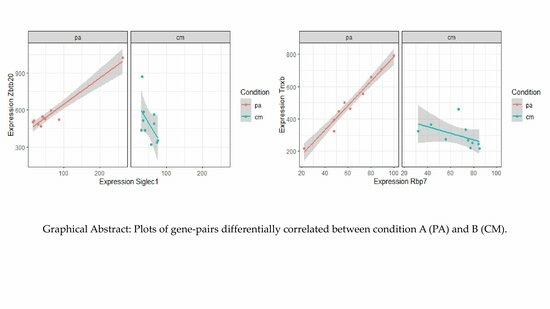
Differential Correlation of Transcriptome Data Reveals Gene Pairs and Pathways Involved in Treatment of Citrobacter rodentium Infection with Bioactive Punicalagin
Abstract
:1. Introduction
2. Results
2.1. Differential Expression of PA vs. CM
2.2. Top Differentially Correlated Genes with Condition A (PA) across Classes +/0 and +/−
2.3. Top Differentially Correlated Gene Pairs from Classes +/0 and +/−
2.4. Module-Based Differential Correlation of Genes between Condition A and Condition B
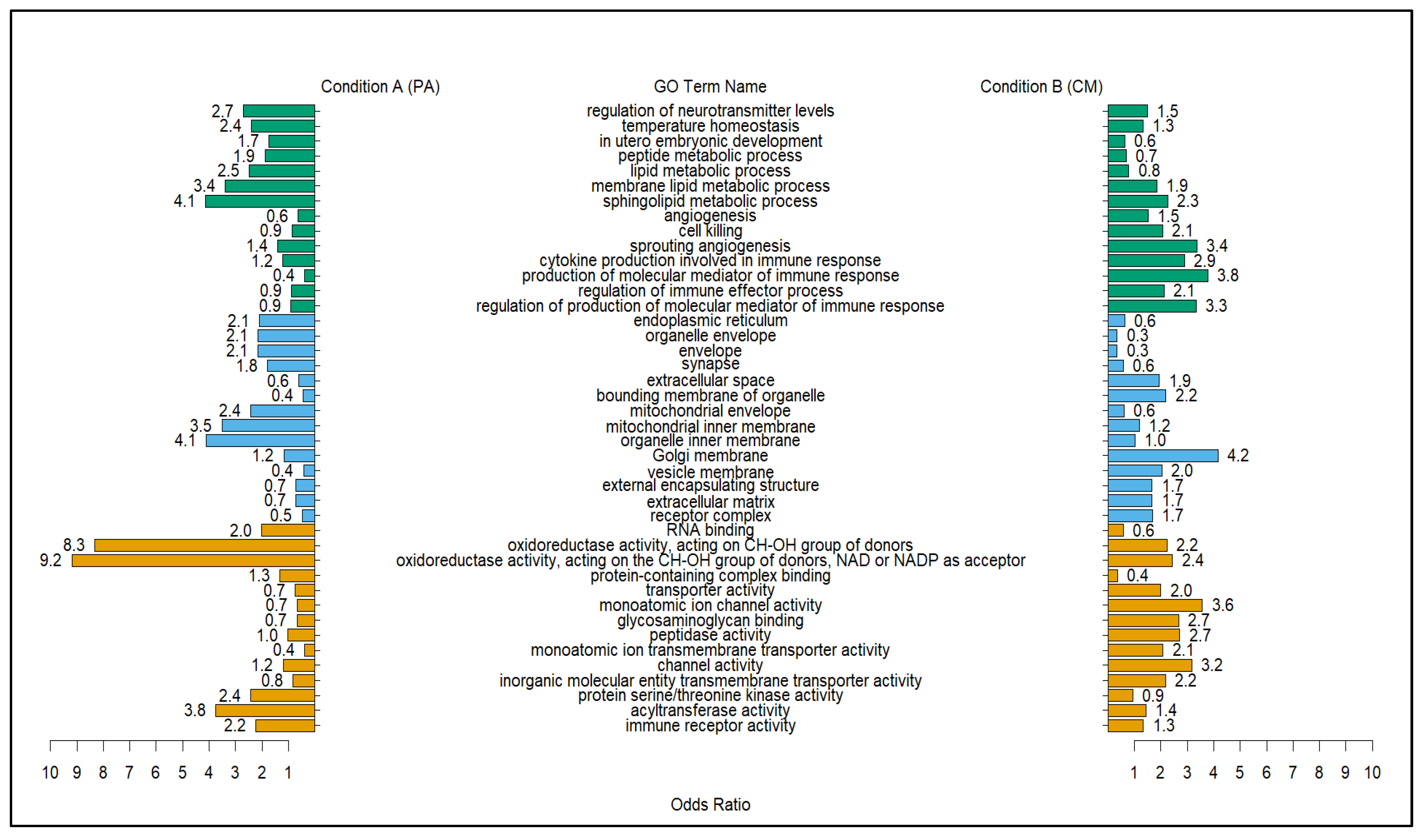
2.5. Top GO Terms Differentially Correlated with Modules with Positive Correlation with PA Treatment
3. Discussion
3.1. Annotated Functions of Positively Correlated Top Genes and Gene Pairs Indicated That Punicalagin-Treated Mice Had a Broad Spectrum-like Impact against C. rodentium
3.2. PA vs. CM Differentially Correlated GO Terms Based on Positively Correlated Gene Modules Further Elucidate the Effects of Punicalagin (PA) against Bacterial Parasites Compared to the Infected Untreated Group (CM)
4. Materials and Methods
4.1. Animals
4.2. RNA Sequencing and Expression Analysis
4.3. Differential Correlation and Gene Ontology Analysis
4.4. Differential Correlation Enrichment of Gene-Specific and Module-Based Gene Ontology (GO)
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Waller, P.J. The Development of Anthelmintic Resistance in Ruminant Livestock. Acta Trop. 1994, 56, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Capela, R.; Moreira, R.; Lopes, F. An Overview of Drug Resistance in Protozoal Diseases. Int. J. Mol. Sci. 2019, 20, 5748. [Google Scholar] [CrossRef]
- Liu, F.; Smith, A.D.; Wang, T.T.Y.; Pham, Q.; Yang, H.; Li, R.W. Ellagitannin Punicalagin Disrupts the Pathways Related to Bacterial Growth and Affects Multiple Pattern Recognition Receptor Signaling by Acting as a Selective Histone Deacetylase Inhibitor. J. Agric. Food Chem. 2023, 71, 5016–5026. [Google Scholar] [CrossRef] [PubMed]
- Ramlagan, P.; Labib, R.M.; Farag, M.A.; Neergheen, V.S. Advances towards the Analysis, Metabolism and Health Benefits of Punicalagin, One of the Largest Ellagitannin from Plants, with Future Perspectives. Phytomed. Plus 2022, 2, 100313. [Google Scholar] [CrossRef]
- Suriyaprom, S.; Mosoni, P.; Leroy, S.; Kaewkod, T.; Desvaux, M.; Tragoolpua, Y. Antioxidants of Fruit Extracts as Antimicrobial Agents against Pathogenic Bacteria. Antioxidants 2022, 11, 602. [Google Scholar] [CrossRef] [PubMed]
