History of Maturation of Prokaryotic Molybdoenzymes—A Personal View
Abstract
1. Introduction
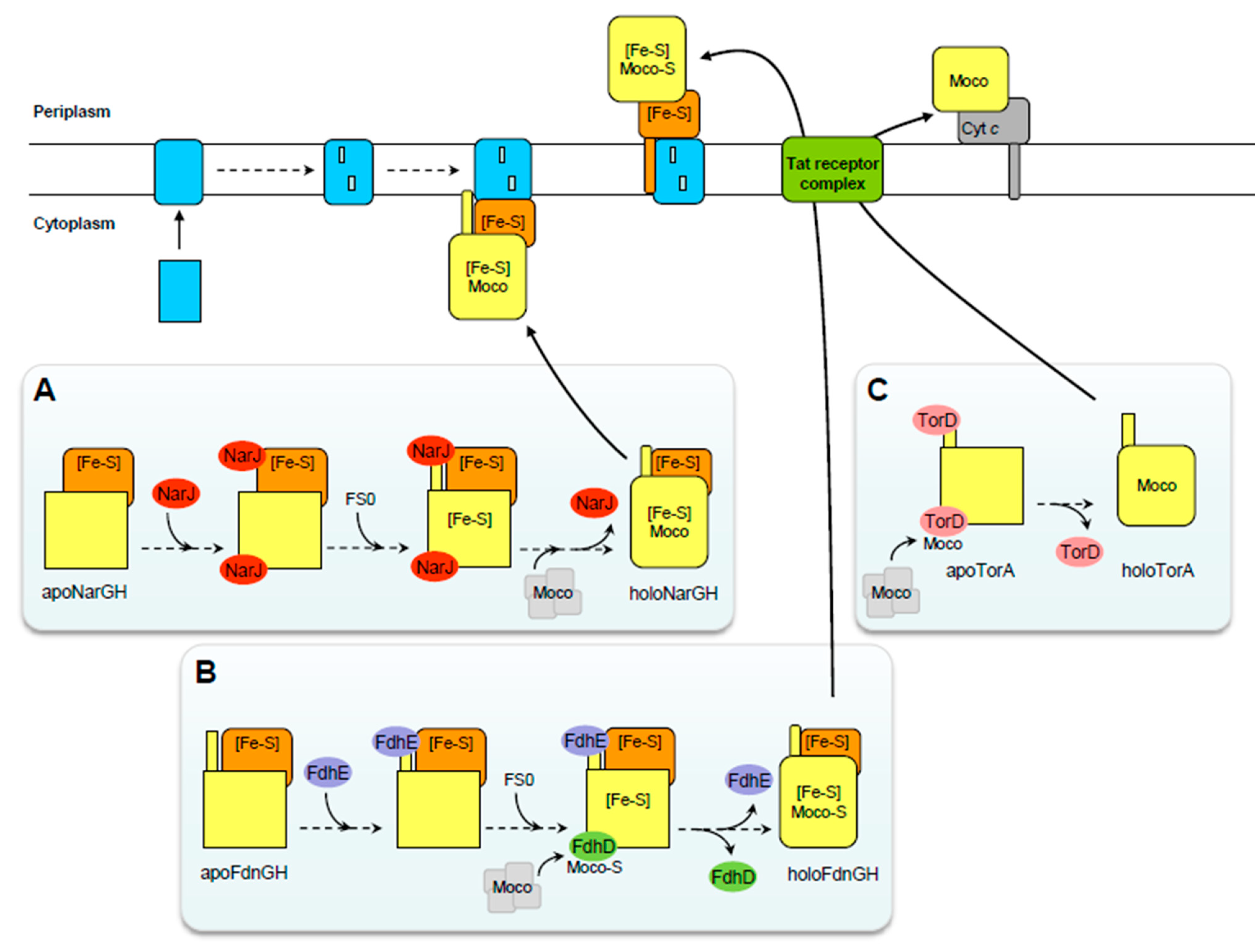
2. Maturation of Mo/W-bisPGD Enzymes Illustrated by the Cytoplasmic and Multimeric Nitrate Reductase
3. Periplasmic and Multimeric Mo-bisPGD Enzymes
3.1. The Case of the DMSO Reductase: A Similar Situation to Nar
3.2. The Case of Formate Dehydrogenases: Two Distinct Chaperones and a Sulfuration Step of the Cofactor
3.3. The Case of Periplasmic Nitrate Reductases: A Distinct Chaperone, a Sulfuration Step of the Cofactor and Open Questions
