Examining the Effect of Notocactus ottonis Cold Vacuum Isolated Plant Cell Extract on Hair Growth in C57BL/6 Mice Using a Combination of Physiological and OMICS Analyses
Abstract
1. Introduction
2. Results
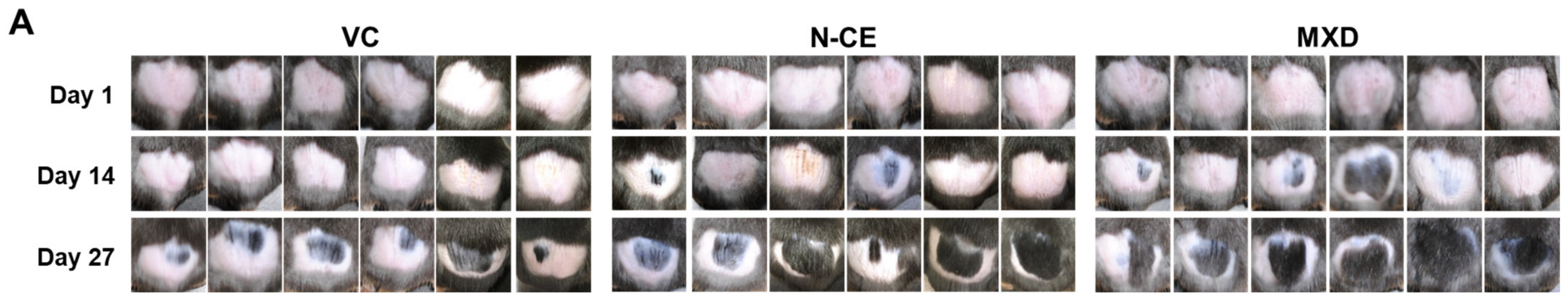
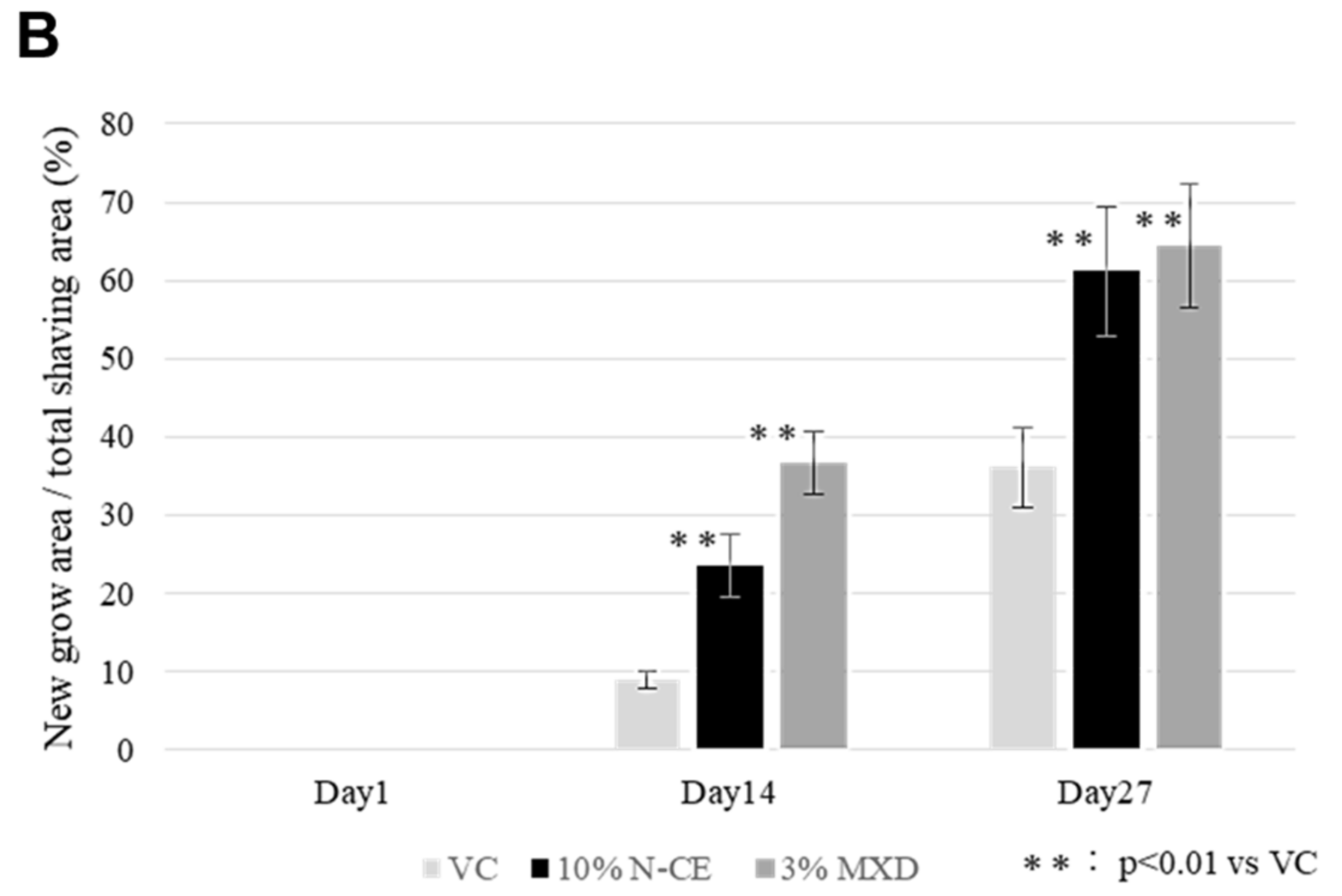
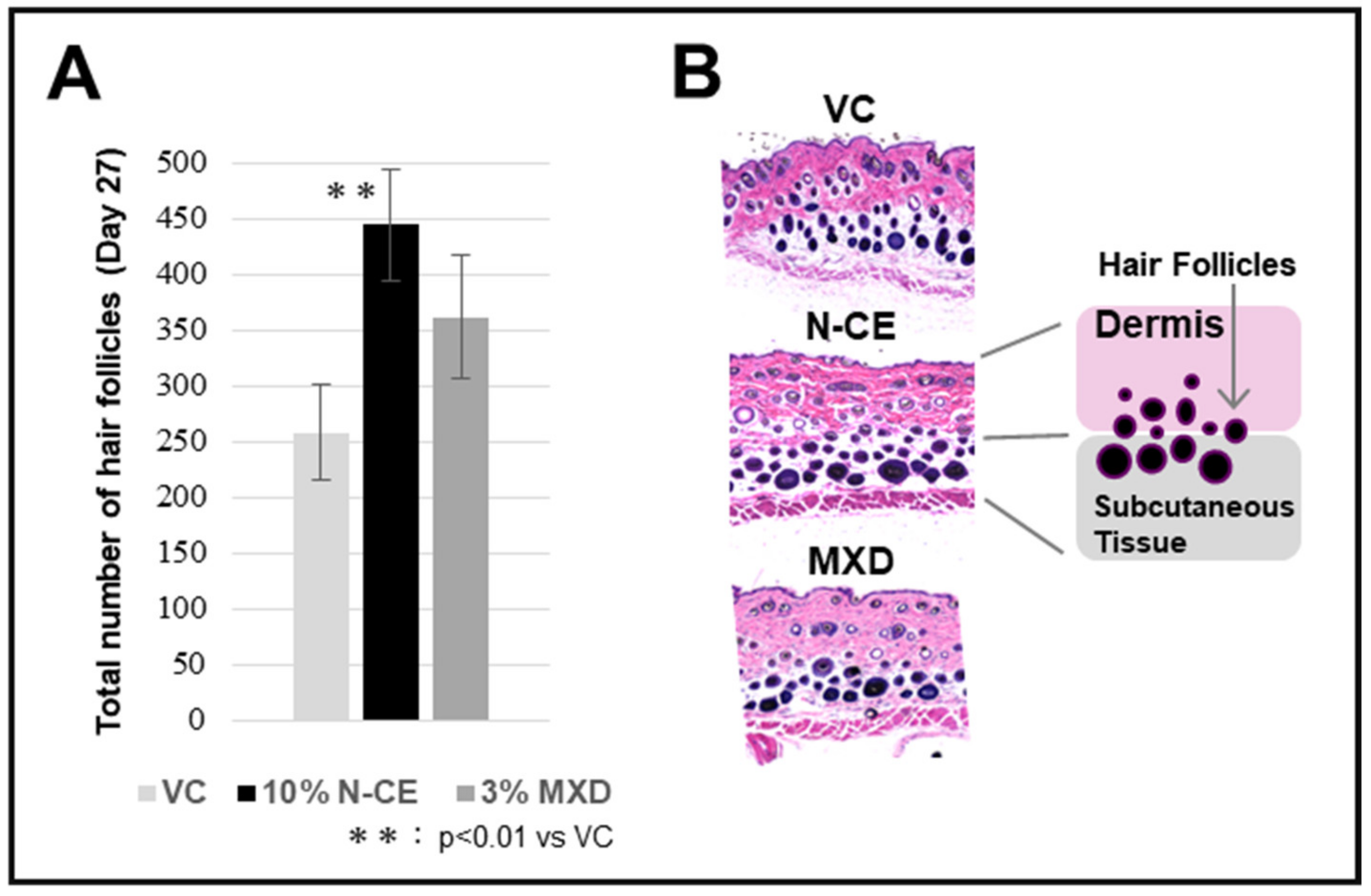
2.1. Hair Growth Promoting Action
2.2. Optical Microscopic Observation with H&E Staining
2.3. Results of the Component Analysis
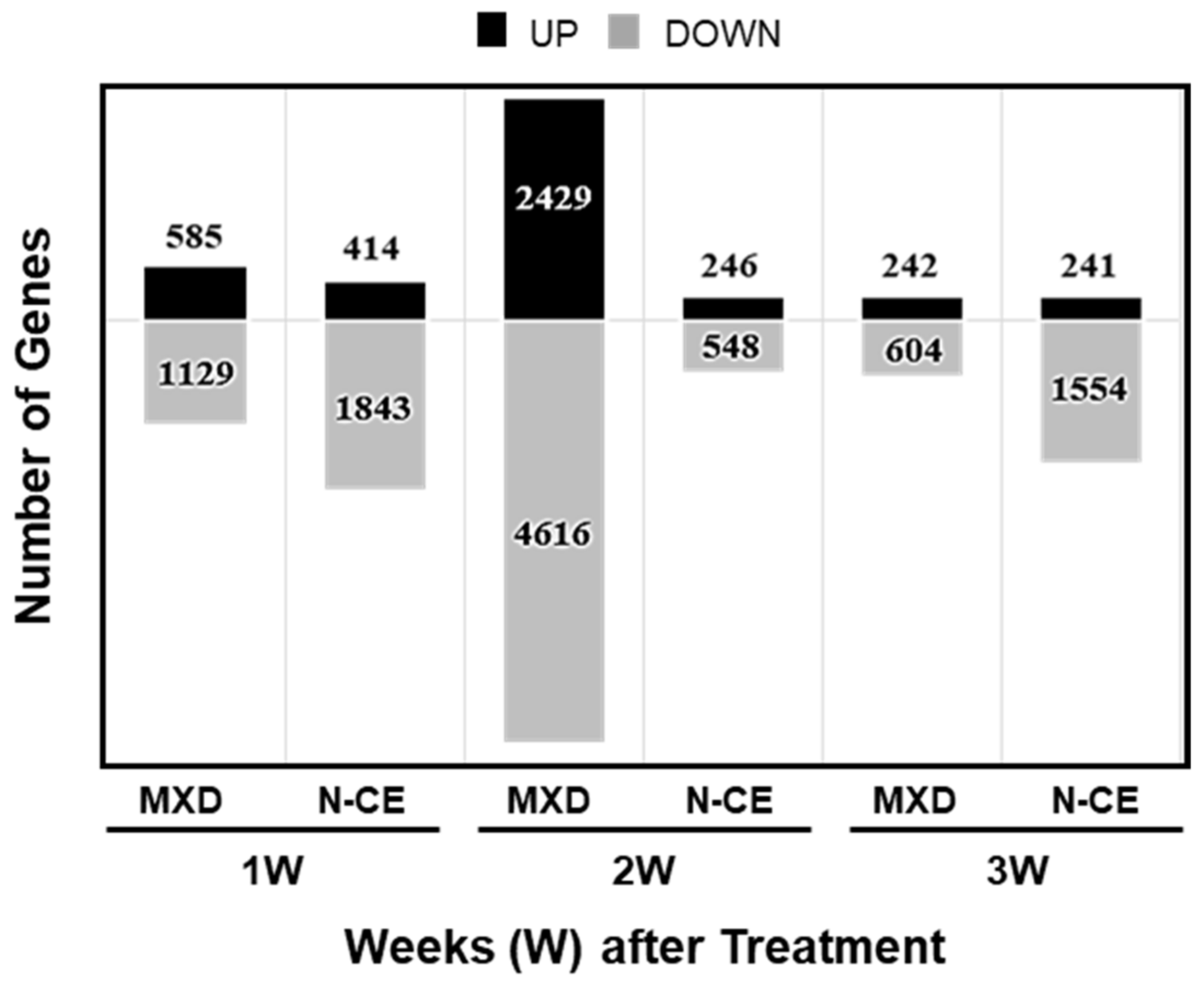
2.4. Results of the DNA Microarray Analysis
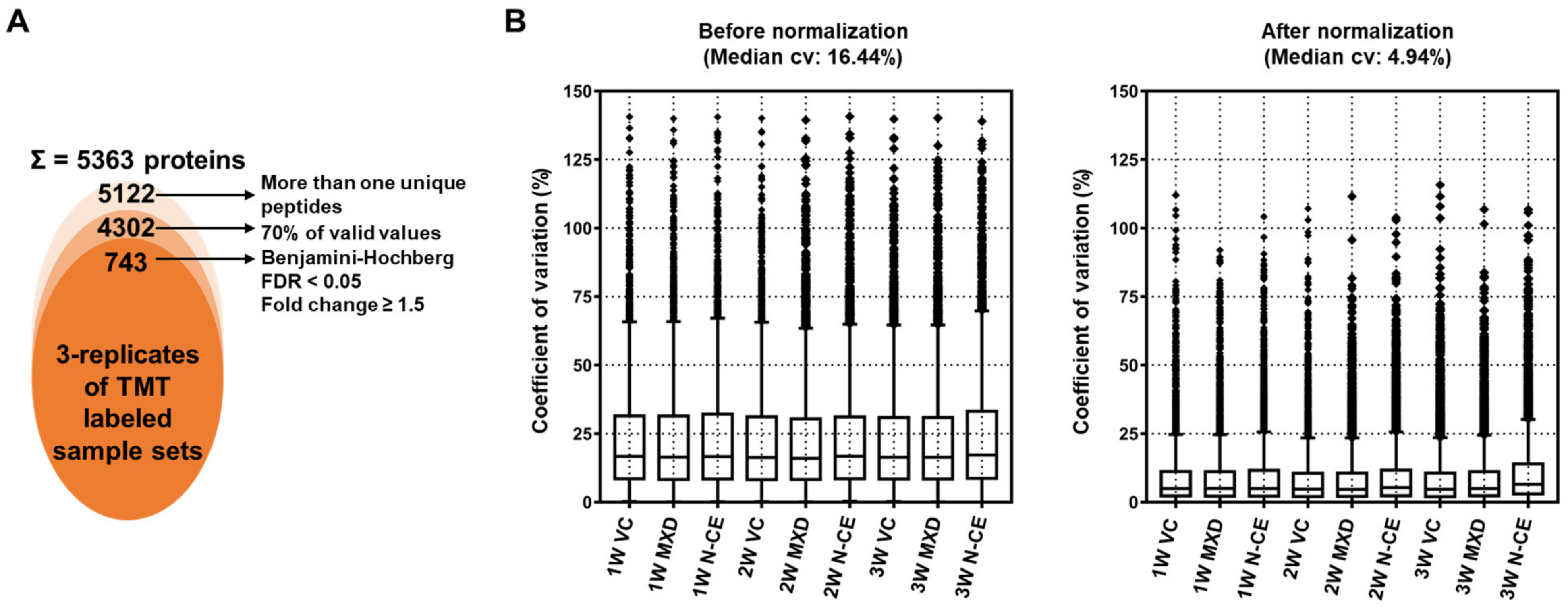
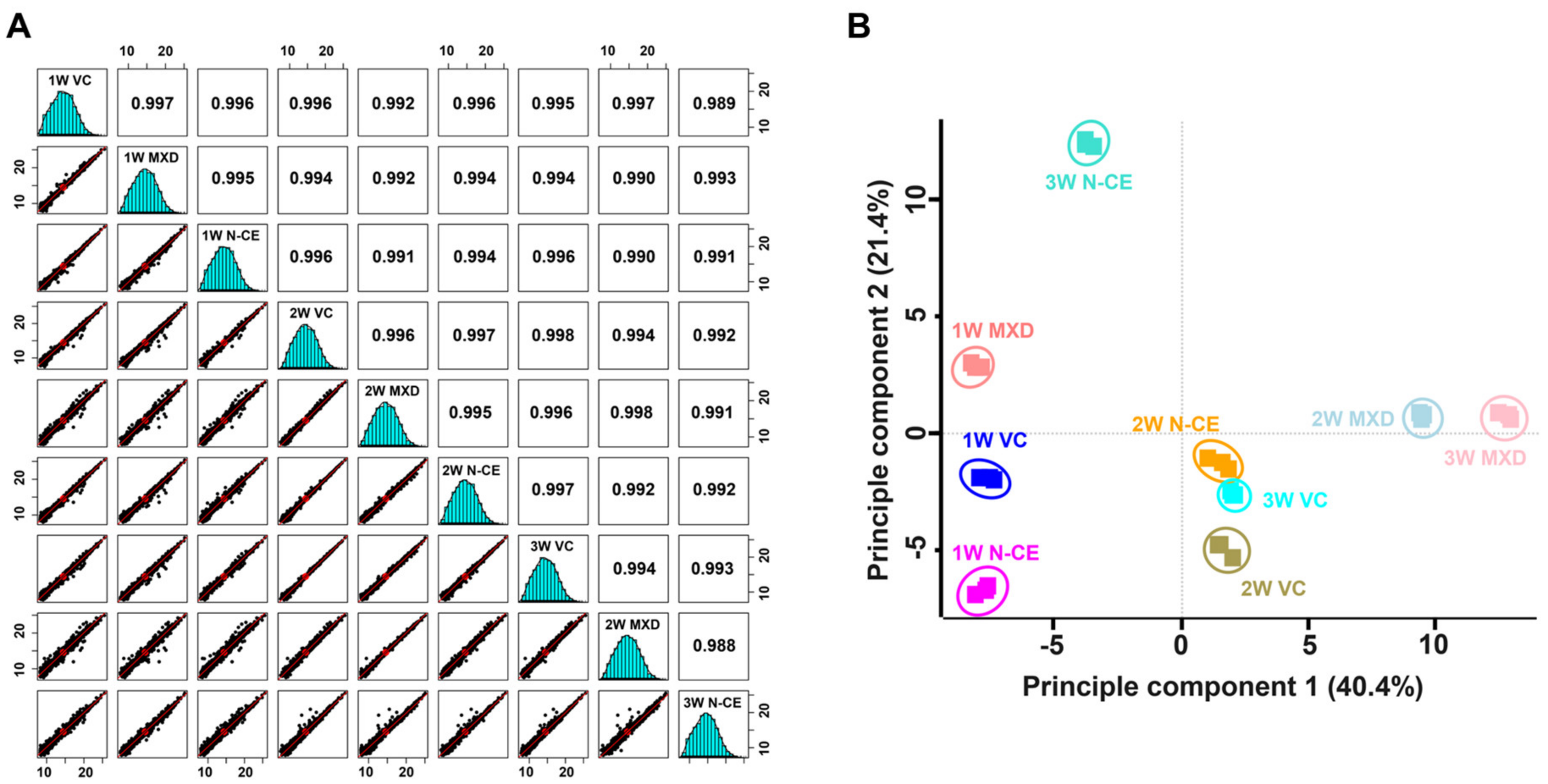
2.5. Proteome Analysis Results
TMT-Based Quantitative Proteomic Analysis
2.6. Biological Functional Enrichment Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Cell Extract by Low-Temperature Vacuum Extraction Method
4.3. Animal Experiment for the Evaluation of Hair Growth Using Mouse Back Skin Model
4.4. Histological Observations
4.5. Component Metabolite Analysis
4.6. DNA Microarray Analysis
4.7. Proteome Analysis
4.7.1. Protein Digestion by Filter-Aided Sample Preparation (FASP)
4.7.2. Peptide Labeling with Tandem Mass Tags (TMT), Desalting, and Basic pH Reversed-Phase (BPRP) Peptide Fractionation Using Stage-Tip
4.7.3. Q-Exactive MS Analysis
4.7.4. Data Processing Using MaxQuant Software and Data Analysis Using Perseus and R Program
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Talavera-Adame, D.; Newman, D.; Newman, N. Conventional and novel stem cell based therapies for androgenic alopecia. Stem Cells Cloning 2017, 10, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Meisheri, K.D.; Cipkus, L.A.; Taylor, C.J. Mechanism of action of minoxidil sulfate-induced vasodilation: A role for increased K+ permeability. J. Pharmacol. Exp. Ther. 1988, 245, 751–760. [Google Scholar] [PubMed]
- Buhl, A.E.; Waldon, D.J.; Conrad, S.J.; Mulholland, M.J.; Shull, K.L.; Kubicek, M.F.; Johnson, G.A.; Brunden, M.N.; Stefanski, K.J.; Stehle, R.G.; et al. Potassium channel conductance: A mechanism affecting hair growth both in vitro and in vivo. J. Investig. Dermatol. 1992, 98, 315–319. [Google Scholar] [CrossRef]
- Lachgar, S.; Charveron, M.; Gall, Y.; Bonafe, J.L. Minoxidil upregulates the expression of vascular endothelial growth factor in human hair dermal papilla cells. Br. J. Dermatol. 1998, 138, 407–411. [Google Scholar] [CrossRef]
- Messenger, A.G.; Rundegren, J. Minoxidil: Mechanisms of action on hair growth. Br. J. Dermatol. 2004, 150, 186–194. [Google Scholar] [CrossRef]
- Mysore, V.; Shashikumar, B.M. Guidelines on the use of finasteride in androgenetic alopecia. Indian J. Dermatol. Venereol. Leprol. 2016, 82, 128–134. [Google Scholar] [CrossRef]
- Melcangi, R.C.; Caruso, D.; Abbiati, F.; Giatti, S.; Calabrese, D.; Piazza, F.; Cavaletti, G. Neuroactive steroid levels are modified in cerebrospinal fluid and plasma of post-finasteride patients showing persistent sexual side effects and anxious/depressive symptomatology. J. Sex. Med. 2013, 10, 2598–2603. [Google Scholar] [CrossRef]
- Kakali, D.; Anu, T.S.; Ashok, M.; Beena, B.; Ramesh, B.; Anand, C.B. Eclipta alba extract with potential for hair growth promoting activity. J. Ethnopharmacol. 2009, 124, 450–456. [Google Scholar]
- Shibato, S.; Takenoya, F.; Hirabayashi, T.; Kimura, A.; Iwasaki, Y.; Toyoda, Y.; Hori, M.; Tamogami, S.; Rakwal, R.; Shioda, S. Towards identification of bioactive compounds in cold vacuum extracted double cherry blossom (Gosen-Sakura) leaves. Plant Signal. Behav. 2019, 14, e1644594. [Google Scholar] [CrossRef]
- El-Mostafa, K.; El Kharrassi, Y.; Badreddine, A.; Andreoletti, P.; Vamecq, J.; El Kebbaj, M.S.; Latruffe, N.; Lizard, G.; Nasser, B.; Cherkaoui-Malki, M. Nopal cactus (Opuntia ficus-indica) as a source of bioactive compounds for nutrition, health and disease. Molecules 2014, 19, 14879–14901. [Google Scholar] [CrossRef]
- Alimi, H.; Hfaiedh, N.; Bouoni, Z.; Hfaiedh, M.; Sakly, M.; Zourgui, L.; Rhouma, K.B. Antioxidant and antiulcerogenic activities of Opuntia ficus indica f. inermis root extract in rats. Phytomedicine 2010, 17, 1120–1126. [Google Scholar] [CrossRef] [PubMed]