- Amer, O.S.O.; Dkhil, M.A.; Hikal, W.M.; Al-Quraishy, S. Antioxidant and Anti-Inflammatory Activities of Pomegranate (Punica granatum) on Eimeria papillata-Induced Infection in Mice. BioMed Res. Int. 2015, 2015, 219670. [Google Scholar] [CrossRef]
- Al-Megrin, W.A. In Vivo Study of Pomegranate (Punica granatum) Peel Extract Efficacy against Giardia Lamblia in Infected Experimental Mice. Asian Pac. J. Trop. Biomed. 2017, 7, 59–63. [Google Scholar] [CrossRef]
- Seeram, N.P.; Adams, L.S.; Henning, S.M.; Niu, Y.; Zhang, Y.; Nair, M.G.; Heber, D. In Vitro Antiproliferative, Apoptotic and Antioxidant Activities of Punicalagin, Ellagic Acid and a Total Pomegranate Tannin Extract Are Enhanced in Combination with Other Polyphenols as Found in Pomegranate Juice. J. Nutr. Biochem. 2005, 16, 360–367. [Google Scholar] [CrossRef]
- Čolić, M.; Mihajlović, D.; Bekić, M.; Marković, M.; Dragišić, B.; Tomić, S.; Miljuš, N.; Šavikin, K.; Škrbić, R. Immunomodulatory Activity of Punicalagin, Punicalin, and Ellagic Acid Differs from the Effect of Pomegranate Peel Extract. Molecules 2022, 27, 7871. [Google Scholar] [CrossRef]
- El Kar, C.; Ferchichi, A.; Attia, F.; Bouajila, J. Pomegranate (Punica granatum) Juices: Chemical Composition, Micronutrient Cations, and Antioxidant Capacity. J. Food Sci. 2011, 76, C795–C800. [Google Scholar] [CrossRef]
- Liu, F.; Smith, A.D.; Wang, T.T.Y.; Pham, Q.; Yang, H.; Li, R.W. Multi-Omics Analysis Detected Multiple Pathways by Which Pomegranate Punicalagin Exerts Its Biological Effects in Modulating Host-Microbiota Interactions in Murine Colitis Models. Food Funct. 2023, 14, 3824–3837. [Google Scholar] [CrossRef] [PubMed]
- Lanusse, C.; Canton, C.; Virkel, G.; Alvarez, L.; Costa-Junior, L.; Lifschitz, A. Strategies to Optimize the Efficacy of Anthelmintic Drugs in Ruminants. Trends Parasitol. 2018, 34, 664–682. [Google Scholar] [CrossRef] [PubMed]
- Stefanou, V.; Tsakni, A.; Timbis, D.; Vougiouka, P.A.; Doumi, I.; Maronikolaki, I.; Siatras, N.; Lougovois, V. Pomegranate as an Antibacterial Agent against Pathogens and at the same Time Advantageous to Beneficial Bacteria: A Review. Int. J. Adv. Res. Microbiol. Immunol. 2020, 2, 1–13. [Google Scholar]
