; Munir, F.; Jamshed, M.B.; Ali, A.; Wu, X.; Shahid, N.; Zaeem, M.; et al. Experimental analysis of T cell epitopes for designing liver cancer vaccine predicted by system-level immunoinformatics approach. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G1055–G1069. [Google Scholar] [CrossRef]
- Altobelli, E.; Rapacchietta, L.; Profeta, V.F.; Fagnano, R. HPV-vaccination and cancer cervical screening in 53 WHO European Countries: An update on prevention programs according to income level. Cancer Med. 2019, 8, 2524–2534. [Google Scholar] [CrossRef]
- Abdelmageed, M.I.; Abdelmoneim, A.H.; Mustafa, M.I.; Elfadol, N.M.; Murshed, N.S.; Shantier, S.W.; Makhawi, A.M. Design of a multiepitope-based peptide vaccine against the E protein of human COVID-19: An immunoinformatics approach. BioMed Res. Int. 2020, 2020, 2683286. [Google Scholar] [CrossRef]
- Bibi, S.; Ullah, I.; Zhu, B.; Adnan, M.; Liaqat, R.; Kong, W.B.; Niu, S. In silico analysis of epitope-based vaccine candidate against tuberculosis using reverse vaccinology. Sci. Rep. 2021, 11, 1–16. [Google Scholar] [CrossRef]
- Rainho-Tomko, J.N.; Pavot, V.; Kishko, M.; Swanson, K.; Edwards, D.; Yoon, H.; Lanza, L.; Alamares-Sapuay, J.; Osei-Bonsu, R.; Mundle, S.T.; et al. Immunogenicity and protective efficacy of RSV G central conserved domain vaccine with a prefusion nanoparticle. Npj Vaccines 2022, 7, 1–9. [Google Scholar] [CrossRef]
- Bappy, S.S.; Sultana, S.; Adhikari, J.; Mahmud, S.; Khan, M.A.; Kibria, K.K.; Rahman, M.M.; Shibly, A.Z. Extensive immunoinformatics study for the prediction of novel peptide-based epitope vaccine with docking confirmation against envelope protein of Chikungunya virus: A computational biology approach. J. Biomol. Struc. Dyn. 2021, 39, 1139–1154. [Google Scholar] [CrossRef]
- Lagousi, T.; Basdeki, P.; Routsias, J.; Spoulou, V. Novel protein-based pneumococcal vaccines: Assessing the use of distinct protein fragments instead of full-length proteins as vaccine antigens. Vaccines 2019, 7, 9. [Google Scholar] [CrossRef]
- Masignani, V.; Pizza, M.; Moxon, E.R. The development of a vaccine against meningococcus B using reverse vaccinology. Front. Immunol. 2019, 10, 751. [Google Scholar] [CrossRef]
- Turner, H.C.; Baussano, I.; Garnett, G.P. Vaccinating women previously exposed to human papillomavirus: A cost-effectiveness analysis of the bivalent vaccine. Soc. Psychiatry 2013, 8, e75552. [Google Scholar] [CrossRef]
- Arbyn, M.; Castellsagué, X.; de Sanjosé, S.; Bruni, L.; Saraiya, M.; Bray, F.; Ferlay, J. Worldwide burden of cervical cancer in 2008. Ann. Oncol. 2011, 22, 2675–2686. [Google Scholar] [CrossRef]