- de Araújo, F.F.; de Paulo Farias, D.; Neri-Numa, I.A.; Pastore, G.M. Underutilized plants of the Cactaceae family: Nutritional aspects and technological applications. Food Chem. 2021, 362, 130196. [Google Scholar] [CrossRef] [PubMed]
- Das, G.; Lim, K.J.; Tantengco, O.A.G.; Carag, H.M.; Gonçalves, S.; Romano, A.; Das, S.K.; Coy-Barrera, E.; Shin, H.S.; Gutiérrez-Grijalva, E.P.; et al. Cactus: Chemical, nutraceutical composition and potential bio-pharmacological properties. Phytother Res. 2021, 35, 1248–1283. [Google Scholar] [CrossRef] [PubMed]
- Oura, H.; Iino, M.; Nakazawa, Y.; Tajima, M.; Ideta, R.; Nakaya, Y.; Arase, S.; Kishimoto, J. Adenosine increases anagen hair growth and thick hairs in Japanese women with female pattern hair loss: A pilot, double-blind, randomized, placebo-controlled trial. J. Dermatol. 2008, 35, 763–767. [Google Scholar] [CrossRef]
- Brzezińska-Wcisło, L. Evaluation of vitamin B6 and calcium pantothenate effectiveness on hair growth from clinical and trichographic aspects for treatment of diffuse alopecia in women. Wiad Lek. 2001, 54, 11–18. [Google Scholar]
- Wang, Z.; Nan, W.; Si, H.; Wang, S.; Haihua Zhang, H.; Li, G. Pantothenic acid promotes dermal papilla cell proliferation in hair follicles of American minks via inhibitor of DNA Binding 3/Notch signaling pathway. Life Sci. 2020, 252, 117667. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, J.; Shi, C.; Huang, Y.; Wang, Y.; Yang, T.; Yang, J. Lef1 contributes to the differentiation of bulge stem cells by nuclear translocation and cross-talk with the Notch signaling pathway. Int. J. Med. Sci. 2013, 10, 738–746. [Google Scholar] [CrossRef]
- Kljuic, A.; Bazzi, H.; Sundberg, J.P.; Martinez-Mir, A.; O’Shaughnessy, R.; Mahoney, M.G.; Levy, M.; Montagutelli, X.; Ahmad, W.; Aita, V.M.; et al. Desmoglein 4 in hair follicle differentiation and epidermal adhesion: Evidence from inherited hypotrichosis and acquired pemphigus vulgaris. Cell 2003, 113, 249–260. [Google Scholar] [CrossRef]
- Hwang, K.-A.; Hwang, Y.-L.; Lee, M.-O.; Kim, N.-R.; Roh, S.S.; Lee, Y.; Kim, C.D.; Lee, J.-H.; Choi, K.-C. Adenosine stimulates growth of dermal papilla and lengthens the anagen phase by increasing the cysteine level via fibroblast growth factors 2 and 7 in an organ culture of mouse vibrissae hair follicles. Int. J. Mol. Med. 2012, 29, 195–201. [Google Scholar]
- Raviv, S.; Bharti, K.; Lencus-Lazar, S.; Chen-Tayar, Y.; Schyr, R.; Evantal, N.; Meshorer, E.; Zilberberg, A.; Idelson, M.; Reubinoff, B.; et al. PAX6 regulates melanogenesis in the retinal pigmented epithelium through feed-forward regulatory interactions with MITF. PLoS Genet. 2014, 10, e1004360. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; Hewapathirana, S.; García-Seisdedos, D.; Kamatchinathan, S.; Kundu, D.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A Hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Bandeira, N.; Sharma, V.; Perez-Riverol, Y.; Carver, J.J.; Kundu, D.J.; García-Seisdedos, D.; Jarnuczak, A.F.; Hewapathirana, S.; Pullman, B.S.; et al. The ProteomeXchange consortium in 2020: Enabling ‘big data’ approaches in proteomics. Nucleic Acids Res. 2020, 48, D1145–D1152. [Google Scholar] [CrossRef]
- Makino, T.; Mizawa, M.; Yoshihisa, Y.; Yamamoto, S.; Tabuchi, Y.; Miyai, M.; Hibino, T.; Sasahara, M.; Shimizu, T. Trichohyalin-like 1 protein plays a crucial role in proliferation and anti-apoptosis of normal human keratinocytes and squamous cell carcinoma cells. Cell Death Discov. 2020, 6, 109. [Google Scholar] [CrossRef]
- Wu, Z.; Latendorf, T.; Meyer-Hoffert, U.; Schröder, J.-M. Identification of trichohyalin-like 1, an s100 fused-type protein selectively expressed in hair follicles. J. Investig. Dermatol. 2011, 131, 1761–1763. [Google Scholar] [CrossRef]