- Smith, A.D.; George, N.S.; Cheung, L.; Bhagavathy, G.V.; Luthria, D.L.; John, K.M.; Bhagwat, A.A. Pomegranate Peel Extract Reduced Colonic Damage and Bacterial Translocation in a Mouse Model of Infectious Colitis Induced by Citrobacter Rodentium. Nutr. Res. 2019, 73, 27–37. [Google Scholar] [CrossRef]
- Cunningham, F.; E Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2021, 50, D988–D995. [Google Scholar] [CrossRef]
- UniProt Consortium; Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bye-A-Jee, H.; et al. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2022, 51, D523–D531. [Google Scholar] [CrossRef]
- Maglott, D.; Ostell, J.; Pruitt, K.D.; Tatusova, T. Entrez Gene: Gene-centered information at NCBI. Nucleic Acids Res. 2010, 39, D52–D57. [Google Scholar] [CrossRef]
- Eppig, J.T. Mouse Genome Informatics (Mgi) Resource: Genetic, Genomic, and Biological Knowledgebase for the Laboratory Mouse. ILAR J. 2017, 58, 17–41. [Google Scholar] [CrossRef]
- Eppig, J.T.; Smith, C.L.; Blake, J.A.; Ringwald, M.; Kadin, J.A.; Richardson, J.E.; Bult, C.J. Mouse Genome Informatics (Mgi): Resources for Mining Mouse Genetic, Genomic, and Biological Data in Support of Primary and Translational Research. Methods Mol. Biol. 2016, 1488, 47–73. [Google Scholar] [CrossRef]
- Charron, A.J.; Sibley, L.D. Host Cells: Mobilizable Lipid Resources for the Intracellular Parasite Toxoplasma gondii. J. Cell Sci. 2002, 115, 3049–3059. [Google Scholar] [CrossRef]
- Vallochi, A.L.; Teixeira, L.; Oliveira, K.d.S.; Maya-Monteiro, C.M.; Bozza, P.T. Lipid Droplet, a Key Player in Host-Parasite Interactions. Front. Immunol. 2018, 9, 1022. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Keen, H.L.; Lu, K.-T.; Liu, X.; Wu, J.; Davis, D.R.; Ibeawuchi, S.-R.C.; Vogel, S.; Quelle, F.W.; Sigmund, C.D. Retinol-Binding Protein 7 Is an Endothelium-Specific Pparγ Cofactor Mediating an Antioxidant Response through Adiponectin. J. Clin. Investig. 2017, 2, e91738. [Google Scholar] [CrossRef] [PubMed]
- Krzyzanowska, A.K.; Haynes II, R.A.; Kovalovsky, D.; Lin, H.-C.; Osorio, L.; Edelblum, K.L.; Corcoran, L.M.; Rabson, A.B.; Denzin, L.K.; Sant’angelo, D.B. Zbtb20 Identifies and Controls a Thymus-Derived Population of Regulatory T Cells That Play a Role in Intestinal Homeostasis. Sci. Immunol. 2022, 7, eabf3717. [Google Scholar] [CrossRef] [PubMed]
- Naim, N.; Amrit, F.R.G.; McClendon, T.B.; Yanowitz, J.L.; Ghazi, A. The Molecular Tug of War between Immunity and Fertility: Emergence of Conserved Signaling Pathways and Regulatory Mechanisms. BioEssays 2020, 42. [Google Scholar] [CrossRef] [PubMed]