- Garçon, N.; Chomez, P.; Van Mechelen, M. GlaxoSmithKline Adjuvant Systems in vaccines: Concepts, achievements and perspectives. Expert Rev. Vaccines 2007, 6, 723–739. [Google Scholar] [CrossRef]
- Greer, C.E.; Wheeler, C.M.; Ladner, M.B.; Beutner, K.; Coyne, M.Y.; Liang, H.; Langenberg, A.; Yen, T.S.; Ralston, R. Human papillomavirus (HPV) type distribution and serological response to HPV type 6 virus-like particles in patients with genital warts. J. Clin. Microbiol. 1995, 33, 2058–2063. [Google Scholar] [CrossRef]
- Barra, F.; Leone Roberti Maggiore, U.; Bogani, G.; Ditto, A.; Signorelli, M.; Martinelli, F.; Chiappa, V.; Lorusso, D.; Raspagliesi, F.; Ferrero, S. New prophylactics human papilloma virus (HPV) vaccines against cervical cancer. J. Obstet. Gynaecol. 2019, 39, 1–10. [Google Scholar] [CrossRef]
- Khan, K.; Alhar, M.S.O.; Abbas, M.N.; Abbas, S.Q.; Kazi, M.; Khan, S.A.; Sadiq, A.; Hassan, S.S.U.; Bungau, S.; Jalal, K. Integrated Bioinformatics-Based Subtractive Genomics Approach to Decipher the Therapeutic Drug Target and Its Possible Intervention against Brucellosis. Bioengineering 2022, 9, 633. [Google Scholar] [CrossRef]
- Paul, K.T. Saving lives: Adapting and adopting human papilloma virus (HPV) vaccination in Austria. Soc. Sci. Med. 2016, 153, 193–200. [Google Scholar] [CrossRef]
- Bishop, J.A.; Ma, X.J.; Wang, H.; Luo, Y.; Illei, P.B.; Begum, S.; Taube, J.M.; Koch, W.M.; Westra, W.H. Detection of transcriptionally active high risk HPV in patients with head and neck squamous cell carcinoma as visualized by a novel E6/E7 mRNA in situ hybridization method. Am. J. Surg. Pathol. 2012, 36, 1874. [Google Scholar] [CrossRef]
- Vinzón, S.E.; Rösl, F. HPV vaccination for prevention of skin cancer. Hum. Vaccines Immunother. 2015, 11, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Setiawan, D.; Luttjeboer, J.; Pouwels, K.B.; Wilschut, J.C.; Postma, M.J. Immunogenicity and safety of human papillomavirus (HPV) vaccination in Asian populations from six countries: A meta-analysis. Jpn. J. Clin. Oncol. 2017, 47, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Hassan, P.M.; Ali, T.; Saber, E.; Asghar, A.; Delaram, D.; Mostafa, S.-V.; Maryam, S.; Maryam, K.; Talieh, S.; Ali, K.M. Potency, toxicity and protection evaluation of PastoCoAd candidate vaccines: Novel preclinical mix and match rAd5 S, rAd5 RBD-N and SOBERANA dimeric-RBD protein. Vaccine 2022, 40, 2856–2868. [Google Scholar]