- Basmanav, F.B.U.; Cau, L.; Tafazzoli, A.; Méchin, M.-C.; Wolf, S.; Romano, M.T.; Valentin, F.; Wiegmann, H.; Huchenq, A.; Kandil, R.; et al. Mutations in three genes encoding proteins involved in hair dhaft formation cause uncombable hair syndrome. Am. J. Hum. Genet. 2016, 99, 1292–1304. [Google Scholar] [CrossRef]
- Patil, K.V.; Mak, K.H.-M.; Genander, M. A hairy cituation—PADIs in regeneration and alopecia. Front. Cell Dev. Biol. 2021, 9, 789676. [Google Scholar] [CrossRef]
- Xu, X.; Mannik, J.; Kudryavtseva, E.; Lin, K.K.; Flanagan, L.A.; Spencer, J.; Soto, A.; Wang, N.; Lu, Z.; Yu, Z.; et al. Co-factors of LIM domains (Clims/Ldb/Nli) regulate corneal homeostasis and maintenance of hair follicle stem cells. Dev. Biol. 2007, 312, 484–500. [Google Scholar] [CrossRef]
- Chen, X.; Xiang, Y.; Yang, L.; Lin, Y. The phenotypic characteristics of patients with athelia and tooth agenesis. Ann. Transl. Med. 2021, 9, 1583. [Google Scholar] [CrossRef]
- Christen, M.; de le Roi, M.; Jagannathan, V.; Becker, K.; Leeb, T. MYO5A Frameshift Variant in a Miniature Dachshund with Coat Color Dilution and Neurological Defects Resembling Human Griscelli Syndrome Type 1. Genes 2021, 12, 1479. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, Z.; Yang, L.; Zhang, Z.; Chen, H.; Ren, J. Novel mutations in the Myo5a gene cause a dilute coat color phenotype in mice. FASEB J. 2021, 35, e21261. [Google Scholar] [CrossRef]
- Sousa, J.B.; Diniz, C. The adenosinergic system as a therapeutic target in the vasculature: New ligands and challenges. Molecules 2017, 22, 752. [Google Scholar] [CrossRef] [PubMed]
- Chai, M.; Jiang, M.; Vergnes, L.; Fu, X.; de Barros, S.C.; Doan, N.B.; Huang, W.; Chu, J.; Jiao, J.; Herschman, H.; et al. Stimulation of hair growth by small molecules that activate autophagy. Cell Rep. 2019, 27, 3413–3421e3. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Wong, D.T.; Hirayama, A.; Soga, T.; Tomita, M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 2010, 6, 78–95. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Rakwal, R.; Shibato, J.; Sawa, C.; Saito, T.; Murayama, A.; Kuwagata, M.; Kageyama, H.; Yagi, M.; Satoh, K.; et al. Seeking gene candidates responsible for developmental origins of health and disease. Congenit Anom. 2011, 51, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Hori, M.; Nakamachi, T.; Rakwal, R.; Shibato, J.; Nakamura, K.; Wada, Y.; Tsuchikawa, D.; Yoshikawa, A.; Tamaki, K.; Shioda, S. Unraveling the ischemic brain transcriptome in a permanent middle cerebral artery occlusion mouse model by DNA microarray analysis. Dis. Model. Mech. 2012, 5, 270–283. [Google Scholar] [CrossRef]
- Hori, M.; Nakamachi, T.; Rakwal, R.; Shibato, J.; Ogawa, T.; Aiuchi, T.; Tsuruyama, T.; Tamaki, K.; Shioda, S. Transcriptomics and proteomics analyses of the PACAP38 influenced ischemic brain in permanent middle cerebral artery occlusion model mice. J. Neuroinflamm. 2012, 23, 256. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Min, C.W.; Hyeon, H.; Gupta, R.; Park, J.; Cheon, Y.E.; Lee, G.H.; Jang, J.W.; Ryu, H.W.; Lee, B.W.; Park, S.U.; et al. Integrated proteomics and metabolomics analysis highlights correlative metabolite-protein networks in soybean seeds subjected to warm-water soaking. J. Agric. Food Chem. 2020, 68, 8057–8067. [Google Scholar] [CrossRef]
- Min, C.W.; Park, J.; Bae, J.W.; Agrawal, G.K.; Rakwal, R.; Kim, Y.; Yang, P.; Kim, S.T.; Gupta, R. In-depth investigation of low-abundance proteins in matured and filling stages seeds of Glycine max employing a combination of protamine sulfate precipitation and TMT-based quantitative proteomic analysis. Cells 2020, 9, 1517. [Google Scholar] [CrossRef]
- Kim, D.K.; Park, J.; Han, D.; Yang, J.; Kim, A.; Woo, J.; Kim, Y.; Mook-Jung, I. Molecular and functional signatures in a novel Alzheimer’s disease mouse model assessed by quantitative proteomics. Mol. Neurodegener. 2018, 13, 1–19. [Google Scholar] [CrossRef]