- Kaiaty, A.M.; Salib, F.A.; El-Gameel, S.M.; Hussien, A.M.; Kamel, M.S. Anthelmintic Activity of Pomegranate Peel Extract (Punica granatum) and Synthetic Anthelmintics against Gastrointestinal Nematodes in Cattle, Sheep, Goats, and Buffalos: In Vivo Study. Parasitol. Res. 2021, 120, 3883–3893. [Google Scholar] [CrossRef]
- Straub, A.C.; Butcher, J.T.; Billaud, M.; Mutchler, S.M.; Artamonov, M.V.; Nguyen, A.T.; Johnson, T.; Best, A.K.; Miller, M.P.; Palmer, L.A.; et al. Hemoglobin α/eNOS Coupling at Myoendothelial Junctions Is Required for Nitric Oxide Scavenging During Vasoconstriction. Arter. Thromb. Vasc. Biol. 2014, 34, 2594–2600. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with Hisat2 and Hisat-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Brown, J.W.; Pirrung, M.; McCue, L.A. Fqc Dashboard: Integrates Fastqc Results into a Web-Based, Interactive, and Extensible Fastq Quality Control Tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for Rna-Seq Data with Deseq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. Feature counts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- McKenzie, A.T.; Katsyv, I.; Song, W.-M.; Wang, M.; Zhang, B. DGCA: A comprehensive R package for Differential Gene Correlation Analysis. BMC Syst. Biol. 2016, 10, 106. [Google Scholar] [CrossRef] [PubMed]
- Song, W.-M.; Zhang, B. Multiscale Embedded Gene Co-expression Network Analysis. PLOS Comput. Biol. 2015, 11, e1004574. [Google Scholar] [CrossRef] [PubMed]

| Genes in Class +/0 | # of DC Pairs | Genes in Class +/− | # of DC Pairs | |
|---|---|---|---|---|
| 1 | Hbb-bt | 658 | Plin1 | 91 |
| 2 | Hba-a1 | 638 | Folr2 | 68 |
| 3 | Hbb-bs | 627 | Gk | 55 |
| 4 | Hba-a2 | 619 | Siglec1 | 54 |
| 5 | Folr2 | 593 | Gpr179 | 51 |
| 6 | Rbp7 | 551 | Hbb-bs | 48 |
| 7 | Tiam1 | 522 | Fabp4 | 46 |
| 8 | Cbfa2t3 | 500 | 1700003D09Rik | 44 |
| 9 | Siglec1 | 489 | Gm20634 | 43 |
| 10 | 1700003D09Rik | 488 | Hbb-bt | 43 |
| 11 | Loxl3 | 482 | Kctd9 | 43 |
| 12 | Vnn1 | 456 | H2ac18 | 41 |
| 13 | Smad5 | 453 | Plin4 | 33 |
| 14 | Wdr35 | 449 | Phtf1os | 31 |
| 15 | Abtb2 | 447 | Zbtb12 | 31 |
| Gene1 | Gene2 | corA (PA) | corA_pVal (PA) | corB (CM) | corB_pVal (CM) | zScoreDiff | pValDiff | pValDiff_adj | Classes |
|---|---|---|---|---|---|---|---|---|---|