- Brown, D.R.; Garland, S.M.; Ferris, D.G.; Joura, E.; Steben, M.; James, M.; Radley, D.; Vuocolo, S.; Garner, E.I.; Haupt, R.M. The humoral response to Gardasil over four years as defined by total IgG and competitive Luminex immunoassay. Hum. Vaccines 2011, 7, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, D.; Earnest-Silveira, L.; Grubor-Bauk, B.; Wijesundara, D.; Boo, I.; Ramsland, P.A.; Vincan, E.; Drummer, H.; Gowans, E.; Torresi, J. Pre-clinical evaluation of a quadrivalent HCV VLP vaccine in pigs following microneedle delivery. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Ewen, C.L.; Rong, J.; Kokaji, A.I.; Bleackley, R.C.; Kane, K.P. Evaluating antigen-specific cytotoxic T lymphocyte responses by a novel mouse granzyme B ELISPOT assay. J. Immunol. Methods 2006, 308, 156–166. [Google Scholar] [CrossRef]
- Ahmad, G.; Zhang, W.; Torben, W.; Ahrorov, A.; Damian, R.T.; Wolf, R.F.; White, G.L.; Carey, D.W.; Mwinzi, P.N.; Ganley-Leal, L. Preclinical prophylactic efficacy testing of Sm-p80–based vaccine in a nonhuman primate model of Schistosoma mansoni infection and immunoglobulin G and E responses to Sm-p80 in human serum samples from an area where Schistosomiasis is endemic. J. Infect. Dis. 2011, 204, 1437–1449. [Google Scholar] [CrossRef]
- Takahashi, Y.; Mine, J.; Kubota, Y.; Yamazaki, E.; Fujiwara, T.J.E. A substantial number of Rasmussen syndrome patients have increased IgG, CD4+ T cells, TNFα, and Granzyme B in CSF. Epilepsia 2009, 50, 1419–1431. [Google Scholar] [CrossRef]
- Patronov, A.; Doytchinova, I. T-cell epitope vaccine design by immunoinformatics. Open Biol. 2013, 3, 120139. [Google Scholar] [CrossRef]
- Jalal, K.; Khan, K.; Basharat, Z.; Abbas, M.N.; Uddin, R.; Ali, F.; Khan, S.A. Reverse vaccinology approach for multi-epitope centered vaccine design against delta variant of the SARS-CoV-2. Environ. Sci. Pollut. Res. 2022, 29, 60035–60053. [Google Scholar] [CrossRef]
- Hou, J.; Liu, Y.; His, J.; Wang, H.; Tao, R.; Shao, Y. Cholera toxin B subunit acts as a potent systemic adjuvant for HIV-1 DNA vaccination intramuscularly in mice. Hum. Vaccines Immunother. 2014, 10, 1274–1283. [Google Scholar] [CrossRef] [PubMed]
- Schellenbacher, C.; Roden, R.; Kirnbauer, R. Chimeric L1-L2 virus-like particles as potential broad-spectrum human papillomavirus vaccines. J. Virol. 2009, 83, 10085–10095. [Google Scholar] [CrossRef] [PubMed]
- Rosales, R.; López-Contreras, M.; Rosales, C.; Magallanes-Molina, J.R.; Gonzalez-Vergara, R.; Arroyo-Cazarez, J.M.; Ricardez-Arenas, A.; del Follo-Valencia, A.; Padilla-Arriaga, S.; Guerrero, M.V.; et al. Regression of human papillomavirus intraepithelial lesions is induced by MVA E2 therapeutic vaccine. Hum. Gene Ther. 2014, 25, 1035–1049. [Google Scholar] [CrossRef] [PubMed]
- Tseng, S.H.; Liu, L.; Peng, S.; Kim, J.; Ferrall, L.; Hung, C.F.; Wu, T.C. Control of spontaneous HPV16 E6/E7 expressing oral cancer in HLA-A2 (AAD) transgenic mice with therapeutic HPV DNA vaccine. J. Biomed. Sci. 2021, 28, 1–12. [Google Scholar] [CrossRef]