- Pajarillo, E.A.B.; Kim, S.H.; Lee, J.Y.; Valeriano, V.D.V.; Kang, D.K. Quantitative proteogenomics and the reconstruction of the metabolic pathway in Lactobacillus mucosae LM1. Korean J. Food Sci. Anim. Resour. 2015, 35, 692–702. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Plubell, D.L.; Wilmarth, P.A.; Zhao, Y.; Fenton, A.M.; Minnier, J.; Reddy, A.P.; Klimek, J.; Yang, X.; David, L.L.; Pamir, N. Extended multiplexing of tandem mass tags (TMT) labeling reveals age and high fat diet specific proteome changes in mouse epididymal adipose tissue. Mol. Cell. Proteomics 2017, 16, 873–890. [Google Scholar] [CrossRef]
| ID | HMT DB | Relative Area | ID | HMT DB | Relative Area | ||
|---|---|---|---|---|---|---|---|
| Compound Name | Cell-Extract | Concentration (μM) | Compound Name | Cell-Extract | Concentration (μM) | ||
| N-CE | N-CE | ||||||
| C_0050 | 1-Methyl-4-imidazoleacetic acid | 3.90 × 10−5 | C_0087 | Kynurenine | 1.50 × 10−5 | ||
| C_0096 | 2’-Deoxyadenosine 5’-Deoxyadenosine | 1.42 × 10−4 | C_0034 | Melamine | 7.97 × 10−5 | ||
| C_0091 | 2’-Deoxycytidine | 6.73 × 10−4 | A_0072 | Mevalonic acid | 1.54 × 10−5 | ||
| C_0099 | 2’-Deoxyguanosine | 1.23 × 10−4 | C_0089 | N-Acetylglucosylamine | 3.36 × 10−5 | ||
| C_0029 | 2-Amino-2-(hydroxymethyl)-1,3-propanediol | 1.46 × 10−4 | A_0057 | N-Acetylmuramic acid | 1.47 × 10−4 | ||
| C_0021 | 3-Amino-2-piperidone | 8.60 × 10−5 | C_0062 | N-Methyltyramine N-Methylphenylethanolamine | 1.20 × 10−4 | ||
| C_0011 | 3-Aminopropane-1,2-diol | 1.90 × 10−4 | C_0079 | N6,N6,N6-Trimethyllysine | 1.29 × 10−5 | ||
| C_0037 | 3-Guanidinopropionic acid | 2.23 × 10−5 | C_0065 | N6-Methyllysine | 3.68 × 10−5 | ||
| C_0063 | 4-(β-Acetylaminoethyl)imidazole | 2.67 × 10−5 | C_0030 | Nicotinic acid | 6.10 × 10−5 | ||
| C_0053 | 4-Guanidinobutyric acid | 3.84 × 10−5 | C_0072 | Noradrenaline 6-Hydroxydopamine | 4.58 × 10−5 | ||
| C_0006 | 4-Methylpyrazole | 2.66 × 10−5 | A_0067 | p-Anisic acid o-Hydroxyphenylacetic acid Mandelic acid Phenoxyacetic acid | 3.48 × 10−5 | ||
| A_0050 | 4-Pyridoxic acid | 1.20 × 10−3 | A_0068 | p-Toluic acid m-Toluic acid o-Toluic acid | 1.46 × 10−5 | ||
| C_0031 | 5-Methylcytosine | 1.98 × 10−5 | A_0058 | Pantothenic acid | 1.30 × 10−5 | ||
| C_0041 | 6-Aminohexanoic acid | 2.31 × 10−4 | C_0097 | Penciclovir | 2.90 × 10−5 | ||
| A_0051 | 6-Hydroxynicotinic acid | 3.43 × 10−5 | A_0059 | Perillic acid | 1.43 × 10−4 | ||
| C_0098 | Adenosine | 1.53 × 10−5 | 0.17 | A_0060 | Phthalic acid | 1.00 × 10−4 | |
| C_0083 | ADMA | 2.33 × 10−5 | A_0061 | Pimelic acid | 1.95 × 10−5 | ||
| C_0047 | Anthranilic acid | 5.11 × 10−5 | 0.56 | A_0062 | Prostaglandin E2 | 3.96 × 10−5 | |
| C_0103 | Argininosuccinic acid | 1.94 × 10−5 | C_0067 | Pterin | 3.27 × 10−5 | ||
| A_0052 | Azelaic acid | 4.14 × 10−5 | C_0070 | Pyridoxal | 3.13 × 10−5 | ||
| C_0081 | Carbendazim | 4.61 × 10−4 | C_0071 | Pyridoxamine | 7.46 × 10−5 | ||
| C_0066 | Carnitine | 1.39 × 10−3 | C_0073 | Pyridoxine | 2.14 × 10−4 | ||
| C_0080 | Castanospermine | 2.10 × 10−4 | C_0107 | Riboflavin | 4.89 × 10−5 | ||
| C_0035 | cis-4-Hydroxyproline | 2.54 × 10−5 | A_0073 | S-Sulfocysteine | 7.82 × 10−6 | ||
| A_0008 | Citric acid | 4.36 × 10−5 | 1.60 | C_0101 | Saccharopine | 1.25 × 10−4 | |
| C_0094 | Cytidine | 2.00 × 10−5 | 0.30 | C_0084 | SDMA | 2.00 × 10−5 | |
| C_0018 | Cytosine | 4.98 × 10−4 | 11.22 | C_0092 | Ser-Glu | 1.30 × 10−5 | |
| A_0053 | Decanoic acid | 2.02 × 10−5 | A_0063 | Suberic acid | 2.88 × 10−5 | ||
| C_0017 | Diethanolamine | 4.49 × 10−4 | C_0069 | Taurocyamine | 4.62 × 10−4 | ||
| C_0002 | Ethanolamine | 3.02 × 10−4 | A_0064 | Terephthalic acid | 7.68 × 10−5 | ||
| A_0069 | Ethyl glucuronide | 3.33 × 10−5 | C_0026 | Thr | 7.14 × 10−5 | 1.21 | |
| A_0054 | Formiminoglutamic acid | 6.04 × 10−5 | C_0093 | Thymidine | 5.22 × 10−4 | 21.33 | |
| A_0055 | Formylanthranilic acid | 4.63 × 10−5 | C_0060 | Triethanolamine | 3.57 × 10−4 | ||
| C_0077 | Galactosamine Glucosamine | 1.93 × 10−4 | C_0001 | Trimethylamine | 1.27 × 10−4 | ||
| C_0106 | Gibberellic acid | 2.47 × 10−4 | C_0004 | Trimethylamine N-oxide | 7.15 × 10−5 | ||
| C_0058 | Glu | 3.21 × 10−5 | 0.72 | C_0095 | Uridine | 1.40 × 10−4 | 6.25 |