4. Periplasmic and Monomeric Mo-bisPGD Enzymes
5. Maturation of Other Prokaryotic Mo/W-Enzymes
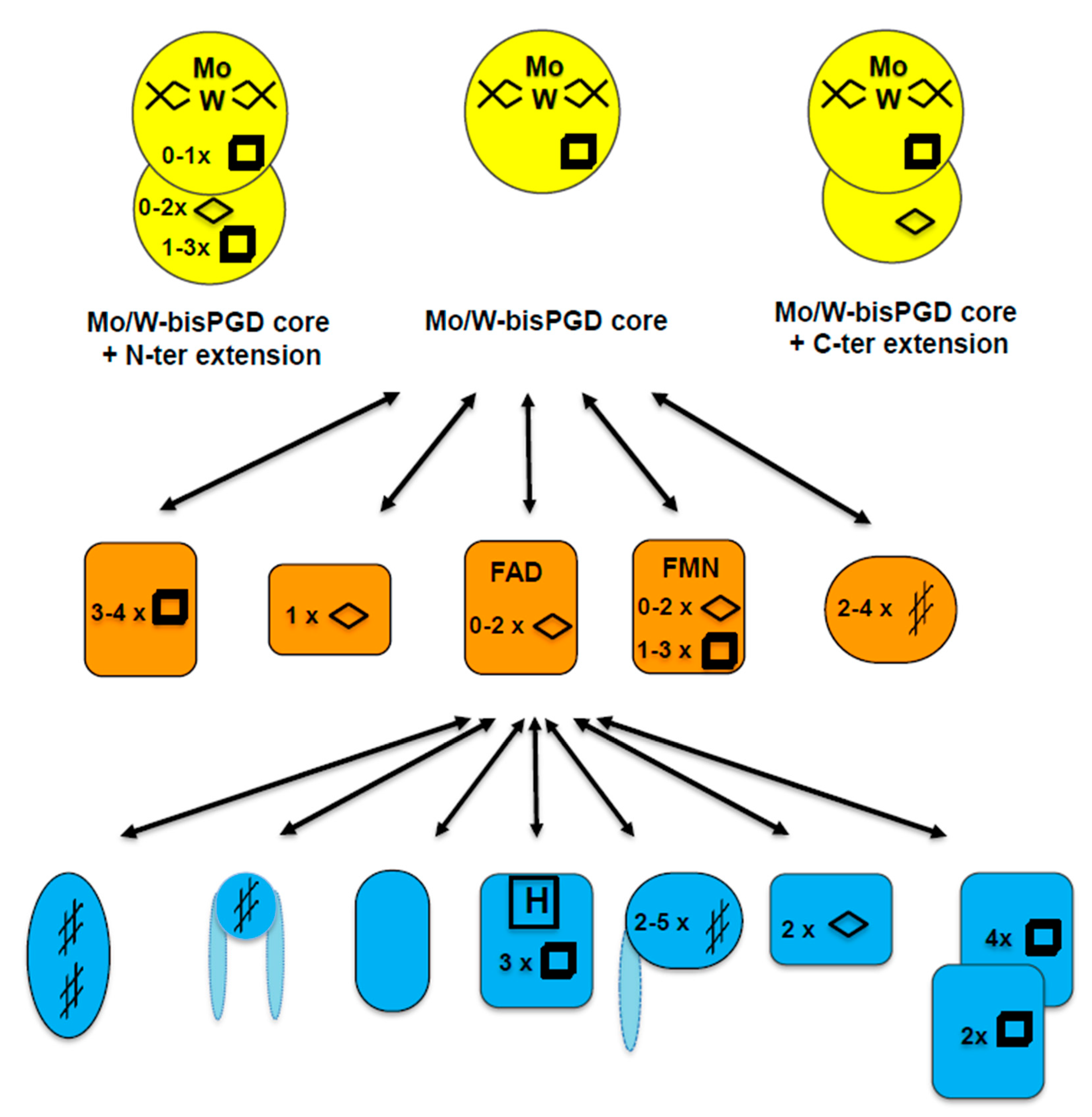
6. Number of Binding Sites and Their Location Both on the Chaperone and the Target Enzyme
7. Specificity versus Promiscuity of the Chaperone?
8. How to Insert Mo/W-bisPGD?
9. Concluding Remarks
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Zhang, Y.; Gladyshev, V.N. Molybdoproteomes and evolution of molybdenum utilization. J. Mol. Biol. 2008, 379, 881–899. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Rump, S.; Gladyshev, V.N. Comparative Genomics and Evolution of Molybdenum Utilization. Coord. Chem. Rev. 2011, 255, 1206–1217. [Google Scholar] [CrossRef] [PubMed]
- Cvetkovic, A.; Menon, A.L.; Thorgersen, M.P.; Scott, J.W.; Poole, F.L., 2nd; Jenney, F.E., Jr.; Lancaster, W.A.; Praissman, J.L.; Shanmukh, S.; Vaccaro, B.J.; et al. Microbial metalloproteomes are largely uncharacterized. Nature 2010, 466, 779–782. [Google Scholar] [CrossRef] [PubMed]
- Wells, M.; Kim, M.; Akob, D.M.; Basu, P.; Stolz, J.F. Impact of the Dimethyl Sulfoxide Reductase Superfamily on the Evolution of Biogeochemical Cycles. Microbiol. Spectr. 2023, 11, e0414522. [Google Scholar] [CrossRef] [PubMed]
- Hagen, W.R. The Development of Tungsten Biochemistry-A Personal Recollection. Molecules 2023, 28, 4017. [Google Scholar] [CrossRef] [PubMed]
- Hille, R. Molybdenum enzymes containing the pyranopterin cofactor: An overview. Met. Ions Biol. Syst. 2002, 39, 187–226. [Google Scholar] [PubMed]
- Hille, R.; Hall, J.; Basu, P. The mononuclear molybdenum enzymes. Chem. Rev. 2014, 114, 3963–4038. [Google Scholar] [CrossRef]
- Grimaldi, S.; Schoepp-Cothenet, B.; Ceccaldi, P.; Guigliarelli, B.; Magalon, A. The prokaryotic Mo/W-bisPGD enzymes family: A catalytic workhorse in bioenergetic. Biochim. Biophys. Acta 2013, 1827, 1048–1085. [Google Scholar] [CrossRef]
- Magalon, A.; Mendel, R.R. Biosynthesis and Insertion of the Molybdenum Cofactor. EcoSal Plus 2015, 6, 10–128. [Google Scholar] [CrossRef]
- Anbar, A.D. Elements and evolution. Science 2008, 322, 1481–1483. [Google Scholar] [CrossRef]
- Finel, M. Organization and evolution of structural elements within complex I. Biochim. Biophys. Acta 1998, 1364, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Rothery, R.A.; Workun, G.J.; Weiner, J.H. The prokaryotic complex iron-sulfur molybdoenzyme family. Biochim. Biophys. Acta 2008, 1778, 1897–1929. [Google Scholar] [CrossRef] [PubMed]
- Yanyushin, M.F.; del Rosario, M.C.; Brune, D.C.; Blankenship, R.E. New class of bacterial membrane oxidoreductases. Biochemistry 2005, 44, 10037–10045. [Google Scholar] [CrossRef] [PubMed]
- Refojo, P.N.; Sousa, F.L.; Teixeira, M.; Pereira, M.M. The alternative complex III: A different architecture using known building modules. Biochim. Biophys. Acta 2010, 1797, 1869–1876. [Google Scholar] [CrossRef] [PubMed]
- Romao, M.J. Molybdenum and tungsten enzymes: A crystallographic and mechanistic overview. Dalton Trans. 2009, 4053–4068. [Google Scholar] [CrossRef] [PubMed]
- Kirk, M.L.; Kc, K. Molybdenum and Tungsten Cofactors and the Reactions They Catalyze. Met. Ions. Life Sci. 2020, 20, 313–342. [Google Scholar]
- Magalon, A.; Ceccaldi, P.; Schoepp-Cothenet, B. The Prokaryotic Mo/W-bisPGD Enzymes Family. In Molybdenum and Tungsten Enzymes: Biochemistry; Hille, R., Schulzke, C., Kirk, M.L., Eds.; Royal Society of Chemistry: London, UK, 2016; Volume 5, pp. 143–191. [Google Scholar]
- Wagner, T.; Ermler, U.; Shima, S. The methanogenic CO2 reducing-and-fixing enzyme is bifunctional and contains 46 [4Fe-4S] clusters. Science 2016, 354, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, H.M.; Righetto, R.D.; Kumar, A.; Wietrzynski, W.; Trischler, R.; Schuller, S.K.; Wagner, J.; Schwarz, F.M.; Engel, B.D.; Muller, V.; et al. Membrane-anchored HDCR nanowires drive hydrogen-powered CO(2) fixation. Nature 2022, 607, 823–830. [Google Scholar] [CrossRef]
- Steinhilper, R.; Hoff, G.; Heider, J.; Murphy, B.J. Structure of the membrane-bound formate hydrogenlyase complex from Escherichia coli. Nat. Commun. 2022, 13, 5395. [Google Scholar] [CrossRef]
- Baymann, F.; Lebrun, E.; Brugna, M.; Schoepp-Cothenet, B.; Giudici-Orticoni, M.T.; Nitschke, W. The redox protein construction kit: Pre-last universal common ancestor evolution of energy-conserving enzymes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003, 358, 267–274. [Google Scholar] [CrossRef]
- Schoepp-Cothenet, B.; van Lis, R.; Atteia, A.; Baymann, F.; Capowiez, L.; Ducluzeau, A.L.; Duval, S.; ten Brink, F.; Russell, M.J.; Nitschke, W. On the universal core of bioenergetics. Biochim. Biophys. Acta 2013, 1827, 79–93. [Google Scholar] [CrossRef]
- Raanan, H.; Pike, D.H.; Moore, E.K.; Falkowski, P.G.; Nanda, V. Modular origins of biological electron transfer chains. Proc. Natl. Acad. Sci. USA 2018, 115, 1280–1285. [Google Scholar] [CrossRef] [PubMed]
- Raanan, H.; Poudel, S.; Pike, D.H.; Nanda, V.; Falkowski, P.G. Small protein folds at the root of an ancient metabolic network. Proc. Natl. Acad. Sci. USA 2020, 117, 7193–7199. [Google Scholar] [CrossRef] [PubMed]
- Santini, C.L.; Iobbi-Nivol, C.; Romane, C.; Boxer, D.H.; Giordano, G. Molybdoenzyme biosynthesis in Escherichia coli: In vitro activation of purified nitrate reductase from a chlB mutant. J. Bacteriol. 1992, 174, 7934–7940. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mandrand-Berthelot, M.A.; Couchoux-Luthaud, G.; Santini, C.L.; Giordano, G. Mutants of Escherichia coli specifically deficient in respiratory formate dehydrogenase activity. Microbiology 1988, 134, 3129–3139. [Google Scholar] [CrossRef] [PubMed]
- Schlindwein, C.; Giordano, G.; Santini, C.L.; Mandrand, M.A. Identification and expression of the Escherichia coli fdhD and fdhE genes, which are involved in the formation of respiratory formate dehydrogenase. J. Bacteriol. 1990, 172, 6112–6121. [Google Scholar] [CrossRef] [PubMed]
- Stewart, V.; Lin, J.T.; Berg, B.L. Genetic evidence that genes fdhD and fdhE do not control synthesis of formate dehydrogenase-N in Escherichia coli K-12. J. Bacteriol. 1991, 173, 4417–4423. [Google Scholar] [CrossRef][Green Version]
- Palmer, T.; Santini, C.L.; Iobbi-Nivol, C.; Eaves, D.J.; Boxer, D.H.; Giordano, G. Involvement of the narJ and mob gene products in distinct steps in the biosynthesis of the molybdoenzyme nitrate reductase in Escherichia coli. Mol. Microbiol. 1996, 20, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Blasco, F.; Dos Santos, J.P.; Magalon, A.; Frixon, C.; Guigliarelli, B.; Santini, C.L.; Giordano, G. NarJ is a specific chaperone required for molybdenum cofactor assembly in nitrate reductase A of Escherichia coli. Mol. Microbiol. 1998, 28, 435–447. [Google Scholar] [CrossRef]