| Gm16185 | Exoc3l4 | 0.975 | 1.75 × 10−6 | −0.725 | 1.76 × 10−2 | −5.79 | 6.92 × 10−9 | 3.74 × 10−7 | +/− |
| Kctd9 | Smim30 | 0.867 | 1.16 × 10−3 | −0.937 | 6.20 × 10−5 | −5.68 | 1.33 × 10−8 | 4.42 × 10−7 | +/− |
| Mgll | Gpr179 | 0.957 | 1.36 × 10−5 | −0.799 | 5.52 × 10−3 | −5.63 | 1.77 × 10−8 | 4.75 × 10−7 | +/− |
| Styx | Foxn3 | 0.909 | 2.63 × 10−4 | −0.902 | 3.65 × 10−4 | −5.62 | 1.90 × 10−8 | 4.75 × 10−7 | +/− |
| Ttc41 | Pald1 | 0.982 | 4.69 × 10−7 | −0.521 | 1.23 × 10−1 | −5.47 | 4.57 × 10−8 | 7.61 × 10−7 | +/0 |
| Rbp7 | Tnxb | 0.983 | 3.62 × 10−7 | −0.483 | 1.57 × 10−1 | −5.44 | 5.46 × 10−8 | 8.41 × 10−7 | +/0 |
| Gm37240 | Mgll | 0.992 | 1.83 × 10−8 | −0.239 | 5.07 × 10−1 | −5.41 | 6.42 × 10−8 | 8.94 × 10−7 | +/0 |
| Boc | Gpr179 | 0.951 | 2.29 × 10−5 | −0.741 | 1.43 × 10−2 | −5.24 | 1.65 × 10−7 | 1.25 × 10−6 | +/− |
| mt-Tt | Mgll | 0.990 | 3.59 × 10−8 | −0.142 | 6.96 × 10−1 | −5.22 | 1.81 × 10−7 | 1.25 × 10−6 | +/0 |
| Coch | Tiam1 | 0.981 | 6.14 × 10−7 | −0.437 | 2.07 × 10−1 | −5.20 | 1.99 × 10−7 | 1.27 × 10−6 | +/0 |
| Adra2a | H2ac18 | 0.796 | 5.93 × 10−3 | −0.929 | 1.01 × 10−4 | −5.12 | 3.01 × 10−7 | 1.43 × 10−6 | +/− |
| Timd4 | Ltk | 0.986 | 1.53 × 10−7 | −0.243 | 4.99 × 10−1 | −5.12 | 3.10 × 10−7 | 1.43 × 10−6 | +/0 |
| Kif12 | Gpr179 | 0.965 | 6.19 × 10−6 | −0.608 | 6.24 × 10−2 | −5.09 | 3.56 × 10−7 | 1.43 × 10−6 | +/0 |
| Nptn | Chp1 | 0.994 | 4.68 × 10−9 | −0.073 | 8.40 × 10−1 | −5.09 | 3.60 × 10−7 | 1.43 × 10−6 | +/0 |
| Smad5 | Ifi27 | 0.960 | 1.03 × 10−5 | −0.633 | 4.95 × 10−2 | −5.05 | 4.53 × 10−7 | 1.51 × 10−6 | +/− |
| Siglec1 | Adam23 | 0.961 | 9.45 × 10−6 | −0.622 | 5.47 × 10−2 | −5.03 | 4.82 × 10−7 | 1.53 × 10−6 | +/0 |
| Folr2 | Gm12216 | 0.938 | 5.90 × 10−5 | −0.745 | 1.34 × 10−2 | −5.02 | 5.09 × 10−7 | 1.57 × 10−6 | +/− |
| Airn | Prkar2b | 0.897 | 4.28 × 10−4 | −0.831 | 2.87 × 10−3 | −4.96 | 7.06 × 10−7 | 1.87 × 10−6 | +/− |
| Cd69 | Eml5 | 0.993 | 1.08 × 10−8 | 0.001 | 9.99 × 10−1 | −4.95 | 7.40 × 10−7 | 1.87 × 10−6 | +/0 |
| Gk | Tns4 | 0.644 | 4.44 × 10−2 | −0.953 | 2.05 × 10−5 | −4.91 | 8.95 × 10−7 | 1.91 × 10−6 | +/− |
| Gpr18 | Tiam1 | 0.974 | 1.98 × 10−6 | −0.433 | 2.11 × 10−1 | −4.91 | 8.98 × 10−7 | 1.91 × 10−6 | +/0 |
| Gm42743 | Ifi27l2a | 0.865 | 1.22 × 10−3 | −0.860 | 1.40 × 10−3 | −4.88 | 1.05 × 10−6 | 2.03 × 10−6 | +/− |
| Celf3 | Fcna | 0.969 | 3.84 × 10−6 | −0.481 | 1.60 × 10−1 | −4.87 | 1.13 × 10−6 | 2.10 × 10−6 | +/0 |