| A_0010 | Gluconic acid | 1.65 × 10−5 | 1.09 | C_0049 | Urocanic acid | 2.34 × 10−4 | |
| C_0082 | Glucosaminic acid | 1.39 × 10−5 | A_0065 | Vanillic acid | 2.85 × 10−5 | ||
| A_0070 | Glucuronic acid-1 Galacturonic acid-1 | 3.16 × 10−5 | A_0074 | XA0003 | 1.29 × 10−5 | ||
| A_0071 | Glucuronic acid-2 Galacturonic acid-2 | 3.29 × 10−5 | C_0076 | Xanthopterin | 7.62 × 10−5 | ||
| C_0012 | Glycerol | 1.37 × 10−3 | A_0066 | Xanthosine | 1.10 × 10−5 | ||
| C_0051 | Histidinol | 3.67 × 10−5 | C_0052 | XC0029 | 3.03 × 10−5 | ||
| C_0033 | Imidazole-4-acetic acid | 6.04 × 10−5 | C_0088 | XC0065 | 3.28 × 10−5 | ||
| C_0005 | Isopropanolamine | 8.90 × 10−5 | C_0054 | γ-Butyrobetaine | 7.07 × 10−5 | ||
| A_0056 | Isovaleric acid Valeric acid | 2.08 × 10−5 |
| MXD 1W | MXD 2W | MXD 3W | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Name | Average Fold | Gene Name | Average Fold | Gene Name | Average Fold | Gene Name | Average Fold | Gene Name | Average Fold | Gene Name | Average Fold |
| Cxcl5 | 89.81 | Myh7 | 0.05 | Kcnk12 | 7.12 | Cyp2a5 | 0.16 | Slc10a4 | 4.00 | Slc22a2 | 0.25 |
| Saa3 | 29.25 | Hamp | 0.13 | Entpd8 | 6.60 | Rab39 | 0.19 | Dppa3 | 3.30 | Ifng | 0.30 |
| Cxcl3 | 14.29 | Myh7 | 0.18 | Syt4 | 5.56 | Trim34a | 0.21 | Krtap19-1 | 3.06 | Dnah17 | 0.32 |
| Sprr2f | 13.23 | Hamp2 | 0.19 | Plb1 | 4.88 | Ifng | 0.22 | Sele | 3.00 | Caln1 | 0.33 |
| Chrng | 10.93 | Apol7a | 0.19 | Gltpd2 | 4.87 | Cpb1 | 0.22 | Krtap19-4 | 2.87 | Slc12a1 | 0.33 |
| Reg1 | 10.35 | Gbp2b | 0.21 | Capn8 | 4.49 | Hs1bp3 | 0.23 | Dhrs2 | 2.76 | Slc5a2 | 0.33 |
| Stfa2l1 | 9.73 | Myl2 | 0.22 | Pax6 | 4.34 | Krt2 | 0.23 | Stfa2l1 | 2.73 | Gzma | 0.34 |
| Gm5483 | 8.99 | Maz | 0.28 | Slc26a3 | 4.31 | Ccdc37 | 0.24 | Nrg1 | 2.71 | Klra7 | 0.35 |
| Sprr2d | 8.70 | Cpb1 | 0.28 | Hcn3 | 4.30 | Pkib | 0.24 | Grp | 2.66 | Cyp2c44 | 0.35 |
| Stfa2 | 8.29 | Serpina3f | 0.29 | Epb4.1 | 4.30 | Lgi1 | 0.24 | Sv2b | 2.61 | Folr4 | 0.36 |
| Ankrd1 | 8.20 | Lix1 | 0.29 | Efr3a | 4.16 | Sox2ot | 0.25 | Stfa2 | 2.60 | Foxc2 | 0.37 |
| Gdf6 | 7.81 | Myl10 | 0.30 | Pcdh8 | 4.04 | Skint11 | 0.25 | Olfr577 | 2.59 | Htra4 | 0.37 |
| Lce3f | 7.44 | Noxa1 | 0.30 | Syce1 | 3.89 | Fam19a3 | 0.26 | Olfr692 | 2.57 | Zc3h6 | 0.37 |
| S100a8 | 7.42 | Tnnt2 | 0.31 | Ret | 3.84 | Serpinb10 | 0.26 | Gpr126 | 2.56 | Olfr644 | 0.37 |
| Stfa1 | 7.17 | Ces2b | 0.31 | Sv2a | 3.81 | Serpinb3a | 0.26 | Nudcd1 | 2.55 | Il1rl1 | 0.37 |
| Krt16 | 6.89 | Reep3 | 0.32 | Foxj1 | 3.80 | Msln | 0.26 | Hsf2 | 2.52 | Esm1 | 0.38 |
| Prg4 | 6.84 | Osbpl6 | 0.33 | Lef1 | 3.76 | Mucl1 | 0.26 | Krtap16-1 | 2.50 | Prr18 | 0.39 |
| Reg3g | 6.81 | Dixdc1 | 0.33 | Cyb5r2 | 3.76 | Krt2 | 0.26 | Spink6 | 2.49 | Col9a3 | 0.39 |
| Myh3 | 6.76 | F2 | 0.33 | Dsg4 | 3.75 | Apof | 0.26 | Pcsk1 | 2.49 | H2-Ob | 0.40 |
| Myh3 | 6.74 | Ap1s3 | 0.34 | Ammecr1 | 3.75 | Gm4788 | 0.27 | Krtap19-1 | 2.47 | Ebf4 | 0.40 |
| N-CE 1W | N-CE 2W | N-CE 3W | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Name | Average Fold | Gene Name | Average Fold | Gene Name | Average Fold | Gene Name | Average Fold | Gene Name | Average Fold | Gene Name | Average Fold |
| Ccr3 | 4.87 | Myh7 | 0.03 | Myh7 | 4.88 | Il10 | 0.30 | Lpp | 6.11 | Slc15a2 | 0.20 |
| Phgr1 | 3.86 | Syt4 | 0.06 | Myh7 | 4.77 | Il10 | 0.33 | Gm7361 | 4.59 | Ccdc37 | 0.28 |
| Misp | 3.82 | Myh7 | 0.06 | Lhx9 | 3.42 | Olfr1384 | 0.35 | Gpatch2 | 3.36 | Hamp | 0.29 |
| Prg2 | 3.81 | Krtap28-10 | 0.09 | Fos | 3.25 | Timd2 | 0.36 | H2afz | 3.15 | Gbp2b | 0.32 |
| Scgb1b27 | 3.72 | Gja3 | 0.09 | Fos | 3.24 | Olfr33 | 0.37 | Sez6 | 3.10 | Paxip1 | 0.33 |
| Olfr619 | 3.55 | Gm7544 | 0.11 | Fos | 3.21 | Kif11 | 0.37 | Rcbtb2 | 3.07 | Serpinb10 | 0.33 |
| Fam163b | 3.51 | Otop2 | 0.11 | Fos | 3.21 | Gpr27 | 0.38 | Pcdhb10 | 2.79 | Ifng | 0.34 |
| Klra2 | 3.43 | Syt4 | 0.11 | Nek5 | 3.19 | Olfr578 | 0.39 | Cyp2j11 | 2.70 | Rhox9 | 0.35 |
| Fabp9 | 3.29 | Krtap4-13 | 0.11 | Fos | 3.18 | Kcnn2 | 0.39 | F12 | 2.63 | Fpr2 | 0.36 |
| Tnfrsf14 | 3.17 | Shh | 0.12 | Fos | 3.15 | Krt2 | 0.40 | Mia2 | 2.56 | Klrc3 | 0.36 |
