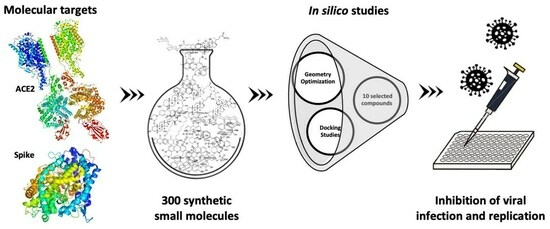
Evaluation of the Cytotoxic and Antiviral Effects of Small Molecules Selected by In Silico Studies as Inhibitors of SARS-CoV-2 Cell Entry
Abstract
:1. Introduction
2. Results
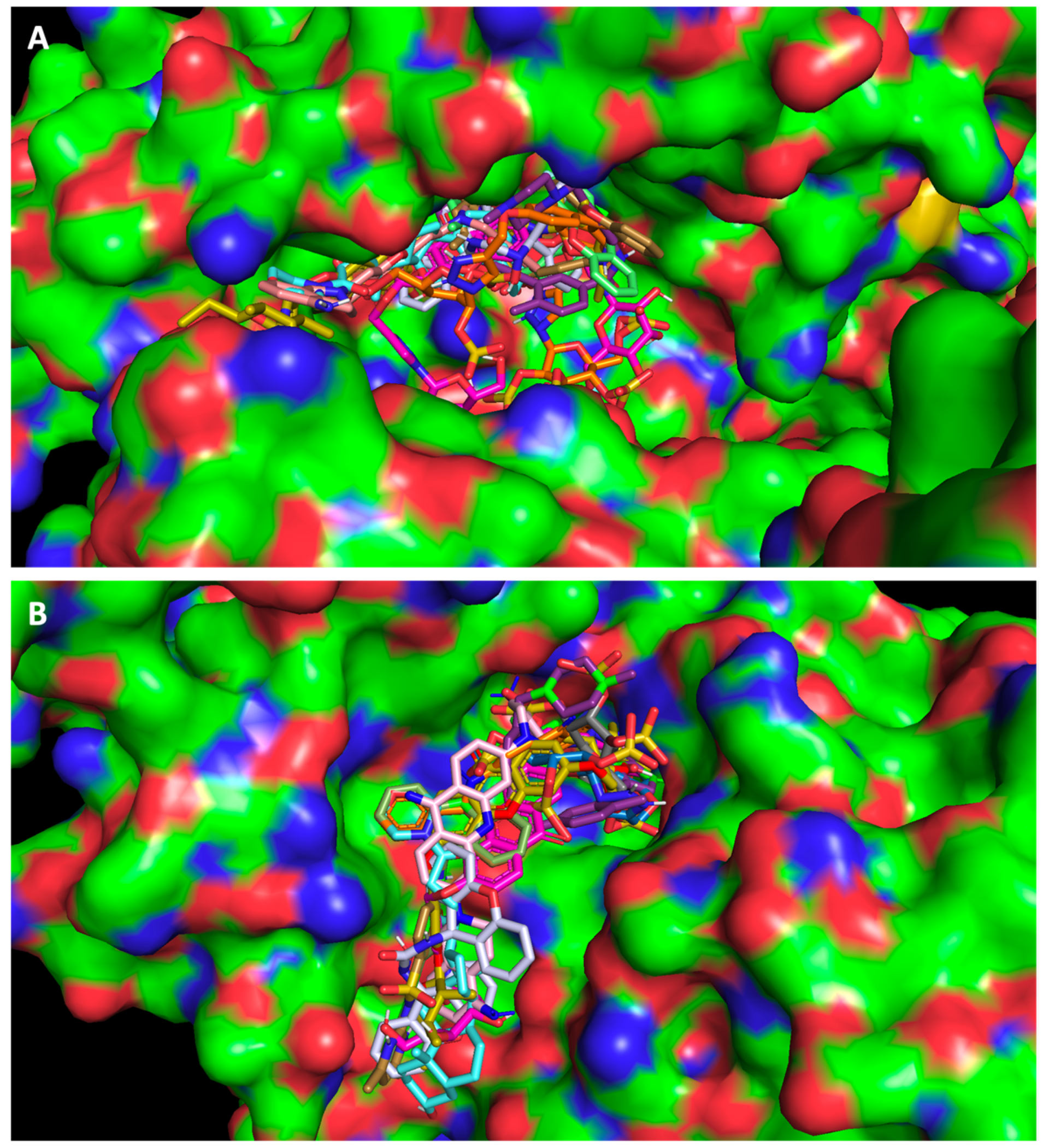
2.1. Screening Studies Revealed Interaction of Several Synthetic Small Molecules with ACE2 Host Receptor and Spike Viral Protein
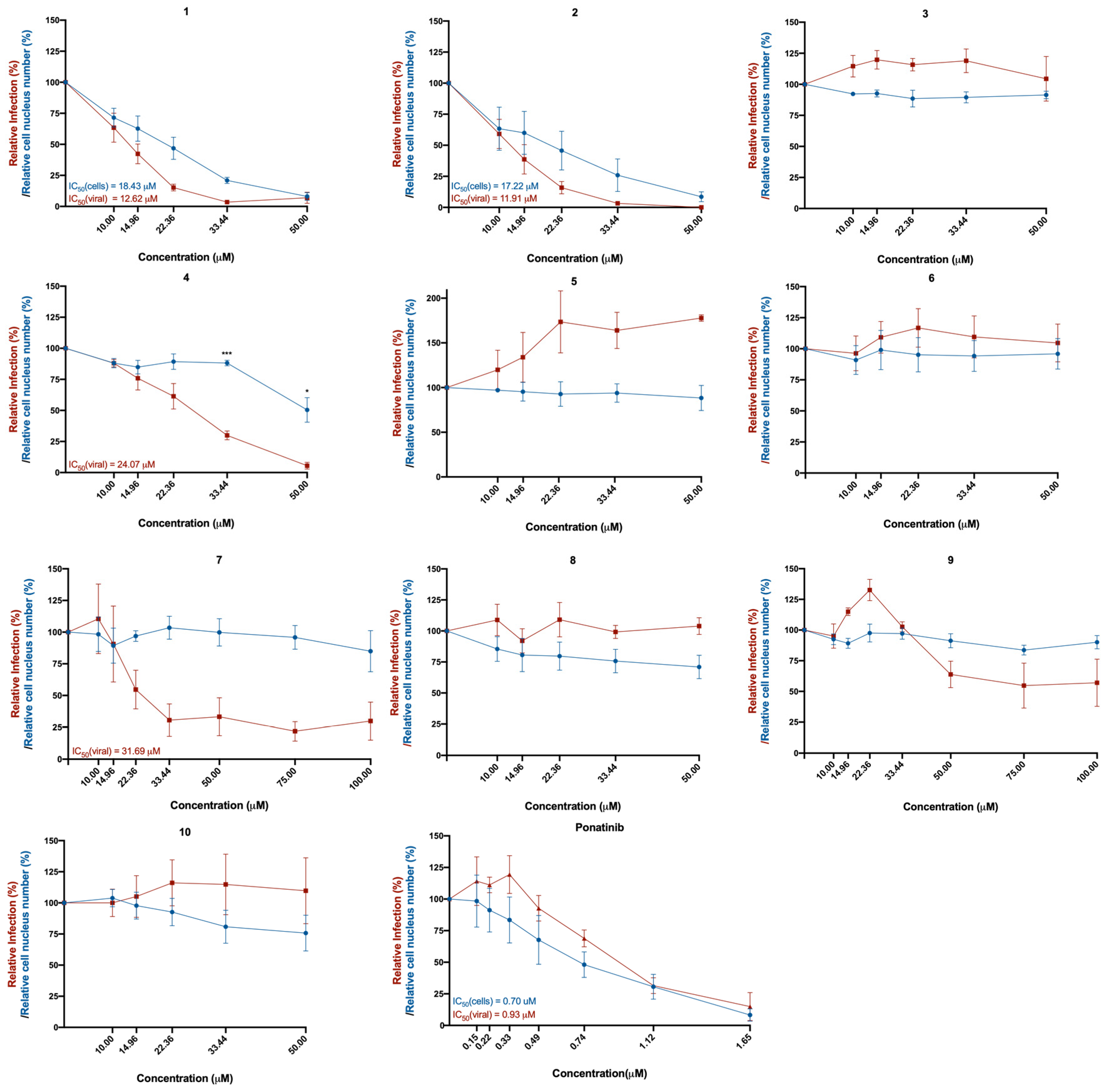
2.2. Five of the Selected Compounds Inhibited SARS-CoV-2 Viral Infection in Vero CCL-81 Cells
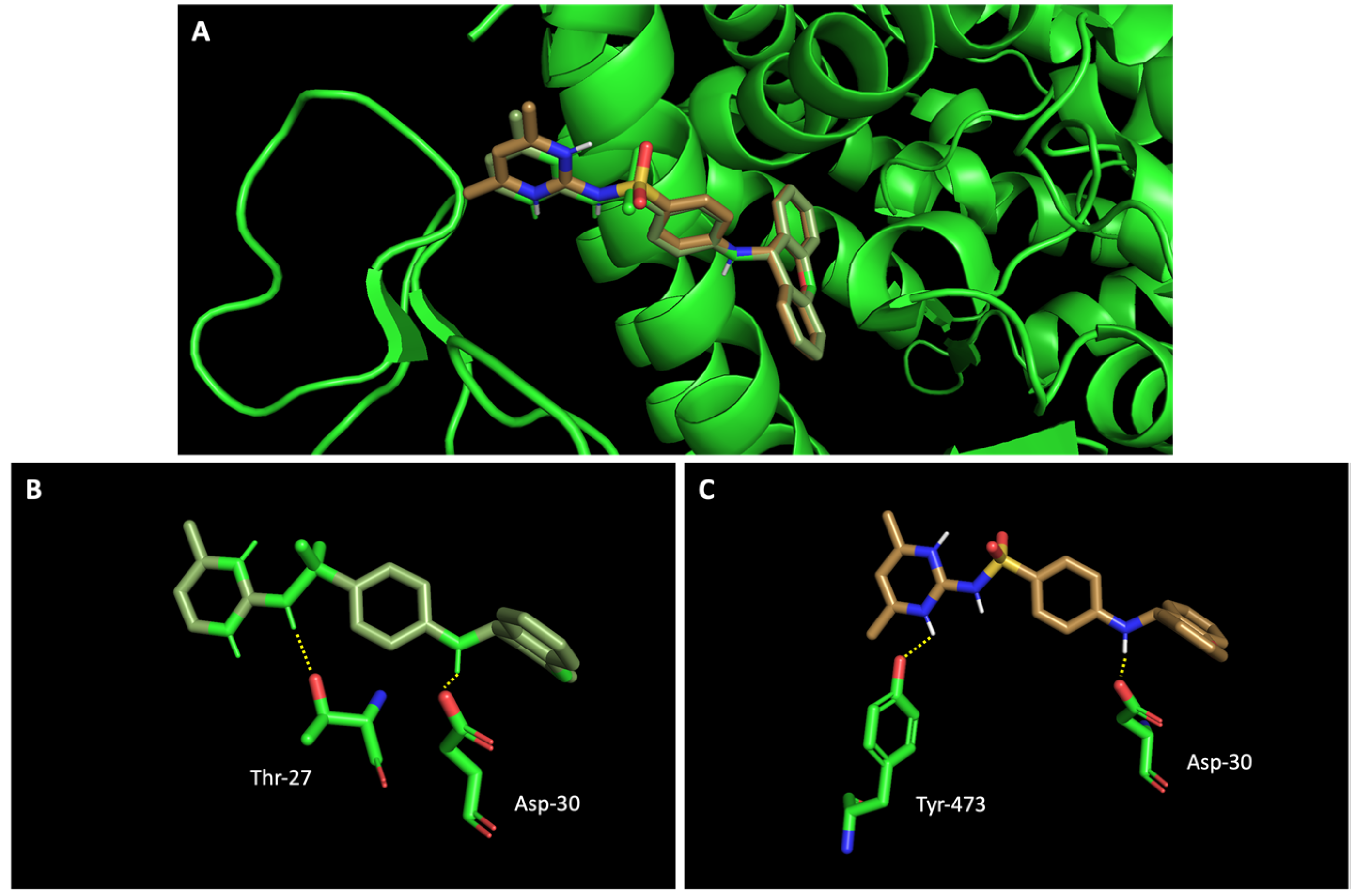
2.3. The Compounds That Exhibited Inhibition of Viral Infection Were Also Predicted to Promisingly Interact with Residues of the Target Proteins
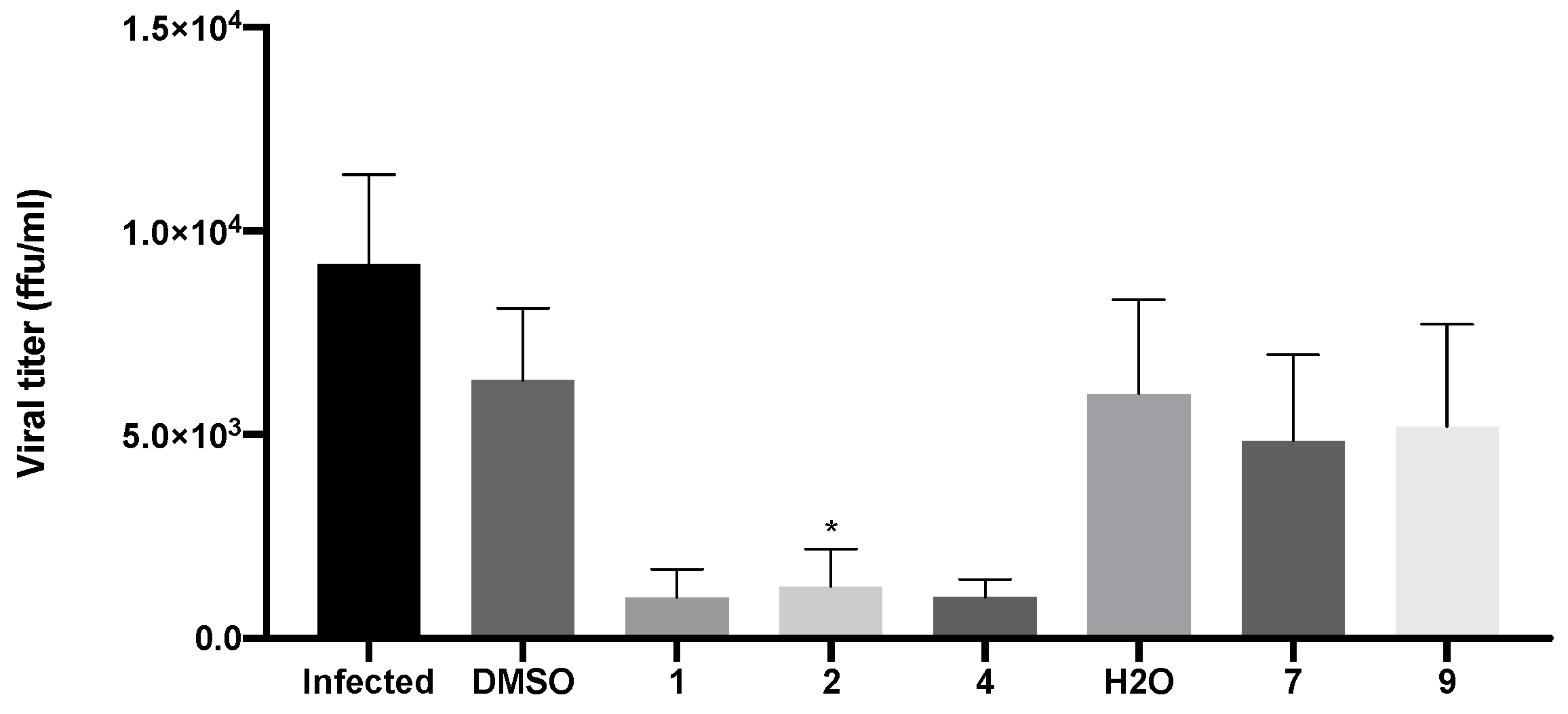
2.4. Compounds 1, 2, and 4 Decreased the Production of New Virions and Their Release from Infected Vero CCL-81 Cells
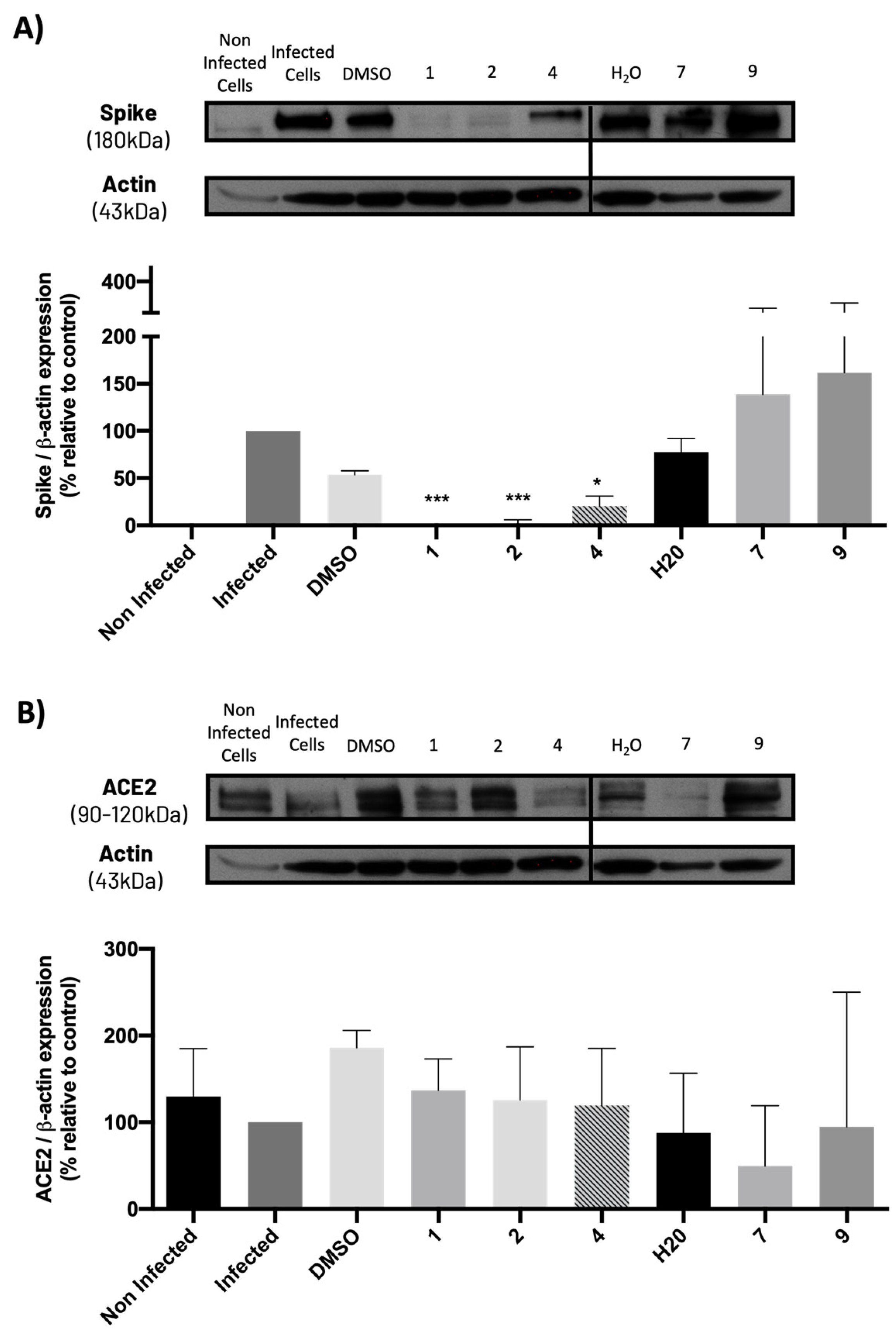
2.5. Compounds 1, 2, and 4 Significantly Decreased Spike Protein Levels in Infected Vero CCL-81 Cells, without Significantly Modifying ACE2 Levels
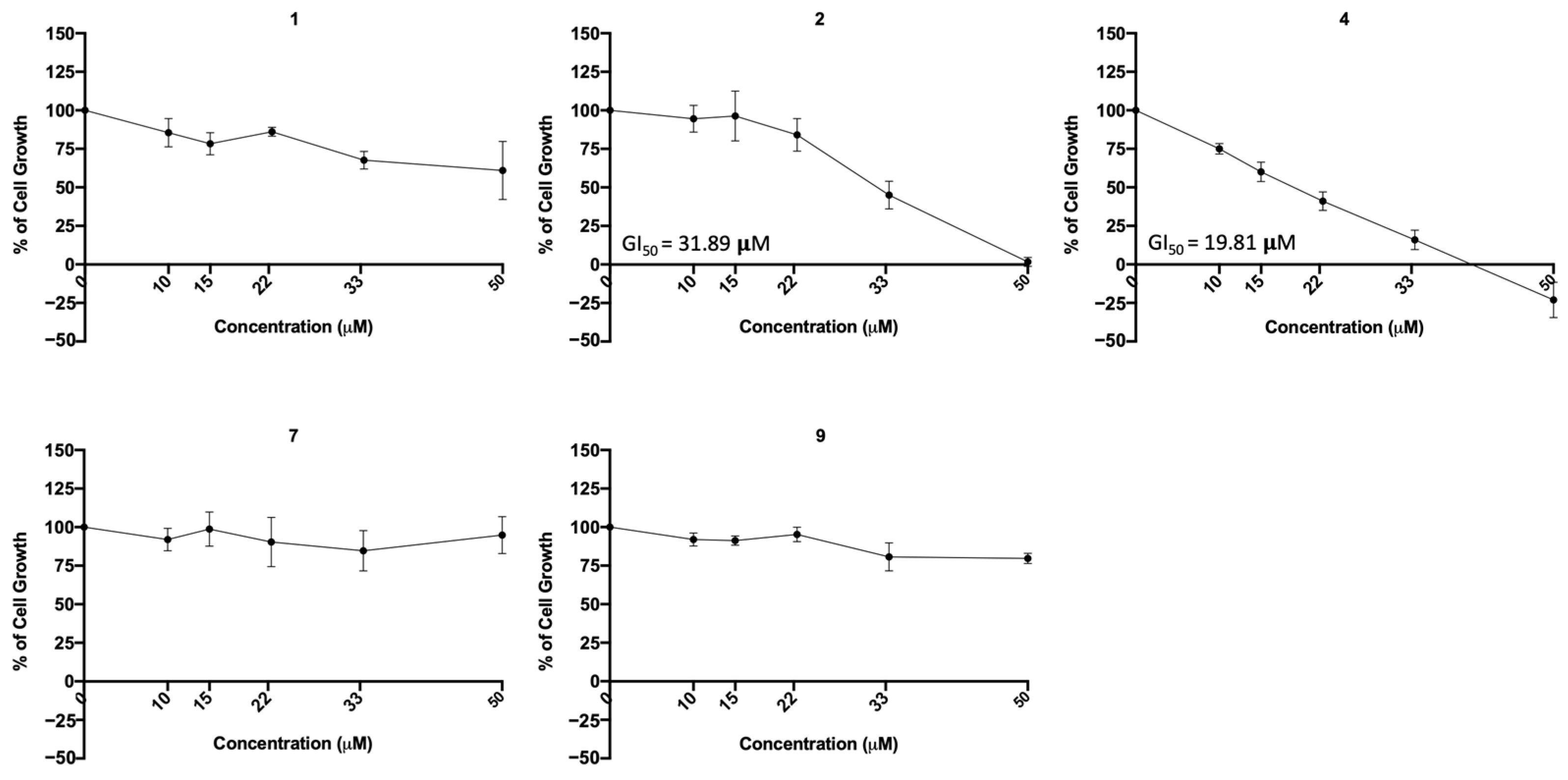
2.6. Compounds 1, 2, 7, and 9 Did Not Exhibit Relevant Toxicity in Human Lung Cells
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culture Conditions
4.2. Virus
4.3. Molecular Docking Virtual Screening
4.4. Tested Compounds
4.5. Effect of the Compounds in Cell Lines
4.5.1. In Non-Infected Cell Lines
4.5.2. In Infected Cell Lines with SARS-CoV-2
4.6. Cytotoxicity Using the Sulforhodamine B (SRB) Assay