- Van Doorslaer, K.; Li, Z.; Xirasagar, S.; Maes, P.; Kaminsky, D.; Liou, D.; Sun, Q.; Kaur, R.; Huyen, Y.; McBride, A.A. The Papillomavirus Episteme: A major update to the papillomavirus sequence database. Nucleic Acids Res. 2017, 45, D499–D506. [Google Scholar] [CrossRef]
- Shen, H.-B.; Chou, K.-C. Virus-mPLoc: A fusion classifier for viral protein subcellular location prediction by incorporating multiple sites. J. Biomol. Struct. Dyn. 2010, 28, 175–186. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 1–7. [Google Scholar] [CrossRef]
- Wakeman, C.A.; Hammer, N.D.; Stauff, D.L.; Attia, A.S.; Anzaldi, L.L.; Dikalov, S.I.; Calcutt, M.W.; Skaar, E.P. Menaquinone biosynthesis potentiates haem toxicity in S taphylococcus aureus. Mol. Microbiol. 2012, 86, 1376–1392. [Google Scholar] [CrossRef]
- Singh, H.; Raghava, G. ProPred1: Prediction of promiscuous MHC Class-I binding sites. Bioinformatics 2003, 19, 1009–1014. [Google Scholar] [CrossRef]
- Camproux, A.; Tuffery, P. Hidden Markov model-derived structural alphabet for proteins: The learning of protein local shapes captures sequence specificity. Biochim. Et Biophys. Acta BBA-Gen. Subj. 2005, 1724, 394–403. [Google Scholar] [CrossRef]
- Zhang, Y.; Rohde, C.; Reinhardt, R.; Voelcker-Rehage, C.; Jeltsch, A. Genome biology. Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Ghufran, M.; Khan, H.A.; Ullah, M.; Ghufran, S.; Ayaz, M.; Siddiq, M.; Hassan, S.S.U.; Bungau, S. In Silico Strategies for Designing of Peptide Inhibitors of Oncogenic K-Ras G12V Mutant: Inhibiting Cancer Growth and Proliferation. Cancers 2022, 14, 4884. [Google Scholar] [CrossRef] [PubMed]
- Ammari, M.G.; Gresham, C.R.; McCarthy, F.M.; Nanduri, B. HPIDB 2.0: A curated database for host–pathogen interactions. Database 2016, 2016, baw103. [Google Scholar] [CrossRef]
- Agallou, M.; Athanasiou, E.; Koutsoni, O.; Dotsika, E.; Karagouni, E. Experimental validation of multi-epitope peptides including promising MHC class I-and II-restricted epitopes of four known Leishmania infantum proteins. Front. Immunol. 2014, 5, 268. [Google Scholar] [CrossRef]
- Zafar, S.; Bai, B.; Guo, J.; Muhammad, S.A.; Naqvi, S.T.Q.; Shabbir, M.N.; Imran, I.; Shaikh, R.S.; Ali, A. Experimental Study of Potential CD8+ Trivalent Synthetic Peptides for Liver Cancer Vaccine Development Using Sprague Dawley Rat Models. Biomed Res. Int. 2022, 2022, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Said, N.M.; Abiola, O. Haematological profile shows that inbred Sprague Dawley rats have exceptional promise for use in biomedical and pharmacological studies. Asian J. Biomed. Pharm. Sci. 2014, 4, 33. [Google Scholar]