| Cftr | 3.15 | Adamts19 | 0.12 | Fos | 3.15 | Vgll1 | 0.40 | Adamts19 | 2.49 | Igj | 0.37 |
| Cabs1 | 3.07 | S100a7a | 0.12 | Fos | 3.14 | Zcwpw1 | 0.41 | Dhrs2 | 2.43 | Tfpi2 | 0.37 |
| Dusp15 | 3.03 | Uox | 0.12 | Fos | 3.14 | Asphd1 | 0.41 | Tdrd12 | 2.40 | Gcsam | 0.37 |
| Zc3h3 | 2.92 | Tchh | 0.12 | Fos | 3.12 | Podnl1 | 0.41 | Bpifb3 | 2.39 | Il10 | 0.37 |
| Sec16b | 2.92 | Adamts18 | 0.12 | Add2 | 3.02 | Ppp1r3e | 0.42 | Olfr128 | 2.38 | Olfr513 | 0.37 |
| Fgf2 | 2.91 | Oca2 | 0.12 | Sim2 | 2.84 | Tcp11l1 | 0.42 | D17H6S53E | 2.37 | Cd28 | 0.38 |
| Angptl3 | 2.90 | Soat2 | 0.13 | Pax6 | 2.77 | Dhx38 | 0.43 | Pbx3 | 2.31 | Syce1l | 0.38 |
| Pde4d | 2.88 | Shh | 0.13 | Pappa | 2.74 | Cyp2c40 | 0.43 | Olfr131 | 2.30 | Wscd2 | 0.38 |
| Dusp15 | 2.87 | Slc7a11 | 0.13 | Gabrg1 | 2.71 | Krt2 | 0.43 | Pard3 | 2.25 | Hgf | 0.38 |
| Cntnap2 | 2.85 | Fam26d | 0.13 | Fam187a | 2.70 | Slc4a4 | 0.43 | Mgat5b | 2.23 | Gzma | 0.38 |
| MXD/Cont (Fold Change Values (LOG2)) | N-CE/Cont (Fold Change Values (LOG2)) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Symbol | 1W | Symbol | 2W | Symbol | 3W | Symbol | 1W | Symbol | 2W | Symbol | 3W |
| S100a9 | 7.03 | Krt27 | 3.11 | Epg5 | 16.10 | Ascc3 | 2.44 | Plcb4 | 3.79 | Vps51 | 28.09 |
| Ngp | 6.16 | Krt25 | 3.03 | Borcs6 | 4.38 | Tcte2 | 2.01 | Fam25c | 3.43 | Dnajc16 | 19.23 |
| Krt6b | 4.21 | Krt28 | 2.96 | Nf1 | 3.37 | Col11a1 | 1.95 | Ect2 | 3.09 | Prkar1a | 5.65 |
| Klf10 | 3.00 | Krt26 | 2.85 | Sptlc2 | 3.36 | Gm45927 | 1.75 | Epg5 | 3.07 | Sil1 | 5.19 |
| Abcb9 | 2.86 | Pinlyp | 2.78 | Krt25 | 2.84 | Ldb1 | 1.74 | Arr3 | 2.73 | Smarcc1 | 5.11 |
| Fgg | 2.52 | Tchh | 2.76 | Krt27 | 2.71 | Fam117b | 1.71 | Timm13 | 2.70 | Cdc42bpg | 3.95 |
| Fgb | 2.42 | Nf1 | 2.64 | Naa40 | 2.59 | Xpo4 | 1.71 | Smc2 | 2.70 | Sptlc2 | 3.73 |
| Fga | 2.19 | Crym | 2.58 | Fbp1 | 2.57 | Gm10964 | 1.66 | Lig3 | 2.64 | Ston1 | 3.67 |
| Vps51 | 2.17 | Fbp1 | 2.29 | Tchh | 2.47 | Hrnr | 1.64 | Uqcc2 | 2.43 | Borcs6 | 3.29 |
| Pla2g4b | 2.14 | Cgnl1 | 2.19 | Pinlyp | 2.43 | Vps35l | 1.63 | Fuom | 2.21 | Smc2 | 2.96 |
| Dnajc16 | 2.09 | Tchhl1 | 2.18 | Crym | 2.36 | Alox8 | 1.63 | Cgn | 2.00 | Manba | 2.75 |
| Myo1e | 1.96 | Padi3 | 2.16 | Tchhl1 | 2.29 | Krt6b | 1.62 | S100a6 | 1.90 | Lig3 | 2.69 |
| Commd3 | 1.78 | Padi1 | 2.12 | Krt28 | 2.26 | Kctd1 | 1.58 | Elmo2 | 1.90 | Myo5a | 2.66 |
| Tmem50a | 1.66 | Padi4 | 2.10 | Krt26 | 2.25 | Pla2g4b | 1.57 | Commd3 | 1.88 | Epb41l2 | 2.61 |
| Prkar1a | 1.63 | Prr9 | 2.09 | Padi4 | 2.22 | Itpkc | 1.55 | Cgnl1 | 1.85 | Borcs5 | 2.53 |
| Pafah1b2 | 1.61 | Map3k10 | 2.08 | Map3k10 | 2.17 | Rad50 | 1.54 | Selenom | 1.85 | Degs1 | 2.51 |
| Ascc3 | 1.59 | Crnn | 2.08 | Cbs | 2.11 | H2ac20 | 1.50 | Slc12a4 | 1.83 | Mfap5 | 2.45 |
| Fn1 | 1.54 | Krtap15 | 2.06 | Krtap15 | 2.10 | Fxyd1 | 1.42 | Agrn | 1.82 | Cstf3 | 2.40 |
| Tcte2 | 1.54 | Cbs | 2.04 | Gnmt | 2.09 | Itgb3 | 1.41 | Ahcyl2 | 1.77 | Ascc3 | 2.38 |
| Lyz2 | 1.53 | Mab21l4 | 2.01 | Rpl24 | 2.07 | Borcs6 | 1.39 | Banf1 | 1.75 | Acp5 | 2.37 |
| Plcb4 | −2.33 | Tnnc2 | −1.76 | Vps51 | −5.64 | Arr3 | −9.32 | Gm10964 | −2.15 | Tmem87a | −2.46 |
| Myl1 | −1.91 | Myl1 | −1.71 | Dnajc16 | −2.80 | Smc2 | −7.52 | Krtap21-1 | −1.52 | Cyfip2 | −2.25 |
| Arr3 | −1.82 | Col11a1 | −1.70 | 4930562C15Rik | −2.08 | Ect2 | −4.86 | Lgals3bp | −1.50 | Scyl1 | −2.16 |
| Tnnc2 | −1.72 | Vps51 | −1.60 | Ston1 | −2.04 | Acp5 | −3.33 | Krt76 | −1.50 | Ephx3 | −2.10 |
| Atp2a1 | −1.65 | 4930562C15Rik | −1.60 | Mybpc1 | −1.97 | Cgn | −3.24 | Gm11567 | −1.49 | Gm45927 | −2.09 |
| Ccdc127 | −1.65 | Kctd1 | −1.56 | Rad50 | −1.91 | Plcb4 | −3.14 | Gm45927 | −1.44 | S100a11 | −2.06 |
| Trdn | −1.63 | Krt76 | −1.56 | Pacsin2 | −1.87 | Slc12a4 | −3.10 | Gm11938 | −1.43 | Sepsecs | −2.04 |
| Spag7 | −1.57 | Nucks1 | −1.46 | Ttn | −1.73 | Lig3 | −2.97 | Pla2g4b | −1.43 | S100a6 | −1.99 |
| Calm3 | −1.55 | Tsc22d1 | −1.44 | Smc5 | −1.73 | Nf1 | −2.70 | Gm11565 | −1.40 | Car13 | −1.98 |
| 4930562C15Rik | −1.54 | Tgfbi | −1.43 | Tnni2 | −1.66 | Fam25c | −2.35 | Rgs13 | −1.40 | Epb41 | −1.97 |