- Blasco, F.; Pommier, J.; Augier, V.; Chippaux, M.; Giordano, G. Involvement of the narJ or narW gene product in the formation of active nitrate reductase in Escherichia coli. Mol. Microbiol. 1992, 6, 221–230. [Google Scholar] [CrossRef]
- Pommier, J.; Mejean, V.; Giordano, G.; Iobbi-Nivol, C. TorD, a cytoplasmic chaperone that interacts with the unfolded trimethylamine N-oxide reductase enzyme (TorA) in Escherichia coli. J. Biol. Chem. 1998, 273, 16615–16620. [Google Scholar] [CrossRef]
- Park, I.S.; Carr, M.B.; Hausinger, R.P. In vitro activation of urease apoprotein and role of UreD as a chaperone required for nickel metallocenter assembly. Proc. Natl. Acad. Sci. USA 1994, 91, 3233–3237. [Google Scholar] [CrossRef] [PubMed]
- Maier, T.; Bock, A. Generation of active [NiFe] hydrogenase in vitro from a nickel-free precursor form. Biochemistry 1996, 35, 10089–10093. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.L.; Leimkuhler, S.; Klipp, W.; Hanson, G.R.; McEwan, A.G. Mutational analysis of the dimethylsulfoxide respiratory (dor) operon of Rhodobacter capsulatus. Microbiology 1999, 145 Pt 6, 1409–1420. [Google Scholar] [CrossRef] [PubMed]
- Oresnik, I.J.; Ladner, C.L.; Turner, R.J. Identification of a twin-arginine leader-binding protein. Mol. Microbiol. 2001, 40, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Ray, N.; Oates, J.; Turner, R.J.; Robinson, C. DmsD is required for the biogenesis of DMSO reductase in Escherichia coli but not for the interaction of the DmsA signal peptide with the Tat apparatus. FEBS Lett. 2003, 534, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.P.; Takio, S.; Satoh, T.; Yamamoto, I. Involvement in denitrification of the napKEFDABC genes encoding the periplasmic nitrate reductase system in the denitrifying phototrophic bacterium Rhodobacter sphaeroides f. sp. denitrificans. Biosci. Biotechnol. Biochem. 1999, 63, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Potter, L.C.; Cole, J.A. Essential roles for the products of the napABCD genes, but not napFGH, in periplasmic nitrate reduction by Escherichia coli K-12. Biochem. J. 1999, 344 Pt 1, 69–76. [Google Scholar] [CrossRef]
- Kern, M.; Mager, A.M.; Simon, J. Role of individual nap gene cluster products in NapC-independent nitrate respiration of Wolinella succinogenes. Microbiology 2007, 153 Pt 11, 3739–3747. [Google Scholar] [CrossRef]
- Coulthurst, S.J.; Dawson, A.; Hunter, W.N.; Sargent, F. Conserved signal peptide recognition systems across the prokaryotic domains. Biochemistry 2012, 51, 1678–1686. [Google Scholar] [CrossRef]
- Jacoby, C.; Eipper, J.; Warnke, M.; Tiedt, O.; Mergelsberg, M.; Stark, H.J.; Daus, B.; Martin-Moldes, Z.; Zamarro, M.T.; Diaz, E.; et al. Four Molybdenum-Dependent Steroid C-25 Hydroxylases: Heterologous Overproduction, Role in Steroid Degradation, and Application for 25-Hydroxyvitamin D(3) Synthesis. MBio 2018, 9, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Leimkuhler, S.; Klipp, W. Role of XDHC in Molybdenum cofactor insertion into xanthine dehydrogenase of Rhodobacter capsulatus. J. Bacteriol. 1999, 181, 2745–2751. [Google Scholar] [CrossRef] [PubMed]
- Magalon, A.; Fedor, J.G.; Walburger, A.; Weiner, J.H. Molybdenum enzymes in bacteria and their maturation. Coord. Chem. Rev. 2011, 255, 1159–1178. [Google Scholar] [CrossRef]
- Mintmier, B.; Nassif, S.; Stolz, J.F.; Basu, P. Functional mononuclear molybdenum enzymes: Challenges and triumphs in molecular cloning, expression, and isolation. J. Biol. Inorg. Chem. 2020, 25, 547–569. [Google Scholar] [CrossRef] [PubMed]
- Sargent, F.; Bogsch, E.G.; Stanley, N.R.; Wexler, M.; Robinson, C.; Berks, B.C.; Palmer, T. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 1998, 17, 3640–3650. [Google Scholar] [CrossRef] [PubMed]
- Weiner, J.H.; Bilous, P.T.; Shaw, G.M.; Lubitz, S.P.; Frost, L.; Thomas, G.H.; Cole, J.A.; Turner, R.J. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell 1998, 93, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Santini, C.L.; Ize, B.; Chanal, A.; Muller, M.; Giordano, G.; Wu, L.F. A novel sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 1998, 17, 101–112. [Google Scholar] [CrossRef]
- Turner, R.J.; Papish, A.L.; Sargent, F. Sequence analysis of bacterial redox enzyme maturation proteins (REMPs). Can. J. Microbiol. 2004, 50, 225–238. [Google Scholar] [CrossRef]
- Bay, D.C.; Chan, C.S.; Turner, R.J. NarJ subfamily system specific chaperone diversity and evolution is directed by respiratory enzyme associations. BMC Evol. Biol. 2015, 15, 110. [Google Scholar] [CrossRef]
- Chan, M.K.; Mukund, S.; Kletzin, A.; Adams, M.W.; Rees, D.C. Structure of a hyperthermophilic tungstopterin enzyme, aldehyde ferredoxin oxidoreductase. Science 1995, 267, 1463–1469. [Google Scholar] [CrossRef]
- Hu, Y.; Faham, S.; Roy, R.; Adams, M.W.; Rees, D.C. Formaldehyde ferredoxin oxidoreductase from Pyrococcus furiosus: The 1.85 A resolution crystal structure and its mechanistic implications. J. Mol. Biol. 1999, 286, 899–914. [Google Scholar] [CrossRef] [PubMed]
- Mathew, L.G.; Haja, D.K.; Pritchett, C.; McCormick, W.; Zeineddine, R.; Fontenot, L.S.; Rivera, M.E.; Glushka, J.; Adams, M.W.W.; Lanzilotta, W.N. An unprecedented function for a tungsten-containing oxidoreductase. J. Biol. Inorg. Chem. 2022, 27, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Loschi, L.; Brokx, S.J.; Hills, T.L.; Zhang, G.; Bertero, M.G.; Lovering, A.L.; Weiner, J.H.; Strynadka, N.C. Structural and biochemical identification of a novel bacterial oxidoreductase. J. Biol. Chem. 2004, 279, 50391–50400. [Google Scholar] [CrossRef] [PubMed]
- Schoepp-Cothenet, B.; van Lis, R.; Philippot, P.; Magalon, A.; Russell, M.J.; Nitschke, W. The ineluctable requirement for the trans-iron elements molybdenum and/or tungsten in the origin of life. Sci. Rep. 2012, 2, 263. [Google Scholar] [CrossRef] [PubMed]
- Guigliarelli, B.; Magalon, A.; Asso, M.; Bertrand, P.; Frixon, C.; Giordano, G.; Blasco, F. Complete coordination of the four Fe-S centers of the beta subunit from Escherichia coli nitrate reductase. Physiological, biochemical, and EPR characterization of site-directed mutants lacking the highest or lowest potential [4Fe-4S] clusters. Biochemistry 1996, 35, 4828–4836. [Google Scholar] [CrossRef] [PubMed]