- Liu, S.; Guo, R.; Liu, F.; Yuan, Q.; Yu, Y.; Ren, F. Gut microbiota regulates depression-like behavior in rats through the neuroendocrine-immune-mitochondrial pathway. Neuropsychiatr. Dis. Treat. 2020, 16, 859. [Google Scholar] [CrossRef]
- Fihiruddin, F.; Artama, W.T.; Wibawa, T.; Mertaniasih, N.M. Expression of immunoglobulin, granzyme-B and perforin against Ag85A and Ag85B proteins of Mycobacterium tuberculosis in Balb/c mice. Afr. J. Infect. Dis. 2019, 13, 13–20. [Google Scholar] [CrossRef]
- Liu, Y.; Li, T.; Alim, A.; Ren, D.; Zhao, Y.; Yang, X. Regulatory effects of stachyose on colonic and hepatic inflammation, gut microbiota dysbiosis, and peripheral CD4+ T cell distribution abnormality in high-fat diet-fed mice. J. Agric. Food Chem. 2019, 67, 11665–11674. [Google Scholar] [CrossRef]
- Strober, W. Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. 2015, 111, A3.B.1–A3.B.3. [Google Scholar] [CrossRef] [PubMed]
- Udén, P.; Robinson, P.; Mateos, G.; Blank, R. Use of replicates in statistical analyses in papers submitted for publication in Animal Feed Science and Technology. Anim. Feed Sci. Technol. 2012, 171, 1–5. [Google Scholar] [CrossRef]
- Khanday, M.; Rafiq, A.; Nazir, K. Mathematical models for drug diffusion through the compartments of blood and tissue medium. Alexandria J. Med. 2017, 53, 245–249. [Google Scholar] [CrossRef]
- Okagbue, H.I.; Oguntunde, P.E.; Obasi, E.C.; Akhmetshin, E.M. Trends and usage pattern of SPSS and Minitab Software in Scientific research. J. Phys. Conf. Ser. 2021, 1734, 1–8. [Google Scholar] [CrossRef]
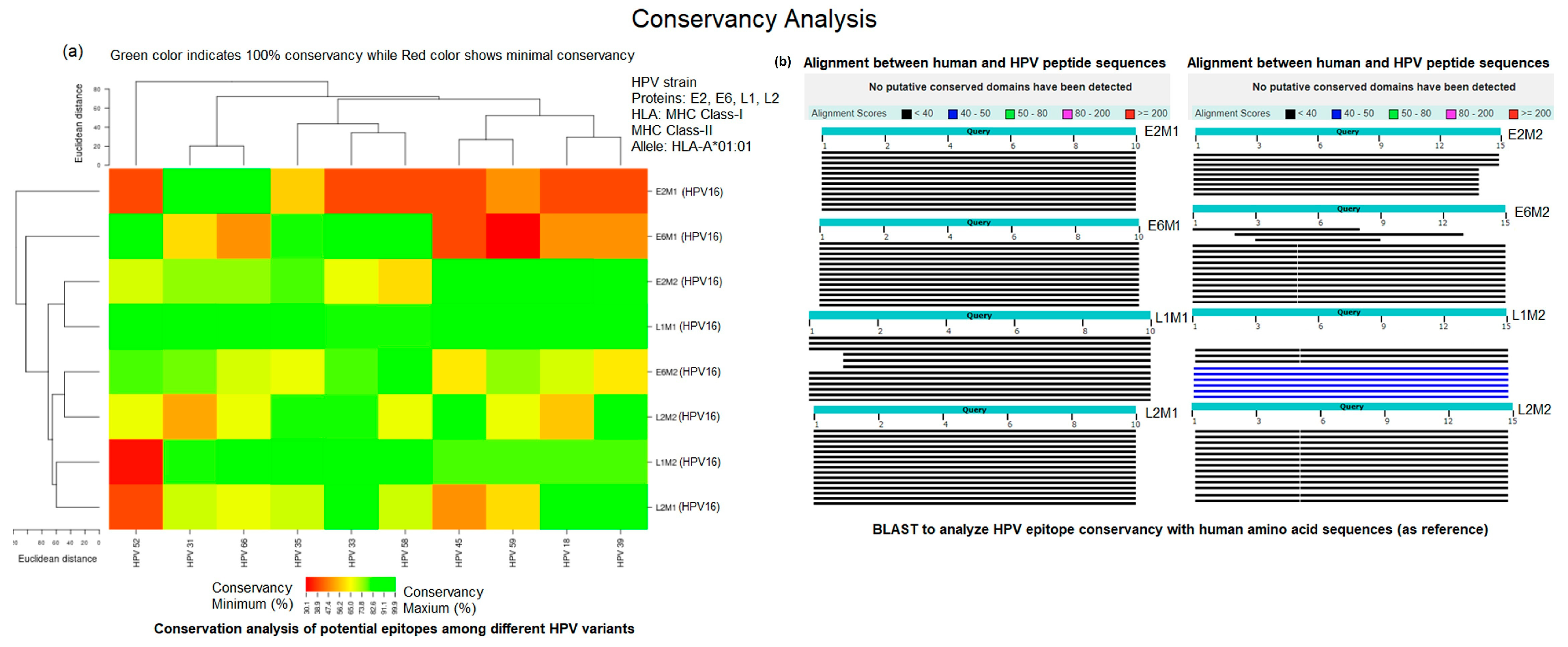


| Protein | Allele | Peptide Code | Peptide Sequence | Percentile Rank | Toxicity Prediction | Hydrop-athicity | Charge | Mol. Weight |
|---|---|---|---|---|---|---|---|---|
| L1 | HLA-A*01:01 | L1M1 | CTSICKYPDY | 0.3 | Non-Toxic | −0.36 | 0.00 | 1192.49 |
| HLA-DRB1*01:01 | L1M2 | NIYYHAGTSRLLAVG | 0.13 | Non-Toxic | 0.25 | 1.50 | 1635.8 | |
| L2 | HLA-A*01:01 | L2M1 | LTSRRTGIRY | 0.2 | Non-Toxic | −0.91 | 3.00 | 1222.55 |
| HLA-DRB1*01:01 | L2M2 | VDPAFVTTPTKLITY | 3.27 | Non-Toxic | 0.44 | 0.00 | 1666.16 | |
| E2 | HLA-A*01:01 | E2M1 | STDLRDHIDY | 0.15 | Non-Toxic | −1.27 | −1.50 | 1234.42 |
| HLA-DRB1*01:01 | E2M2 | EMGFKHINHQVVPTL | 7.93 | Non-Toxic | −0.14 | 1.00 | 1750.29 | |
| E6 | HLA-A*01:01 | E6M1 | KISEYRHYCY | 0.55 | Non-Toxic | −0.09 | 1.50 | 1361.67 |
| HLA-DRB1*01:01 | E6M2 | RHYCYSLYGTTLEQQ | 2.91 | Non-Toxic | −0.97 | 0.50 | 1862.27 |
| Sr. No. | Name | Use | Links |
|---|---|---|---|
| 1 | Papillomavirus Episteme | Retrieval of HPV Genome | https://pave.niaid.nih.gov/#home (accessed on 5 April 2018) |