4.7. Protein Expression Analysis by Western Blotting
4.7.1. In Non-Infected Cell Lines
4.7.2. In Cell Lines Infected with SARS-CoV-2
4.8. Viral Titration
4.9. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Vasconcelos, M.H.; Alcaro, S.; Arechavala-Gomeza, V.; Baumbach, J.; Borges, F.; Brevini, T.A.L.; Rivas, J.D.L.; Devaux, Y.; Hozak, P.; Keinänen-Toivola, M.M.; et al. Joining European Scientific Forces to Face Pandemics. Trends Microbiol. 2021, 29, 92–97. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Gitman, M.R.; Shaban, M.V.; Paniz-Mondolfi, A.E.; Sordillo, E.M. Laboratory Diagnosis of SARS-CoV-2 Pneumonia. Diagnostics 2021, 11, 1270. [Google Scholar] [CrossRef] [PubMed]
- Pizzato, M.; Baraldi, C.; Boscato Sopetto, G.; Finozzi, D.; Gentile, C.; Gentile, M.D.; Marconi, R.; Paladino, D.; Raoss, A.; Riedmiller, I.; et al. SARS-CoV-2 and the Host Cell: A Tale of Interactions. Front. Virol. 2022, 1, 815388. [Google Scholar] [CrossRef]
- Chaudhry, F.; Lavandero, S.; Xie, X.; Sabharwal, B.; Zheng, Y.-Y.; Correa, A.; Narula, J.; Levy, P. Manipulation of ACE2 expression in COVID-19. Open Heart 2020, 7, e001424. [Google Scholar] [CrossRef]
- Verdecchia, P.; Cavallini, C.; Spanevello, A.; Angeli, F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020, 76, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Neptune, E.; Cui, H. Targeting ACE2 for COVID-19 Therapy: Opportunities and Challenges. Am. J. Respir. Cell. Mol. Biol. 2021, 64, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Dutta, K. Allosteric Site of ACE-2 as a Drug Target for COVID-19. ACS Pharmacol. Transl. Sci. 2022, 5, 179–182. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.-Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e899. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Aquino, R.S.; Park, P.W. Glycosaminoglycans and infection. Front. Biosci. 2016, 21, 1260–1277. [Google Scholar] [CrossRef]
- Cagno, V.; Tseligka, E.D.; Jones, S.T.; Tapparel, C. Heparan Sulfate Proteoglycans and Viral Attachment: True Receptors or Adaptation Bias? Viruses 2019, 11, 596. [Google Scholar] [CrossRef]
- Chittum, J.E.; Sankaranarayanan, N.V.; O’Hara, C.P.; Desai, U.R. On the Selectivity of Heparan Sulfate Recognition by SARS-CoV-2 Spike Glycoprotein. ACS Med. Chem. Lett. 2021, 12, 1710–1717. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, V.; Tandon, R.; Sankaranarayanan, N.V.; Beer, J.C.; Kohlmeir, E.K.; Swanson-Mungerson, M.; Desai, U.R. Preferential recognition and antagonism of SARS-CoV-2 spike glycoprotein binding to 3-O-sulfated heparan sulfate. bioRxiv 2020. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444. [Google Scholar] [CrossRef] [PubMed]
- Emeny, J.M.; Morgan, M.J. Regulation of the interferon system: Evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 1979, 43, 247–252. [Google Scholar] [CrossRef]
- Sène, M.-A.; Xia, Y.; Kamen, A.A. Overview of recent advances in Vero cells genomic characterization and engineering for high-throughput vaccine manufacturing. Clin. Transl. Discov. 2022, 2, e40. [Google Scholar] [CrossRef]