| Gm37033 | Siglec1 | 0.872 | 9.91 × 10−4 | −0.848 | 1.95 × 10−3 | −4.85 | 1.25 × 10−6 | 2.14 × 10−6 | +/− |
| Siglec1 | Zbtb20 | 0.964 | 7.19 × 10−6 | −0.533 | 1.13 × 10−1 | −4.85 | 1.26 × 10−6 | 2.14 × 10−6 | +/0 |
| Rbp7 | Slc16a7 | 0.984 | 2.76 × 10−7 | −0.175 | 6.29 × 10−1 | −4.84 | 1.28 × 10−6 | 2.14 × 10−6 | +/0 |
| Gbe1 | Nras | 0.781 | 7.69 × 10−3 | −0.911 | 2.51 × 10−4 | −4.82 | 1.41 × 10−6 | 2.18 × 10−6 | +/− |
| Enpep | Gpr179 | 0.905 | 3.21 × 10−4 | −0.790 | 6.59 × 10−3 | −4.80 | 1.55 × 10−6 | 2.23 × 10−6 | +/− |
| Samd5 | Etfbkmt | 0.871 | 1.02 × 10−3 | −0.840 | 2.33 × 10−3 | −4.79 | 1.65 × 10−6 | 2.24 × 10−6 | +/− |
| Ccdc69 | Tiam1 | 0.933 | 8.32 × 10−5 | −0.706 | 2.24 × 10−2 | −4.79 | 1.70 × 10−6 | 2.24 × 10−6 | +/− |
| Module | Size | MeDC | pVal | Top_GOC | Top_LOC |
|---|---|---|---|---|---|
| c1_80 | 50 | 1.07 | 0 | Gpr179, Fcna, Gm37240, Hoxb5os, Rgs11, Znf41-ps, 9230105E05Rik, Gm43254, Ccdc125, Fcgr4 | |
| c1_104 | 30 | 0.86 | 0 | Hba-a1, Hba-a2, Fosb, Dstyk, Pkmyt1, 2700054A10Rik, Apod, Mcub, Vsig10 | |
| c1_207 | 80 | 0.86 | 0.38 | Tiam1, Basp1, Serpina3g, Memo1 | Mt4 |
| c1_123 | 75 | 0.70 | 0 | Hbb-bs, Hbb-bt, Ifi27l2a, H2ac8, Ybx2, Pde1a, Mdfi, Smim6, Prss27, Fam161a | Zmiz1 |
| c1_39 | 129 | 0.60 | 0 | Hbb-bs, Hbb-bt, Ifi27l2a, H2ac8, Klra2, Ybx2, Pde1a, Mdfi, Prss27, Cercam | Zmiz1 |
| c1_202 | 58 | −1.65 | 0 | Slc4a11 | Igkv1-110, Igkv1-117, Slc16a12, Jchain, Gli1, Maob, Ces2d-ps, Rab11fip4, Igkv5-45, Chst1 |
| c1_49 | 41 | −1.51 | 0 | Cdc42ep2, Nfatc1, Birc5, Thop1, Eno1b, Omp, Gm6311, Rps2-ps13, Gm12366, Gm5786 | |
| c1_27 | 41 | −1.49 | 0 | Atf3, Tacc2, Gpat3, Tmcc3, Tube1, Dusp10, Cdk6, Brca2, Schip1, Rasgrf2 | |
| c1_24 | 33 | −1.45 | 0 | Atg10, Dlst, Col6a5, Ric8b, Kif5c, Msi2, Acsl3, Abcb1b, Atp2b1, Heg1 | |
| c1_212 | 35 | −1.40 | 0 | Gm12320, Pou2f2, Igkv10-94, Cd180, Slc40a1, Gdpd1, Ano10, Zfp608, Phf21a, H2bc24 |
| Module | Size | MeDC | Pval | Top GOC in Condition A (PA) |
|---|---|---|---|---|
| C1_80 | 50 | 1.07 | 0 | Gpr179, Fcna, Gm37240, Hoxb5os, Rgs11, Znf41ps, 9230105E05Rik, Gm43254, Ccdc125, Fcgr4 |
| C1_104 | 30 | 0.86 | 0 | Hba-a1, Hba-a2, Fosb, Dstyk, Pkmyt1, 2700054A10Rik, Apod, Mcub, Vsig10 |
| C1_207 | 80 | 0.86 | 0.38 | Tiam1, Basp1, Serpina3g, Memo1 |
| C1_123 | 75 | 0.70 | 0 | Hbb-bs, Hbb-bt, Ifi27l2a, H2ac8, Ybx2, Pde1a, Mdfi, Smim6, Prss27, Fam161a |