- Lanciano, P.; Savoyant, A.; Grimaldi, S.; Magalon, A.; Guigliarelli, B.; Bertrand, P. New method for the spin quantitation of [4Fe-4S](+) clusters with S = (3)/(2). Application to the FS0 center of the NarGHI nitrate reductase from Escherichia coli. J. Phys. Chem. B 2007, 111, 13632–13637. [Google Scholar] [CrossRef] [PubMed]
- Magalon, A.; Lemesle-Meunier, D.; Rothery, R.A.; Frixon, C.; Weiner, J.H.; Blasco, F. Heme axial ligation by the highly conserved His residues in helix II of cytochrome b (NarI) of Escherichia coli nitrate reductase A. J. Biol. Chem. 1997, 272, 25652–25658. [Google Scholar] [CrossRef]
- Bertero, M.G.; Rothery, R.A.; Palak, M.; Hou, C.; Lim, D.; Blasco, F.; Weiner, J.H.; Strynadka, N.C. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat. Struct. Mol. Biol. 2003, 10, 681–687. [Google Scholar] [CrossRef]
- Vergnes, A.; Pommier, J.; Toci, R.; Blasco, F.; Giordano, G.; Magalon, A. NarJ chaperone binds on two distinct sites of the aponitrate reductase of Escherichia coli to coordinate molybdenum cofactor insertion and assembly. J. Biol. Chem. 2006, 281, 2170–2176. [Google Scholar] [CrossRef]
- Lanciano, P.; Vergnes, A.; Grimaldi, S.; Guigliarelli, B.; Magalon, A. Biogenesis of a respiratory complex is orchestrated by a single accessory protein. J. Biol. Chem. 2007, 282, 17468–17474. [Google Scholar] [CrossRef]
- Rothery, R.A.; Bertero, M.G.; Cammack, R.; Palak, M.; Blasco, F.; Strynadka, N.C.; Weiner, J.H. The Catalytic Subunit of Escherichia coli Nitrate Reductase A Contains a Novel [4Fe-4S] Cluster with a High-Spin Ground State. Biochemistry 2004, 43, 5324–5333. [Google Scholar] [CrossRef] [PubMed]
- Rothery, R.A.; Bertero, M.G.; Spreter, T.; Bouromand, N.; Strynadka, N.C.; Weiner, J.H. Protein crystallography reveals a Role for the FS0 cluster of Escherichia coli nitrate reductase A (NarGHI) in enzyme maturation. J. Biol. Chem. 2010, 285, 8801–8807. [Google Scholar] [CrossRef] [PubMed]
- Hanzelmann, P.; Dobbek, H.; Gremer, L.; Huber, R.; Meyer, O. The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase. J. Mol. Biol. 2000, 301, 1221–1235. [Google Scholar] [CrossRef] [PubMed]
- Temple, C.A.; Rajagopalan, K.V. Mechanism of assembly of the Bis(Molybdopterin guanine dinucleotide)molybdenum cofactor in Rhodobacter sphaeroides dimethyl sulfoxide reductase. J. Biol. Chem. 2000, 275, 40202–40210. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.S.; Howell, J.M.; Workentine, M.L.; Turner, R.J. Twin-arginine translocase may have a role in the chaperone function of NarJ from Escherichia coli. Biochem. Biophys. Res. Commun. 2006, 343, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Jack, R.L.; Buchanan, G.; Dubini, A.; Hatzixanthis, K.; Palmer, T.; Sargent, F. Coordinating assembly and export of complex bacterial proteins. EMBO J. 2004, 23, 3962–3972. [Google Scholar] [CrossRef] [PubMed]
- Ize, B.; Coulthurst, S.J.; Hatzixanthis, K.; Caldelari, I.; Buchanan, G.; Barclay, E.C.; Richardson, D.J.; Palmer, T.; Sargent, F. Remnant signal peptides on non-exported enzymes: Implications for the evolution of prokaryotic respiratory chains. Microbiology 2009, 155 Pt 12, 3992–4004. [Google Scholar] [CrossRef] [PubMed]
- Zafra, O.; Ramirez, S.; Castan, P.; Moreno, R.; Cava, F.; Valles, C.; Caro, E.; Berenguer, J. A cytochrome c encoded by the nar operon is required for the synthesis of active respiratory nitrate reductase in Thermus thermophilus. FEBS Lett. 2002, 523, 99–102. [Google Scholar] [CrossRef]
- Zafra, O.; Cava, F.; Blasco, F.; Magalon, A.; Berenguer, J. Membrane-associated maturation of the heterotetrameric nitrate reductase of Thermus thermophilus. J. Bacteriol. 2005, 187, 3990–3996. [Google Scholar] [CrossRef]
- Arias-Cartin, R.; Ceccaldi, P.; Schoepp-Cothenet, B.; Frick, K.; Blanc, J.M.; Guigliarelli, B.; Walburger, A.; Grimaldi, S.; Friedrich, T.; Receveur-Brechot, V.; et al. Redox cofactors insertion in prokaryotic molybdoenzymes occurs via a conserved folding mechanism. Sci. Rep. 2016, 6, 37743. [Google Scholar] [CrossRef]
- Vergnes, A.; Gouffi-Belhabich, K.; Blasco, F.; Giordano, G.; Magalon, A. Involvement of the molybdenum cofactor biosynthetic machinery in the maturation of the Escherichia coli nitrate reductase A. J. Biol. Chem. 2004, 279, 41398–41403. [Google Scholar] [CrossRef]
- Magalon, A.; Frixon, C.; Pommier, J.; Giordano, G.; Blasco, F. In vivo interactions between gene products involved in the final stages of molybdenum cofactor biosynthesis in Escherichia coli. J. Biol. Chem. 2002, 277, 48199–48204. [Google Scholar] [CrossRef] [PubMed]
- Schroder, I.; Rech, S.; Krafft, T.; Macy, J.M. Purification and characterization of the selenate reductase from Thauera selenatis. J. Biol. Chem. 1997, 272, 23765–23768. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Espinosa, R.M.; Dridge, E.J.; Bonete, M.J.; Butt, J.N.; Butler, C.S.; Sargent, F.; Richardson, D.J. Look on the positive side! The orientation, identification and bioenergetics of ’Archaeal’ membrane-bound nitrate reductases. FEMS Microbiol. Lett. 2007, 276, 129–139. [Google Scholar] [CrossRef]
- Thorell, H.D.; Stenklo, K.; Karlsson, J.; Nilsson, T. A gene cluster for chlorate metabolism in Ideonella dechloratans. Appl. Environ. Microbiol. 2003, 69, 5585–5592. [Google Scholar] [CrossRef] [PubMed]
- Barnum, T.P.; Coates, J.D. An uncharacterized clade in the DMSO reductase family of molybdenum oxidoreductases is a new type of chlorate reductase. Environ. Microbiol. Rep. 2020, 12, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Bender, K.S.; Shang, C.; Chakraborty, R.; Belchik, S.M.; Coates, J.D.; Achenbach, L.A. Identification, characterization, and classification of genes encoding perchlorate reductase. J. Bacteriol. 2005, 187, 5090–5096. [Google Scholar] [CrossRef] [PubMed]
- Clark, I.C.; Youngblut, M.; Jacobsen, G.; Wetmore, K.M.; Deutschbauer, A.; Lucas, L.; Coates, J.D. Genetic dissection of chlorate respiration in Pseudomonas stutzeri PDA reveals syntrophic (per)chlorate reduction. Environ. Microbiol. 2016, 18, 3342–3354. [Google Scholar] [CrossRef]
- Krafft, T.; Macy, J.M. Purification and characterization of the respiratory arsenate reductase of Chrysiogenes arsenatis. Eur. J. Biochem. 1998, 255, 647–653. [Google Scholar] [CrossRef]
- Afkar, E.; Lisak, J.; Saltikov, C.; Basu, P.; Oremland, R.S.; Stolz, J.F. The respiratory arsenate reductase from Bacillus selenitireducens strain MLS10. FEMS Microbiol. Lett. 2003, 226, 107–112. [Google Scholar] [CrossRef]
- James, M.J.; Coulthurst, S.J.; Palmer, T.; Sargent, F. Signal peptide etiquette during assembly of a complex respiratory enzyme. Mol. Microbiol. 2013, 90, 400–414. [Google Scholar] [CrossRef]