- Yuan, C.; Ma, Z.; Xie, J.; Li, W.; Su, L.; Zhang, G.; Xu, J.; Wu, Y.; Zhang, M.; Liu, W. The role of cell death in SARS-CoV-2 infection. Signal Transduct. Target. Ther. 2023, 8, 357. [Google Scholar] [CrossRef]
- Li, L.; Liao, H.; Meng, Y.; Li, W.; Han, P.; Liu, K.; Wang, Q.; Li, D.; Zhang, Y.; Wang, L.; et al. Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1. Cell 2022, 185, 2952–2960.e2910. [Google Scholar] [CrossRef]
- Durmaz, V.; Köchl, K.; Krassnigg, A.; Parigger, L.; Hetmann, M.; Singh, A.; Nutz, D.; Korsunsky, A.; Kahler, U.; König, C.; et al. Structural bioinformatics analysis of SARS-CoV-2 variants reveals higher hACE2 receptor binding affinity for Omicron B.1.1.529 spike RBD compared to wild type reference. Sci. Rep. 2022, 12, 14534. [Google Scholar] [CrossRef]
- Hidayat, S.; Ibrahim, F.; Pratama, K.; Muchtaridi, M. The interaction of alpha-mangostin and its derivatives against main protease enzyme in COVID-19 using in silico methods. J. Adv. Pharm. Technol. Res. 2021, 12, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X.; et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 2020, 10, 766–788. [Google Scholar] [CrossRef]
- Dean, B.; Cooper, G.; Shivkumar, M.; Snape, T.J. Hydroxy-xanthones as promising antiviral agents: Synthesis and biological evaluation against human coronavirus OC43. Bioorg. Med. Chem. Lett. 2023, 84, 129211. [Google Scholar] [CrossRef]