| Epg5 | −1.53 | Ncdn | −1.43 | Serpinb7 | −1.66 | Rfc4 | −2.19 | Dera | −1.40 | Gng12 | −1.94 |
| Rgs13 | −1.52 | Rad50 | −1.43 | Plcl1 | −1.64 | Agrn | −2.08 | Tcte2 | −1.39 | Get4 | −1.93 |
| Pvalb | −1.52 | Gm45927 | −1.42 | Atp2a1 | −1.63 | Vps33b | −2.07 | Rad50 | −1.38 | Ccdc127 | −1.93 |
| Selenom | −1.50 | Slc25a4 | −1.41 | Atp2a3 | −1.60 | Manba | −1.99 | Cnbp | −1.36 | Tsc22d1 | −1.92 |
| Rpl24 | −1.49 | Mcpt4 | −1.40 | Eno1 | −1.59 | Krt76 | −1.93 | Col11a1 | −1.35 | Cav3 | −1.89 |
| Scd1 | −1.48 | Apobec2 | −1.40 | Lmod3 | −1.58 | Armt1 | −1.92 | Eno2 | −1.34 | Plcl1 | −1.88 |
| Plcl1 | −1.48 | Trim16 | −1.40 | Slc25a4 | −1.58 | Borcs5 | −1.89 | Sdhc | −1.32 | Eno2 | −1.84 |
| Sdhc | −1.47 | Mapt | −1.40 | Uggt1 | −1.58 | Epg5 | −1.87 | Sos1 | −1.31 | Itpkc | −1.83 |
| Fabp3 | −1.46 | Calm3 | −1.38 | Ndufa3 | −1.57 | Hmcn1 | −1.85 | Kctd1 | −1.30 | Hook1 | −1.80 |
| Atp2a3 | −1.45 | Tnni2 | −1.38 | Myh1 | −1.57 | Fam83h | −1.84 | Mb | −1.30 | Commd9 | −1.79 |
| MXD—UP: Microarray | MXD—UP: Proteome | ||||
|---|---|---|---|---|---|
| 1W | 2W | 3W | 1W | 2W | 3W |
| Keratin | Keratin | Keratin | Keratin | Keratin | Keratin |
| Disulfide bond | Cell division | Melanin biosynthesis | Intermediate filament | Intermediate filament | Intermediate filament |
| Keratinization | Cell cycle | Albinism | Lysosome | Lipid biosynthesis | Fatty acid metabolism |
| Collagen | Intermediate filament | Kinetochore | Lipid metabolism | Lipid metabolism | Protein phosphatase |
| Inflammatory response | Citrullination | Acyltransferase | Fatty acid metabolism | Lysosome | |
| Chemotaxis | Microtubule | Cell adhesion | Cell cycle | Fatty acid biosynthesis | |
| Cell adhesion | Centromere | Hemostasis | Acyltransferase | Lipid metabolism | |
| Myosin | Melanin biosynthesis | Blood coagulation | Lysosome | Pyridoxal phosphate | |
| Intermediate filament | Disease mutation | Cysteine biosynthesis | Fatty acid biosynthesis | Lipid biosynthesis | |
| Cytokine | Wnt signaling pathway | Cytoskeleton | Pyridoxal phosphate | Cysteine biosynthesis | |
| Melanin biosynthesis | Keratinization | Fatty acid metabolism | Protein phosphatase | Cell junction | |
| Acute phase | DNA repair | Disulfide bond | Cell junction | ||
| Albinism | Actin-binding | Amino-acid biosynthesis | Cysteine biosynthesis | ||
| Amidation | Homeobox | Cytoskeleton | |||
| Proteoglycan | Albinism | Heparin-binding | |||
| Antimicrobial | Lipid-binding | Cell division | |||
| Zinc transport | Endoplasmic reticulum | ||||
| Tyrosine-protein kinase | Proteoglycan | ||||
| MXD—Down: Microarray | MXD—Down: Proteome | ||||
| 1W | 2W | 3W | 1W | 2W | 3W |
| Muscle protein | Disulfide bond | Disulfide bond | Muscle protein | Muscle protein | Muscle protein |
| Lipid metabolism | Muscle protein | Muscle protein | Keratin | Endocytosis | Calmodulin-binding |
| Lipid biosynthesis | Sarcoplasmic reticulum | Actin-binding | Intermediate filament | Sarcoplasmic reticulum | Myosin |
| Thick filament | Lectin | Sarcoplasmic reticulum | Myosin | Thick filament | Respiratory chain |
| Fatty acid metabolism | Actin-binding | Cell adhesion | Thick filament | Calmodulin-binding | Sarcoplasmic reticulum |
| Calmodulin-binding | Cytokine | Collagen | Calmodulin-binding | Myosin | Thick filament |
| Oxidoreductase | Inflammatory response | Thick filament | Actin-binding | Lysosome | Actin-binding |
| Sarcoplasmic reticulum | Myogenesis | Growth factor | Metal-binding | Actin-binding | Cytoskeleton |
| Actin-binding | Calmodulin-binding | Sugar | Endoplasmic reticulum | Mitochondrion | Metal-binding |
| Myosin | Lipoprotein | Heme | Endocytosis | Cytoskeleton | Lysosome |
| Fatty acid biosynthesis | Thick filament | Calmodulin-binding | Cell division | Prenylation | |
| Heme | Growth factor | Iron | Disulfide bond | ||
| Growth factor | Heme | Lysosome | |||
| Lipid | Lipid metabolism | ||||
| Disulfide bond | Myosin | ||||
| Acylferase | Chemotaxis | ||||
| Lipoprotein | Osteogenesis | ||||