- Kniemeyer, O.; Heider, J. Ethylbenzene dehydrogenase, a novel hydrocarbon-oxidizing molybdenum/iron-sulfur/heme enzyme. J. Biol. Chem. 2001, 276, 21381–21386. [Google Scholar] [CrossRef] [PubMed]
- Dermer, J.; Fuchs, G. Molybdoenzyme that catalyzes the anaerobic hydroxylation of a tertiary carbon atom in the side chain of cholesterol. J. Biol. Chem. 2012, 287, 36905–36916. [Google Scholar] [CrossRef] [PubMed]
- McDevitt, C.A.; Hugenholtz, P.; Hanson, G.R.; McEwan, A.G. Molecular analysis of dimethyl sulphide dehydrogenase from Rhodovulum sulfidophilum: Its place in the dimethyl sulphoxide reductase family of microbial molybdopterin-containing enzymes. Mol. Microbiol. 2002, 44, 1575–1587. [Google Scholar] [CrossRef] [PubMed]
- Richey, C.; Chovanec, P.; Hoeft, S.E.; Oremland, R.S.; Basu, P.; Stolz, J.F. Respiratory arsenate reductase as a bidirectional enzyme. Biochem. Biophys. Res. Commun. 2009, 382, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Szyttenholm, J.; Chaspoul, F.; Bauzan, M.; Ducluzeau, A.L.; Chehade, M.H.; Pierrel, F.; Denis, Y.; Nitschke, W.; Schoepp-Cothenet, B. The controversy on the ancestral arsenite oxidizing enzyme; deducing evolutionary histories with phylogeny and thermodynamics. Biochim. Biophys. Acta Bioenerg. 2020, 1861, 148252. [Google Scholar] [CrossRef] [PubMed]
- Stolz, J.F.; Basu, P.; Santini, J.M.; Oremland, R.S. Arsenic and selenium in microbial metabolism. Ann. Rev. Microbiol. 2006, 60, 107–130. [Google Scholar] [CrossRef] [PubMed]
- Duval, S.; Ducluzeau, A.L.; Nitschke, W.; Schoepp-Cothenet, B. Enzyme phylogenies as markers for the oxidation state of the environment: The case of respiratory arsenate reductase and related enzymes. BMC Evol. Biol. 2008, 8, 206. [Google Scholar] [CrossRef] [PubMed]
- van Lis, R.; Nitschke, W.; Duval, S.; Schoepp-Cothenet, B. Arsenics as bioenergetic substrates. Biochim. Biophys. Acta 2013, 1827, 176–188. [Google Scholar] [CrossRef]
- Guymer, D.; Maillard, J.; Sargent, F. A genetic analysis of in vivo selenate reduction by Salmonella enterica serovar Typhimurium LT2 and Escherichia coli K12. Arch. Microbiol. 2009, 191, 519–528. [Google Scholar] [CrossRef]
- Lubitz, S.P.; Weiner, J.H. The Escherichia coli ynfEFGHI operon encodes polypeptides which are paralogues of dimethyl sulfoxide reductase (DmsABC). Arch. Biochem. Biophys. 2003, 418, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Winstone, T.L.; Workentine, M.L.; Sarfo, K.J.; Binding, A.J.; Haslam, B.D.; Turner, R.J. Physical nature of signal peptide binding to DmsD. Arch. Biochem. Biophys. 2006, 455, 89–97. [Google Scholar] [CrossRef]
- Sambasivarao, D.; Turner, R.J.; Simala-Grant, J.L.; Shaw, G.; Hu, J.; Weiner, J.H. Multiple roles for the twin arginine leader sequence of dimethyl sulfoxide reductase of Escherichia coli. J. Biol. Chem. 2000, 275, 22526–22531. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Rothery, R.A.; Voss, J.E.; Weiner, J.H. Correct assembly of iron-sulfur cluster FS0 into Escherichia coli dimethyl sulfoxide reductase (DmsABC) is a prerequisite for molybdenum cofactor insertion. J. Biol. Chem. 2011, 286, 15147–15154. [Google Scholar] [CrossRef] [PubMed]
- Rothery, R.A.; Grant, J.L.; Johnson, J.L.; Rajagopalan, K.V.; Weiner, J.H. Association of molybdopterin guanine dinucleotide with Escherichia coli dimethyl sulfoxide reductase: Effect of tungstate and a mob mutation. J. Bacteriol. 1995, 177, 2057–2063. [Google Scholar] [CrossRef]
- Sambasivarao, D.; Turner, R.J.; Bilous, P.T.; Rothery, R.A.; Shaw, G.; Weiner, J.H. Differential effects of a molybdopterin synthase sulfurylase (moeB) mutation on Escherichia coli molybdoenzyme maturation. Biochem. Cell Biol. 2002, 80, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Stanley, N.R.; Sargent, F.; Buchanan, G.; Shi, J.; Stewart, V.; Palmer, T.; Berks, B.C. Behaviour of topological marker proteins targeted to the Tat protein transport pathway. Mol. Microbiol. 2002, 43, 1005–1021. [Google Scholar] [CrossRef]
- Ramasamy, S.K.; Clemons, W.M., Jr. Structure of the twin-arginine signal-binding protein DmsD from Escherichia coli. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009, 65 Pt 8, 746–750. [Google Scholar] [CrossRef]
- Stevens, C.M.; Winstone, T.M.; Turner, R.J.; Paetzel, M. Structural analysis of a monomeric form of the twin-arginine leader peptide binding chaperone Escherichia coli DmsD. J. Mol. Biol. 2009, 389, 124–133. [Google Scholar] [CrossRef]
- Qiu, Y.; Zhang, R.; Binkowski, T.A.; Tereshko, V.; Joachimiak, A.; Kossiakoff, A. The 1.38 A crystal structure of DmsD protein from Salmonella typhimurium, a proofreading chaperone on the Tat pathway. Proteins 2008, 71, 525–533. [Google Scholar] [CrossRef]
- Sarfo, K.J.; Winstone, T.L.; Papish, A.L.; Howell, J.M.; Kadir, H.; Vogel, H.J.; Turner, R.J. Folding forms of Escherichia coli DmsD, a twin-arginine leader binding protein. Biochem. Biophys. Res. Commun. 2004, 315, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Palmer, T.; Stansfeld, P.J. Targeting of proteins to the twin-arginine translocation pathway. Mol. Microbiol. 2020, 113, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.S.; Bay, D.C.; Leach, T.G.; Winstone, T.M.; Kuzniatsova, L.; Tran, V.A.; Turner, R.J. ‘Come into the fold’: A comparative analysis of bacterial redox enzyme maturation protein members of the NarJ subfamily. Biochim. Biophys. Acta (BBA) Biomembr. 2014, 1838, 2971–2984. [Google Scholar] [CrossRef] [PubMed]
- Papish, A.L.; Ladner, C.L.; Turner, R.J. The twin-arginine leader-binding protein, DmsD, interacts with the TatB and TatC subunits of the Escherichia coli twin-arginine translocase. J. Biol. Chem. 2003, 278, 32501–32506. [Google Scholar] [CrossRef] [PubMed]
- Kostecki, J.S.; Li, H.; Turner, R.J.; DeLisa, M.P. Visualizing interactions along the Escherichia coli twin-arginine translocation pathway using protein fragment complementation. PLoS ONE 2010, 5, e9225. [Google Scholar] [CrossRef]
- Kuzniatsova, L.; Winstone, T.M.; Turner, R.J. Identification of protein-protein interactions between the TatB and TatC subunits of the twin-arginine translocase system and respiratory enzyme specific chaperones. Biochim. Biophys. Acta (BBA) Biomembr. 2016, 1858, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Bageshwar, U.K.; DattaGupta, A.; Musser, S.M. Influence of the TorD signal peptide chaperone on Tat-dependent protein translocation. PLoS ONE 2021, 16, e0256715. [Google Scholar] [CrossRef]