- Weisberg, E.; Parent, A.; Yang, P.L.; Sattler, M.; Liu, Q.; Liu, Q.; Wang, J.; Meng, C.; Buhrlage, S.J.; Gray, N.; et al. Repurposing of Kinase Inhibitors for Treatment of COVID-19. Pharm Res. 2020, 37, 167. [Google Scholar] [CrossRef] [PubMed]
- Huntington, K.E.; Carlsen, L.; So, E.Y.; Piesche, M.; Liang, O.; El-Deiry, W.S. Integrin/TGF-β1 inhibitor GLPG-0187 blocks SARS-CoV-2 Delta and Omicron pseudovirus infection of airway epithelial cells which could attenuate disease severity. medRxiv 2022. [Google Scholar] [CrossRef] [PubMed]
- Brevini, T.; Maes, M.; Webb, G.J.; John, B.V.; Fuchs, C.D.; Buescher, G.; Wang, L.; Griffiths, C.; Brown, M.L.; Scott, W.E.; et al. FXR inhibition may protect from SARS-CoV-2 infection by reducing ACE2. Nature 2023, 615, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Chen, X.; Wu, J.; Duan, X.; Men, K. Small molecules in the treatment of COVID-19. Signal Transduct. Target. Ther. 2022, 7, 387. [Google Scholar] [CrossRef]
- Magalhães, A.C.; Ricardo, S.; Moreira, A.C.; Nunes, M.; Tavares, M.; Pinto, R.J.; Gomes, M.S.; Pereira, L. InfectionCMA: A Cell MicroArray Approach for Efficient Biomarker Screening in In Vitro Infection Assays. Pathogens 2022, 11, 313. [Google Scholar] [CrossRef]
- Sussman, J.L.; Lin, D.; Jiang, J.; Manning, N.O.; Prilusky, J.; Ritter, O.; Abola, E.E. Protein Data Bank (PDB): Database of three-dimensional structural information of biological macromolecules. Acta Crystallogr. D Biol. Crystallogr. 1998, 54, 1078–1084. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Wang, L.; Liao, S.; Xiao, X.; Qu, J.; Wu, C.; Zhu, Y.; Tai, W.; Huang, Y.; Wang, P.; et al. A Retinol Derivative Inhibits SARS-CoV-2 Infection by Interrupting Spike-Mediated Cellular Entry. mBio 2022, 13, e0148522. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Maia, M.; Durães, F.; Resende, D.I.S.P.; Szemerédi, N.; Gales, L.; Martins-da-Costa, P.; Pinto, M.; Spengler, G.; Sousa, E. Xanthene Derivatives Targeting Bacterial Efflux Pumps, Quorum-Sensing, and Biofilm Formation. Drugs Drug Candidates 2022, 1, 29–42. [Google Scholar] [CrossRef]
- Almeida, J.R.; Correia-da-Silva, M.; Sousa, E.; Antunes, J.; Pinto, M.; Vasconcelos, V.; Cunha, I. Antifouling potential of Nature-inspired sulfated compounds. Sci. Rep. 2017, 7, 42424. [Google Scholar] [CrossRef]
- Vilas-Boas, C.; Neves, A.R.; Carvalhal, F.; Pereira, S.; Calhorda, M.J.; Vasconcelos, V.; Pinto, M.; Sousa, E.; Almeida, J.R.; Silva, E.R.; et al. Multidimensional characterization of a new antifouling xanthone: Structure-activity relationship, environmental compatibility, and immobilization in marine coatings. Ecotoxicol. Environ. Saf. 2021, 228, 112970. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef]
- Branco, H.; Oliveira, J.; Antunes, C.; Santos, L.L.; Vasconcelos, M.H.; Xavier, C.P.R. Pirfenidone Sensitizes NCI-H460 Non-Small Cell Lung Cancer Cells to Paclitaxel and to a Combination of Paclitaxel with Carboplatin. Int. J. Mol. Sci. 2022, 23, 3631. [Google Scholar] [CrossRef]