| C1_39 | 129 | 0.60 | 0 | Hbb-bs, Hbb-bt, Ifi27l2a, H2ac8, Klra2, Ybx2, Pde1a, Mdfi, Prss27, Cercam |
| C1_10 | 56 | 0.57 | 0 | Ttc41, Klhl4, Oxnad1, Per3, Plscr2, Dbp, 5830444B04Rik, Pald1, Prxl2a, Il18r1 |
| C1_44 | 31 | 0.48 | 0.04 | C4b, Tle2, Slc9a5, Tie1, Slc22a17, Zfp72 |
| C1_149 | 159 | 0.41 | 0.54 | Tiam1, Abtb2, Socs1, Memo1, Tchh |
| C1_45 | 47 | 0.36 | 0.12 | Gm47798, Cdk5r1, Trafd1, Slc18a1 |
| C1_48 | 40 | 0.34 | 0.08 | Gm49322, Rac3, Fam177a, 2700038G22Rik, Il4ra |
| C1_151 | 48 | 0.32 | 0.08 | Dclk2, Elavl1, Ptpro, Snx32, Mill2, Syt8, Six5, Vstm5, Septin3 |
| C1_20 | 92 | 0.32 | 0.34 | 1700003D09Rik, Gm20634, H2ac18, Hba-a1, Hba-a2, Ablim2, Apod, Pkmyt1 |
| C1_22 | 42 | 0.28 | 0.18 | Loxl3, Tmcc2, 9830144P21Rik |
| C1_67 | 171 | 0.26 | 0.32 | Siglec1, Plin1, Igkv4-74, Batf3, Ntng2, Car3, Timd4, Plin4, Il6ra, Exoc3l4 |
| C1_9 | 89 | 0.26 | 0.26 | Gpr179, Prkar2b, Fcna, Gm37240, Hoxb5os, Fcgr4, Znf41-ps, Rgs11, Ccdc125, mt-Tt |
| C1_25 | 106 | 0.24 | 0.26 | Folr2, Cd209f, Hspa12b, Thrsp, Dcn, Gm21188, Plxna3, Snai1, Cavin2, Tbx21 |
| C1_47 | 44 | 0.09 | 0.5 | Meox1, Gm15675, BC034090, Ccr5, Scara5, Dcp1a, 5830408C22Rik, Gm43980 |
| C1_43 | 59 | 0.07 | 0.72 | Fabp4, Cd36, Gm36161, Ifit2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fleming, D.S.; Liu, F.; Li, R.W. Differential Correlation of Transcriptome Data Reveals Gene Pairs and Pathways Involved in Treatment of Citrobacter rodentium Infection with Bioactive Punicalagin. Molecules 2023, 28, 7369. https://doi.org/10.3390/molecules28217369
Fleming DS, Liu F, Li RW. Differential Correlation of Transcriptome Data Reveals Gene Pairs and Pathways Involved in Treatment of Citrobacter rodentium Infection with Bioactive Punicalagin. Molecules. 2023; 28(21):7369. https://doi.org/10.3390/molecules28217369
Chicago/Turabian StyleFleming, Damarius S., Fang Liu, and Robert W. Li. 2023. "Differential Correlation of Transcriptome Data Reveals Gene Pairs and Pathways Involved in Treatment of Citrobacter rodentium Infection with Bioactive Punicalagin" Molecules 28, no. 21: 7369. https://doi.org/10.3390/molecules28217369
APA StyleFleming, D. S., Liu, F., & Li, R. W. (2023). Differential Correlation of Transcriptome Data Reveals Gene Pairs and Pathways Involved in Treatment of Citrobacter rodentium Infection with Bioactive Punicalagin. Molecules, 28(21), 7369. https://doi.org/10.3390/molecules28217369