- Li, H.; Chang, L.; Howell, J.M.; Turner, R.J. DmsD, a Tat system specific chaperone, interacts with other general chaperones and proteins involved in the molybdenum cofactor biosynthesis. Biochim. Biophys. Acta (BBA) Biomembr. 2010, 1804, 1301–1309. [Google Scholar] [CrossRef][Green Version]
- Wells, M.; Kanmanii, N.J.; Al Zadjali, A.M.; Janecka, J.E.; Basu, P.; Oremland, R.S.; Stolz, J.F. Methane, arsenic, selenium and the origins of the DMSO reductase family. Sci. Rep. 2020, 10, 10946. [Google Scholar] [CrossRef]
- Cerqueira, N.M.; Gonzalez, P.J.; Fernandes, P.A.; Moura, J.J.; Ramos, M.J. Periplasmic Nitrate Reductase and Formate Dehydrogenase: Similar Molecular Architectures with Very Different Enzymatic Activities. Acc. Chem. Res. 2015, 48, 2875–2884. [Google Scholar] [CrossRef]
- Maia, L.B.; Moura, J.J.; Moura, I. Molybdenum and tungsten-dependent formate dehydrogenases. J. Biol. Inorg. Chem. 2015, 20, 287–309. [Google Scholar] [CrossRef] [PubMed]
- Jormakka, M.; Tornroth, S.; Byrne, B.; Iwata, S. Molecular basis of proton motive force generation: Structure of formate dehydrogenase-N. Science 2002, 295, 1863–1868. [Google Scholar] [CrossRef] [PubMed]
- Boyington, J.C.; Gladyshev, V.N.; Khangulov, S.V.; Stadtman, T.C.; Sun, P.D. Crystal structure of formate dehydrogenase H: Catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science 1997, 275, 1305–1308. [Google Scholar] [CrossRef] [PubMed]
- Pommier, J.; Mandrand, M.A.; Holt, S.E.; Boxer, D.H.; Giordano, G. A second phenazine methosulphate-linked formate dehydrogenase isoenzyme in Escherichia coli. Biochim. Biophys. Acta (BBA) Biomembr. 1992, 1107, 305–313. [Google Scholar] [CrossRef]
- Abaibou, H.; Pommier, J.; Benoit, S.; Giordano, G.; Mandrand-Berthelot, M.A. Expression and characterization of the Escherichia coli fdo locus and a possible physiological role for aerobic formate dehydrogenase. J. Bacteriol. 1995, 177, 7141–7149. [Google Scholar] [CrossRef] [PubMed]
- Arnoux, P.; Ruppelt, C.; Oudouhou, F.; Lavergne, J.; Siponen, M.I.; Toci, R.; Mendel, R.R.; Bittner, F.; Pignol, D.; Magalon, A.; et al. Sulphur shuttling across a chaperone during molybdenum cofactor maturation. Nat. Commun. 2015, 6, 6148. [Google Scholar] [CrossRef] [PubMed]
- Luke, I.; Butland, G.; Moore, K.; Buchanan, G.; Lyall, V.; Fairhurst, S.A.; Greenblatt, J.F.; Emili, A.; Palmer, T.; Sargent, F. Biosynthesis of the respiratory formate dehydrogenases from Escherichia coli: Characterization of the FdhE protein. Arch. Microbiol. 2008, 190, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Thome, R.; Gust, A.; Toci, R.; Mendel, R.; Bittner, F.; Magalon, A.; Walburger, A. A sulfurtransferase is essential for activity of formate dehydrogenases in Escherichia coli. J. Biol. Chem. 2012, 287, 4671–4678. [Google Scholar] [CrossRef]
- Raaijmakers, H.C.; Romao, M.J. Formate-reduced E. coli formate dehydrogenase H: The reinterpretation of the crystal structure suggests a new reaction mechanism. J. Biol. Inorg. Chem. 2006, 11, 849–854. [Google Scholar] [CrossRef]
- Coelho, C.; Gonzalez, P.J.; Moura, J.G.; Moura, I.; Trincao, J.; Joao Romao, M. The crystal structure of Cupriavidus necator nitrate reductase in oxidized and partially reduced states. J. Mol. Biol. 2011, 408, 932–948. [Google Scholar] [CrossRef]
- Hartmann, T.; Leimkuhler, S. The oxygen-tolerant and NAD+-dependent formate dehydrogenase from Rhodobacter capsulatus is able to catalyze the reduction of CO2 to formate. FEBS J. 2013, 280, 6083–6096. [Google Scholar] [CrossRef] [PubMed]
- Bohmer, N.; Hartmann, T.; Leimkuhler, S. The chaperone FdsC for Rhodobacter capsulatus formate dehydrogenase binds the bis-molybdopterin guanine dinucleotide cofactor. FEBS Lett. 2014, 588, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Schwanhold, N.; Iobbi-Nivol, C.; Lehmann, A.; Leimkuhler, S. Same but different: Comparison of two system-specific molecular chaperones for the maturation of formate dehydrogenases. PLoS ONE 2018, 13, e0201935. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sieprawska-Lupa, M.; Whitman, W.B.; White, R.H. Cysteine is not the sulfur source for iron-sulfur cluster and methionine biosynthesis in the methanogenic archaeon Methanococcus maripaludis. J. Biol. Chem. 2010, 285, 31923–31929. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Beer, L.L.; Whitman, W.B. Sulfur metabolism in archaea reveals novel processes. Environ. Microbiol. 2012, 14, 2632–2644. [Google Scholar] [CrossRef] [PubMed]
- Garcia, P.S.; D’Angelo, F.; Ollagnier de Choudens, S.; Dussouchaud, M.; Bouveret, E.; Gribaldo, S.; Barras, F. An early origin of iron-sulfur cluster biosynthesis machineries before Earth oxygenation. Nat. Ecol. Evol. 2022, 6, 1564–1572. [Google Scholar] [CrossRef] [PubMed]
- Hitchcock, A.; Hall, S.J.; Myers, J.D.; Mulholland, F.; Jones, M.A.; Kelly, D.J. Roles of the twin-arginine translocase and associated chaperones in the biogenesis of the electron transport chains of the human pathogen Campylobacter jejuni. Microbiology 2010, 156 Pt 10, 2994–3010. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Maillard, J.; Spronk, C.A.; Buchanan, G.; Lyall, V.; Richardson, D.J.; Palmer, T.; Vuister, G.W.; Sargent, F. Structural diversity in twin-arginine signal peptide-binding proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 15641–15646. [Google Scholar] [CrossRef]
- Grahl, S.; Maillard, J.; Spronk, C.A.; Vuister, G.W.; Sargent, F. Overlapping transport and chaperone-binding functions within a bacterial twin-arginine signal peptide. Mol. Microbiol. 2012, 83, 1254–1267. [Google Scholar] [CrossRef]
- Dow, J.M.; Grahl, S.; Ward, R.; Evans, R.; Byron, O.; Norman, D.G.; Palmer, T.; Sargent, F. Characterization of a periplasmic nitrate reductase in complex with its biosynthetic chaperone. FEBS J. 2014, 281, 246–260. [Google Scholar] [CrossRef]
- Zakian, S.; Lafitte, D.; Vergnes, A.; Pimentel, C.; Sebban-Kreuzer, C.; Toci, R.; Claude, J.B.; Guerlesquin, F.; Magalon, A. Basis of recognition between the NarJ chaperone and the N-terminus of the NarG subunit from Escherichia coli nitrate reductase. FEBS J. 2010, 277, 1886–1895. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.S.; Chang, L.; Winstone, T.M.; Turner, R.J. Comparing system-specific chaperone interactions with their Tat dependent redox enzyme substrates. FEBS Lett. 2010, 584, 4553–4558. [Google Scholar] [CrossRef] [PubMed]