| Glucose metabolism | Heparin-binding | ||||
| N-CE—UP: Microarray | N-CE—UP: Proteome | ||||
|---|---|---|---|---|---|
| 1W | 2W | 3W | 1W | 2W | 3W |
| Immunity | Homeobox | Citrullination | Keratin | Keratin | Keratin |
| Lectin | Disease mutation | Cell adhesion | Intermediate filament | Lysosome | Intermediate filament |
| Disulfide bond | Developmental protein | Disease mutation | Metal-binding | Lipid biosynthesis | Lysosome |
| Innate immunity | Spermatogenesis | Acetylation | Calmodulin-binding | Cytoskeleton | Cytoskeleton |
| Cytokine | Zymogen | Homeobox | Hemostasis | Cell adhesion | Keratinization |
| Inflammatory response | Blood coagulation | Lipid metabolism | Calmodulin-binding | ||
| Chemotaxis | Muscle protein | Fatty acid metabolism | Cysteine biosynthesis | ||
| Antiviral defense | Actin-binding | Cell cycle | Proteoglycan | ||
| Acute phase | Cytoskeleton | Sarcoplasmic reticulum | Myosin | ||
| Golgi apparatus | Disease mutation | Cysteine biosynthesis | Actin-binding | ||
| Myosin | Intermediate filament | Acyltransferase | |||
| N-CE—Down: Microarray | Endocytosis | Proteoglycan | Aminopeptidase | ||
| 1W | 2W | 3W | Lipid-binding | Myosin | Protein phosphatase |
| Keratin | Lectin | G-protein coupled | Prenylation | ||
| Cell cycle | Disulfide bond | Disulfide bond | Cell division | ||
| Mitosis | Muscle protein | Olfaction | |||
| Cell division | Pheromone-binding | Sensory transduction | N-CE—Down: Proteome | ||
| Kinetochore | Calmodulin-binding | Cell adhesion | 1W | 2W | 3W |
| Intermediate filament | Oxygen | Sulfation | Keratin | Keratin | Muscle protein |
| Microtubule | Lipoprotein | Lectin | Fatty acid metabolism | Intermediate filament | Metal-binding |
| DNA repair | Adaptive immunity | Lysosome | Muscle protein | Sarcoplasmic reticulum | |
| DNA replication | Heme | Intermediate filament | Lipid-binding | Fatty acid metabolism | |
| Melanin biosynthesis | Hormone | Lipid biosynthesis | Metal-binding | Fatty acid biosynthesis | |
| Calmodulin-binding | Immunity | Muscle protein | Myosin | Lipid biosynthesis | |
| Citrullination | Iron transport | Fatty acid biosynthesis | Endocytosis | Lipid metabolism | |
| Albinism | Homeobox | Lipid metabolism | Keratinization | Endocytosis | |
| Wnt ing pathway | Myogenesis | Cysteine biosynthesis | Actin-binding | Keratin | |
| Helicase | Cytokine | Cell cycle | Calmodulin-binding | ||
| Osteogenesis | Zymogen | Cell adhesion | |||
| Muscle protein | Actin-binding | Proteoglycan | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shibato, J.; Takenoya, F.; Kimura, A.; Min, C.W.; Yamashita, M.; Gupta, R.; Kim, S.T.; Rakwal, R.; Shioda, S. Examining the Effect of Notocactus ottonis Cold Vacuum Isolated Plant Cell Extract on Hair Growth in C57BL/6 Mice Using a Combination of Physiological and OMICS Analyses. Molecules 2023, 28, 1565. https://doi.org/10.3390/molecules28041565
Shibato J, Takenoya F, Kimura A, Min CW, Yamashita M, Gupta R, Kim ST, Rakwal R, Shioda S. Examining the Effect of Notocactus ottonis Cold Vacuum Isolated Plant Cell Extract on Hair Growth in C57BL/6 Mice Using a Combination of Physiological and OMICS Analyses. Molecules. 2023; 28(4):1565. https://doi.org/10.3390/molecules28041565
Chicago/Turabian StyleShibato, Junko, Fumiko Takenoya, Ai Kimura, Cheol Woo Min, Michio Yamashita, Ravi Gupta, Sun Tae Kim, Randeep Rakwal, and Seiji Shioda. 2023. "Examining the Effect of Notocactus ottonis Cold Vacuum Isolated Plant Cell Extract on Hair Growth in C57BL/6 Mice Using a Combination of Physiological and OMICS Analyses" Molecules 28, no. 4: 1565. https://doi.org/10.3390/molecules28041565
APA StyleShibato, J., Takenoya, F., Kimura, A., Min, C. W., Yamashita, M., Gupta, R., Kim, S. T., Rakwal, R., & Shioda, S. (2023). Examining the Effect of Notocactus ottonis Cold Vacuum Isolated Plant Cell Extract on Hair Growth in C57BL/6 Mice Using a Combination of Physiological and OMICS Analyses. Molecules, 28(4), 1565. https://doi.org/10.3390/molecules28041565