- Vichai, V.; Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef]
- Payne, A.F.; Binduga-Gajewska, I.; Kauffman, E.B.; Kramer, L.D. Quantitation of flaviviruses by fluorescent focus assay. J. Virol. Methods 2006, 134, 183–189. [Google Scholar] [CrossRef]


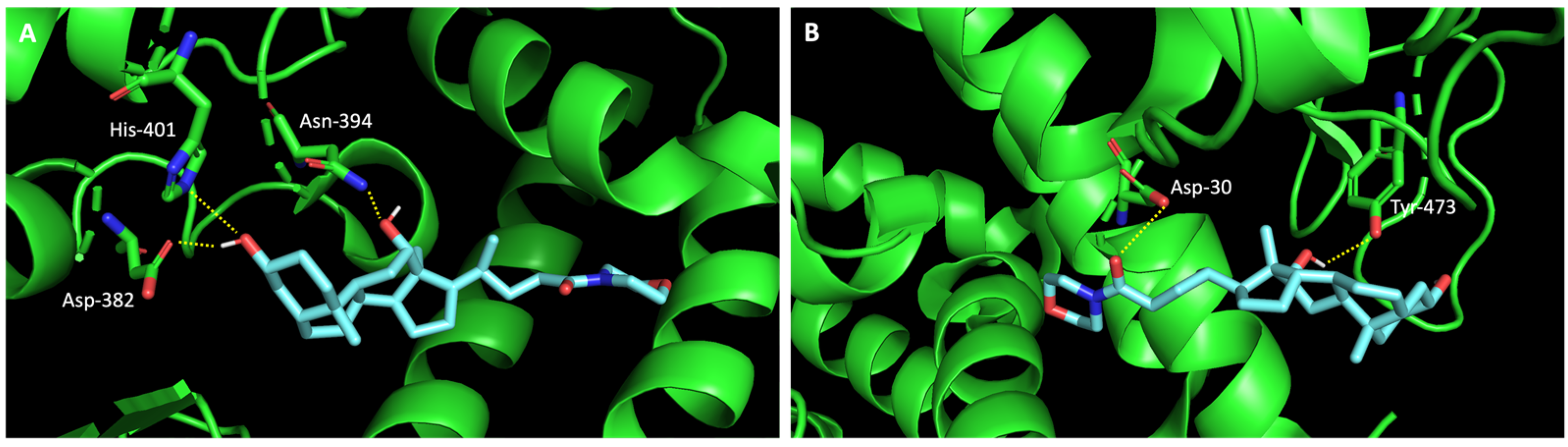


| Code | Structure | Docking Score ACE2 (kcal/mol) | Docking Score RBD of Spike (kcal/mol) |
|---|---|---|---|
| 1 | 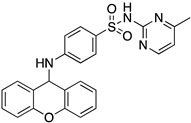 | −8.7 | −9.0 |
| 2 | 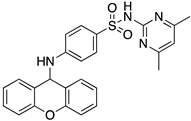 | −8.9 | −9.1 |
| 3 |  | −8.7 | −7.8 |
| 4 | 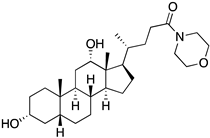 | −8.9 | −7.4 |
| 5 | 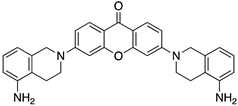 | −9.8 | −9.4 |
| 6 | 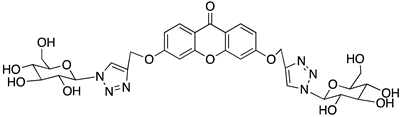 | −9.4 | −9.4 |
| 7 |  | −9.3 | −9.0 |
| 8 |  | −9.1 | −8.7 |
| 9 |  | −8.7 | −7.8 |
| 10 | 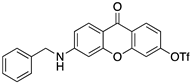 | −8.3 | −9.1 |
| Ponatinib | 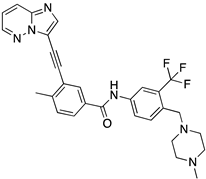 | −8.3 | −8.1 |
| Compounds | Antiviral IC50 (Vero CCL-81 Monkey Cells) | Cytotoxic IC50 (Vero CCL-81 Monkey Cells) | Viral Titer and Spike Levels (Vero CCL-81 Monkey Cells) | ACE2 Levels (Vero CCL-81 Monkey Cells) | Cytotoxic IC50 (A549 Human Lung Cells) |
|---|---|---|---|---|---|
| 1 | 12.62 μM | 18.43 μM | ↓↓ | − | n.d. |
| 2 | 11.91 μM | 17.22 μM | ↓↓ | − | 31.89 μM |
| 4 | 24.07 μM | n.d. | ↓↓ | − | 19.81 μM |
| 7 | 31.69 μM | n.d. | − | − | n.d. |