- Brondijk, T.H.; Fiegen, D.; Richardson, D.J.; Cole, J.A. Roles of NapF, NapG and NapH, subunits of the Escherichia coli periplasmic nitrate reductase, in ubiquinol oxidation. Mol. Microbiol. 2002, 44, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Kern, M.; Simon, J. Periplasmic nitrate reduction in Wolinella succinogenes: Cytoplasmic NapF facilitates NapA maturation and requires the menaquinol dehydrogenase NapH for membrane attachment. Microbiology 2009, 155 Pt 8, 2784–2794. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reyes, F.; Gavira, M.; Castillo, F.; Moreno-Vivian, C. Periplasmic nitrate-reducing system of the phototrophic bacterium Rhodobacter sphaeroides DSM 158: Transcriptional and mutational analysis of the napKEFDABC gene cluster. Biochem. J. 1998, 331 Pt 3, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Olmo-Mira, M.F.; Gavira, M.; Richardson, D.J.; Castillo, F.; Moreno-Vivian, C.; Roldan, M.D. NapF is a cytoplasmic iron-sulfur protein required for Fe-S cluster assembly in the periplasmic nitrate reductase. J. Biol. Chem. 2004, 279, 49727–49735. [Google Scholar] [CrossRef] [PubMed]
- Nilavongse, A.; Brondijk, T.H.; Overton, T.W.; Richardson, D.J.; Leach, E.R.; Cole, J.A. The NapF protein of the Escherichia coli periplasmic nitrate reductase system: Demonstration of a cytoplasmic location and interaction with the catalytic subunit, NapA. Microbiology 2006, 152 Pt 11, 3227–3237. [Google Scholar] [CrossRef] [PubMed]
- Pittman, M.S.; Elvers, K.T.; Lee, L.; Jones, M.A.; Poole, R.K.; Park, S.F.; Kelly, D.J. Growth of Campylobacter jejuni on nitrate and nitrite: Electron transport to NapA and NrfA via NrfH and distinct roles for NrfA and the globin Cgb in protection against nitrosative stress. Mol. Microbiol. 2007, 63, 575–590. [Google Scholar] [CrossRef]
- Neumann, M.; Schulte, M.; Junemann, N.; Stocklein, W.; Leimkuhler, S. Rhodobacter capsulatus XdhC is involved in molybdenum cofactor binding and insertion into xanthine dehydrogenase. J. Biol. Chem. 2006, 281, 15701–15708. [Google Scholar] [CrossRef]
- Neumann, M.; Stocklein, W.; Walburger, A.; Magalon, A.; Leimkuhler, S. Identification of a Rhodobacter capsulatus L-cysteine desulfurase that sulfurates the molybdenum cofactor when bound to XdhC and before its insertion into xanthine dehydrogenase. Biochemistry 2007, 46, 9586–9595. [Google Scholar] [CrossRef]
- Mendel, R.R. The History of the Molybdenum Cofactor-A Personal View. Molecules 2022, 27, 4934. [Google Scholar] [CrossRef] [PubMed]
- Silvestro, A.; Pommier, J.; Pascal, M.C.; Giordano, G. The inducible trimethylamine N-oxide reductase of Escherichia coli K12: Its localization and inducers. Biochim. Biophys. Acta (BBA) Protein Struct. Mol. Enzym. 1989, 999, 208–216. [Google Scholar] [CrossRef]
- Mejean, V.; Iobbi-Nivol, C.; Lepelletier, M.; Giordano, G.; Chippaux, M.; Pascal, M.C. TMAO anaerobic respiration in Escherichia coli: Involvement of the tor operon. Mol. Microbiol. 1994, 11, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Genest, O.; Ilbert, M.; Mejean, V.; Iobbi-Nivol, C. TorD, an essential chaperone for TorA molybdoenzyme maturation at high temperature. J. Biol. Chem. 2005, 280, 15644–15648. [Google Scholar] [CrossRef] [PubMed]
- Genest, O.; Seduk, F.; Ilbert, M.; Mejean, V.; Iobbi-Nivol, C. Signal peptide protection by specific chaperone. Biochem. Biophys. Res. Commun. 2006, 339, 991–995. [Google Scholar] [CrossRef]
- Genest, O.; Seduk, F.; Theraulaz, L.; Mejean, V.; Iobbi-Nivol, C. Chaperone protection of immature molybdoenzyme during molybdenum cofactor limitation. FEMS Microbiol. Lett. 2006, 265, 51–55. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Redelberger, D.; Genest, O.; Arabet, D.; Mejean, V.; Ilbert, M.; Iobbi-Nivol, C. Quality control of a molybdoenzyme by the Lon protease. FEBS Lett. 2013, 587, 3935–3942. [Google Scholar] [CrossRef]
- Lemaire, O.N.; Honore, F.A.; Jourlin-Castelli, C.; Mejean, V.; Fons, M.; Iobbi-Nivol, C. Efficient respiration on TMAO requires TorD and TorE auxiliary proteins in Shewanella oneidensis. Res. Microbiol. 2016, 167, 630–637. [Google Scholar] [CrossRef]
- Gon, S.; Patte, J.C.; Mejean, V.; Iobbi-Nivol, C. The torYZ (yecK bisZ) operon encodes a third respiratory trimethylamine N-oxide reductase in Escherichia coli. J. Bacteriol. 2000, 182, 5779–5786. [Google Scholar] [CrossRef]
- Ilbert, M.; Mejean, V.; Giudici-Orticoni, M.T.; Samama, J.P.; Iobbi-Nivol, C. Involvement of a mate chaperone (TorD) in the maturation pathway of molybdoenzyme TorA. J. Biol. Chem. 2003, 278, 28787–28792. [Google Scholar] [CrossRef]
- Hatzixanthis, K.; Clarke, T.A.; Oubrie, A.; Richardson, D.J.; Turner, R.J.; Sargent, F. Signal peptide-chaperone interactions on the twin-arginine protein transport pathway. Proc. Natl. Acad. Sci. USA 2005, 102, 8460–8465. [Google Scholar] [CrossRef]
- Li, S.Y.; Chang, B.Y.; Lin, S.C. Coexpression of TorD enhances the transport of GFP via the TAT pathway. J. Biotechnol. 2006, 122, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Tranier, S.; Mortier-Barriere, I.; Ilbert, M.; Birck, C.; Iobbi-Nivol, C.; Mejean, V.; Samama, J.P. Characterization and multiple molecular forms of TorD from Shewanella massilia, the putative chaperone of the molybdoenzyme TorA. Protein Sci. 2002, 11, 2148–2157. [Google Scholar] [CrossRef] [PubMed]
- Tranier, S.; Iobbi-Nivol, C.; Birck, C.; Ilbert, M.; Mortier-Barriere, I.; Mejean, V.; Samama, J.P. A novel protein fold and extreme domain swapping in the dimeric TorD chaperone from Shewanella massilia. Structure 2003, 11, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Ilbert, M.; Mejean, V.; Iobbi-Nivol, C. Functional and structural analysis of members of the TorD family, a large chaperone family dedicated to molybdoproteins. Microbiology 2004, 150 Pt 4, 935–943. [Google Scholar] [CrossRef] [PubMed]
- Guymer, D.; Maillard, J.; Agacan, M.F.; Brearley, C.A.; Sargent, F. Intrinsic GTPase activity of a bacterial twin-arginine translocation proofreading chaperone induced by domain swapping. FEBS J. 2010, 277, 511–525. [Google Scholar] [CrossRef] [PubMed]
- Dow, J.M.; Gabel, F.; Sargent, F.; Palmer, T. Characterization of a pre-export enzyme-chaperone complex on the twin-arginine transport pathway. Biochem. J. 2013, 452, 57–66. [Google Scholar] [CrossRef]
- Genest, O.; Neumann, M.; Seduk, F.; Stocklein, W.; Mejean, V.; Leimkuhler, S.; Iobbi-Nivol, C. Dedicated metallochaperone connects apoenzyme and molybdenum cofactor biosynthesis components. J. Biol. Chem. 2008, 283, 21433–21440. [Google Scholar] [CrossRef]
- Bittner, F.; Oreb, M.; Mendel, R.R. ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 40381–40384. [Google Scholar] [CrossRef]
- Leimkuhler, S.; Iobbi-Nivol, C. Bacterial molybdoenzymes: Old enzymes for new purposes. FEMS Microbiol. Rev. 2016, 40, 1–18. [Google Scholar] [CrossRef]