| 2 | Virus-mPLoc | Subcellular Localization | http://www.csbio.sjtu.edu.cn/bioinf/virus-multi/ (accessed on 15 January 2018) |
| 3 | MvirDB | Virulence Factor Prediction | http://mvirdb.llnl.gov/ (accessed on 16 January 2018) |
| 4 | Vaxijen server | Antigenicity Prediction | http://www.jenner.ac.uk/VaxiJen (accessed on 3 February 2018) |
| 5 | Immune Epitope Database | Epitopes Prediction | http://tools.iedb.org/main/ (accessed on 7 February 2019) |
| 6 | Protein information resource | Mol. Weight Prediction | http://pir.georgetown.edu/cgi-bin/comp_mw.pl (accessed on 8 February 2018) |
| 7 | Immune Epitope Database | MHC-peptide binding | http://tools.iedb.org/main/ (accessed on 1 March 2019) |
| 8 | Immune Epitope Database | Epitope Conservation | http://tools.iedb.org/main/analysis-tools/ (accessed on 5 March 2020) |
| 9 | PEP-FOLD | Peptide modeling | http://bioserv.rpbs.univ-paris-diderot.fr/PEP-FOLD/ (accessed on 15 March 2019) |
| 10 | Uniprot | Protein Information | http://www.uniprot.org/ (accessed on 7 January 2018) |
| 11 | NCBI | Genomic/Proteomic data | https://www.ncbi.nlm.nih.gov/ (accessed on 2 January 2018) |
| 12 | MOE | Docking | https://www.chemcomp.com (accessed on 7 August 2019) |
| 13 | TASSER | Polylinker Design | https://zhanglab.ccmb.med.umich.edu/I-TASSER/ (accessed on 1 September 2020) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ismail, M.; Bai, B.; Guo, J.; Bai, Y.; Sajid, Z.; Muhammad, S.A.; Shaikh, R.S. Experimental Validation of MHC Class I and II Peptide-Based Potential Vaccine Candidates for Human Papilloma Virus Using Sprague-Dawly Models. Molecules 2023, 28, 1687. https://doi.org/10.3390/molecules28041687
Ismail M, Bai B, Guo J, Bai Y, Sajid Z, Muhammad SA, Shaikh RS. Experimental Validation of MHC Class I and II Peptide-Based Potential Vaccine Candidates for Human Papilloma Virus Using Sprague-Dawly Models. Molecules. 2023; 28(4):1687. https://doi.org/10.3390/molecules28041687
Chicago/Turabian StyleIsmail, Mehreen, Baogang Bai, Jinlei Guo, Yuhui Bai, Zureesha Sajid, Syed Aun Muhammad, and Rehan Sadiq Shaikh. 2023. "Experimental Validation of MHC Class I and II Peptide-Based Potential Vaccine Candidates for Human Papilloma Virus Using Sprague-Dawly Models" Molecules 28, no. 4: 1687. https://doi.org/10.3390/molecules28041687
APA StyleIsmail, M., Bai, B., Guo, J., Bai, Y., Sajid, Z., Muhammad, S. A., & Shaikh, R. S. (2023). Experimental Validation of MHC Class I and II Peptide-Based Potential Vaccine Candidates for Human Papilloma Virus Using Sprague-Dawly Models. Molecules, 28(4), 1687. https://doi.org/10.3390/molecules28041687