| 9 | n.d. | n.d. | − | − | n.d. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carvalhal, F.; Magalhães, A.C.; Rebelo, R.; Palmeira, A.; Resende, D.I.S.P.; Durães, F.; Maia, M.; Xavier, C.P.R.; Pereira, L.; Sousa, E.; et al. Evaluation of the Cytotoxic and Antiviral Effects of Small Molecules Selected by In Silico Studies as Inhibitors of SARS-CoV-2 Cell Entry. Molecules 2023, 28, 7204. https://doi.org/10.3390/molecules28207204
Carvalhal F, Magalhães AC, Rebelo R, Palmeira A, Resende DISP, Durães F, Maia M, Xavier CPR, Pereira L, Sousa E, et al. Evaluation of the Cytotoxic and Antiviral Effects of Small Molecules Selected by In Silico Studies as Inhibitors of SARS-CoV-2 Cell Entry. Molecules. 2023; 28(20):7204. https://doi.org/10.3390/molecules28207204
Chicago/Turabian StyleCarvalhal, Francisca, Ana Cristina Magalhães, Rita Rebelo, Andreia Palmeira, Diana I. S. P. Resende, Fernando Durães, Miguel Maia, Cristina P. R. Xavier, Luísa Pereira, Emília Sousa, and et al. 2023. "Evaluation of the Cytotoxic and Antiviral Effects of Small Molecules Selected by In Silico Studies as Inhibitors of SARS-CoV-2 Cell Entry" Molecules 28, no. 20: 7204. https://doi.org/10.3390/molecules28207204
APA StyleCarvalhal, F., Magalhães, A. C., Rebelo, R., Palmeira, A., Resende, D. I. S. P., Durães, F., Maia, M., Xavier, C. P. R., Pereira, L., Sousa, E., Correia-da-Silva, M., & Vasconcelos, M. H. (2023). Evaluation of the Cytotoxic and Antiviral Effects of Small Molecules Selected by In Silico Studies as Inhibitors of SARS-CoV-2 Cell Entry. Molecules, 28(20), 7204. https://doi.org/10.3390/molecules28207204