- Gennaris, A.; Ezraty, B.; Henry, C.; Agrebi, R.; Vergnes, A.; Oheix, E.; Bos, J.; Leverrier, P.; Espinosa, L.; Szewczyk, J.; et al. Repairing oxidized proteins in the bacterial envelope using respiratory chain electrons. Nature 2015, 528, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Hille, R.; Nishino, T.; Bittner, F. Molybdenum enzymes in higher organisms. Coord. Chem. Rev. 2011, 255, 1179–1205. [Google Scholar] [CrossRef] [PubMed]
- Leimkuhler, S.; Kern, M.; Solomon, P.S.; McEwan, A.G.; Schwarz, G.; Mendel, R.R.; Klipp, W. Xanthine dehydrogenase from the phototrophic purple bacterium Rhodobacter capsulatus is more similar to its eukaryotic counterparts than to prokaryotic molybdenum enzymes. Mol. Microbiol. 1998, 27, 853–869. [Google Scholar] [CrossRef] [PubMed]
- Truglio, J.J.; Theis, K.; Leimkuhler, S.; Rappa, R.; Rajagopalan, K.V.; Kisker, C. Crystal Structures of the Active and Alloxanthine-Inhibited Forms of Xanthine Dehydrogenase from Rhodobacter capsulatus. Structure 2002, 10, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Neumann, M.; Stocklein, W.; Leimkuhler, S. Transfer of the molybdenum cofactor synthesized by Rhodobacter capsulatus MoeA to XdhC and MobA. J. Biol. Chem. 2007, 282, 28493–28500. [Google Scholar] [CrossRef] [PubMed]
- Neumann, M.; Mittelstadt, G.; Iobbi-Nivol, C.; Saggu, M.; Lendzian, F.; Hildebrandt, P.; Leimkuhler, S. A periplasmic aldehyde oxidoreductase represents the first molybdopterin cytosine dinucleotide cofactor containing molybdo-flavoenzyme from Escherichia coli. FEBS J. 2009, 276, 2762–2774. [Google Scholar] [CrossRef] [PubMed]
- Schumann, S.; Saggu, M.; Moller, N.; Anker, S.D.; Lendzian, F.; Hildebrandt, P.; Leimkuhler, S. The mechanism of assembly and cofactor insertion into Rhodobacter capsulatus xanthine dehydrogenase. J. Biol. Chem. 2008, 283, 16602–16611. [Google Scholar] [CrossRef]
- Chan, C.S.; Winstone, T.M.; Chang, L.; Stevens, C.M.; Workentine, M.L.; Li, H.; Wei, Y.; Ondrechen, M.J.; Paetzel, M.; Turner, R.J. Identification of Residues in DmsD for Twin-Arginine Leader Peptide Binding, Defined through Random and Bioinformatics-Directed Mutagenesis. Biochemistry 2008, 47, 2749–2759. [Google Scholar] [CrossRef]
- Lorenzi, M.; Sylvi, L.; Gerbaud, G.; Mileo, E.; Halgand, F.; Walburger, A.; Vezin, H.; Belle, V.; Guigliarelli, B.; Magalon, A. Conformational selection underlies recognition of a molybdoenzyme by its dedicated chaperone. PLoS ONE 2012, 7, e49523. [Google Scholar] [CrossRef]
- Winstone, T.M.; Turner, R.J. Thermodynamic characterization of the DmsD binding site for the DmsA twin-arginine motif. Biochemistry 2015, 54, 2040–2051. [Google Scholar] [CrossRef]
- Niedzialkowska, E.; Mrugala, B.; Rugor, A.; Czub, M.P.; Skotnicka, A.; Cotelesage, J.J.H.; George, G.N.; Szaleniec, M.; Minor, W.; Lewinski, K. Optimization of overexpression of a chaperone protein of steroid C25 dehydrogenase for biochemical and biophysical characterization. Protein Expr. Purif. 2017, 134, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Pierro, A.; Bonucci, A.; Normanno, D.; Ansaldi, M.; Pilet, E.; Ouari, O.; Guigliarelli, B.; Etienne, E.; Gerbaud, G.; Magalon, A.; et al. Probing the Structural Dynamics of a Bacterial Chaperone in Its Native Environment by Nitroxide-Based EPR Spectroscopy. Chemistry 2022, 28, e202202249. [Google Scholar] [CrossRef] [PubMed]
- Stevens, C.M.; Okon, M.; McIntosh, L.P.; Paetzel, M. (1)H, (1)(3)C and (1)(5)N resonance assignments and peptide binding site chemical shift perturbation mapping for the Escherichia coli redox enzyme chaperone DmsD. Biomol. NMR Assign. 2013, 7, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Winstone, T.M.; Tran, V.A.; Turner, R.J. The hydrophobic region of the DmsA twin-arginine leader peptide determines specificity with chaperone DmsD. Biochemistry 2013, 52, 7532–7541. [Google Scholar] [CrossRef]
- Tzeng, S.R.; Kalodimos, C.G. Protein dynamics and allostery: An NMR view. Curr. Opin. Struct. Biol. 2011, 21, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Shomar, H.; Bokinsky, G. Towards a Synthetic Biology Toolset for Metallocluster Enzymes in Biosynthetic Pathways: What We Know and What We Need. Molecules 2021, 26, 6930. [Google Scholar] [CrossRef] [PubMed]
- Blasco, F.; Nunzi, F.; Pommier, J.; Brasseur, R.; Chippaux, M.; Giordano, G. Formation of active heterologous nitrate reductases between nitrate reductases A and Z of Escherichia coli. Mol. Microbiol. 1992, 6, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Pinchbeck, B.J.; Soriano-Laguna, M.J.; Sullivan, M.J.; Luque-Almagro, V.M.; Rowley, G.; Ferguson, S.J.; Roldan, M.D.; Richardson, D.J.; Gates, A.J. A dual functional redox enzyme maturation protein for respiratory and assimilatory nitrate reductases in bacteria. Mol. Microbiol. 2019, 111, 1592–1603. [Google Scholar] [CrossRef]
- Hensel, M.; Hinsley, A.P.; Nikolaus, T.; Sawers, G.; Berks, B.C. The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol. Microbiol. 1999, 32, 275–287. [Google Scholar] [CrossRef]
- Rubio, L.M.; Rangaraj, P.; Homer, M.J.; Roberts, G.P.; Ludden, P.W. Cloning and Mutational Analysis of the gamma Gene from Azotobacter vinelandii Defines a New Family of Proteins Capable of Metallocluster Binding and Protein Stabilization. J. Biol. Chem. 2002, 277, 14299–14305. [Google Scholar] [CrossRef]
- Haja, D.K.; Wu, C.H.; Ponomarenko, O.; Poole, F.L., 2nd; George, G.N.; Adams, M.W.W. Improving Arsenic Tolerance of Pyrococcus furiosus by Heterologous Expression of a Respiratory Arsenate Reductase. Appl. Environ. Microbiol. 2020, 86, e01728-20. [Google Scholar] [CrossRef]
- Mintmier, B.; McGarry, J.M.; Sparacino-Watkins, C.E.; Sallmen, J.; Fischer-Schrader, K.; Magalon, A.; McCormick, J.R.; Stolz, J.F.; Schwarz, G.; Bain, D.J.; et al. Molecular cloning, expression and biochemical characterization of periplasmic nitrate reductase from Campylobacter jejuni. FEMS Microbiol. Lett. 2018, 365, fny151. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magalon, A. History of Maturation of Prokaryotic Molybdoenzymes—A Personal View. Molecules 2023, 28, 7195. https://doi.org/10.3390/molecules28207195
Magalon A. History of Maturation of Prokaryotic Molybdoenzymes—A Personal View. Molecules. 2023; 28(20):7195. https://doi.org/10.3390/molecules28207195
Chicago/Turabian StyleMagalon, Axel. 2023. "History of Maturation of Prokaryotic Molybdoenzymes—A Personal View" Molecules 28, no. 20: 7195. https://doi.org/10.3390/molecules28207195
APA StyleMagalon, A. (2023). History of Maturation of Prokaryotic Molybdoenzymes—A Personal View. Molecules, 28(20), 7195. https://doi.org/10.3390/molecules28207195




