Recent Computational Advances Regarding Amyloid-β and Tau Membrane Interactions in Alzheimer’s Disease
Abstract
:1. Introduction
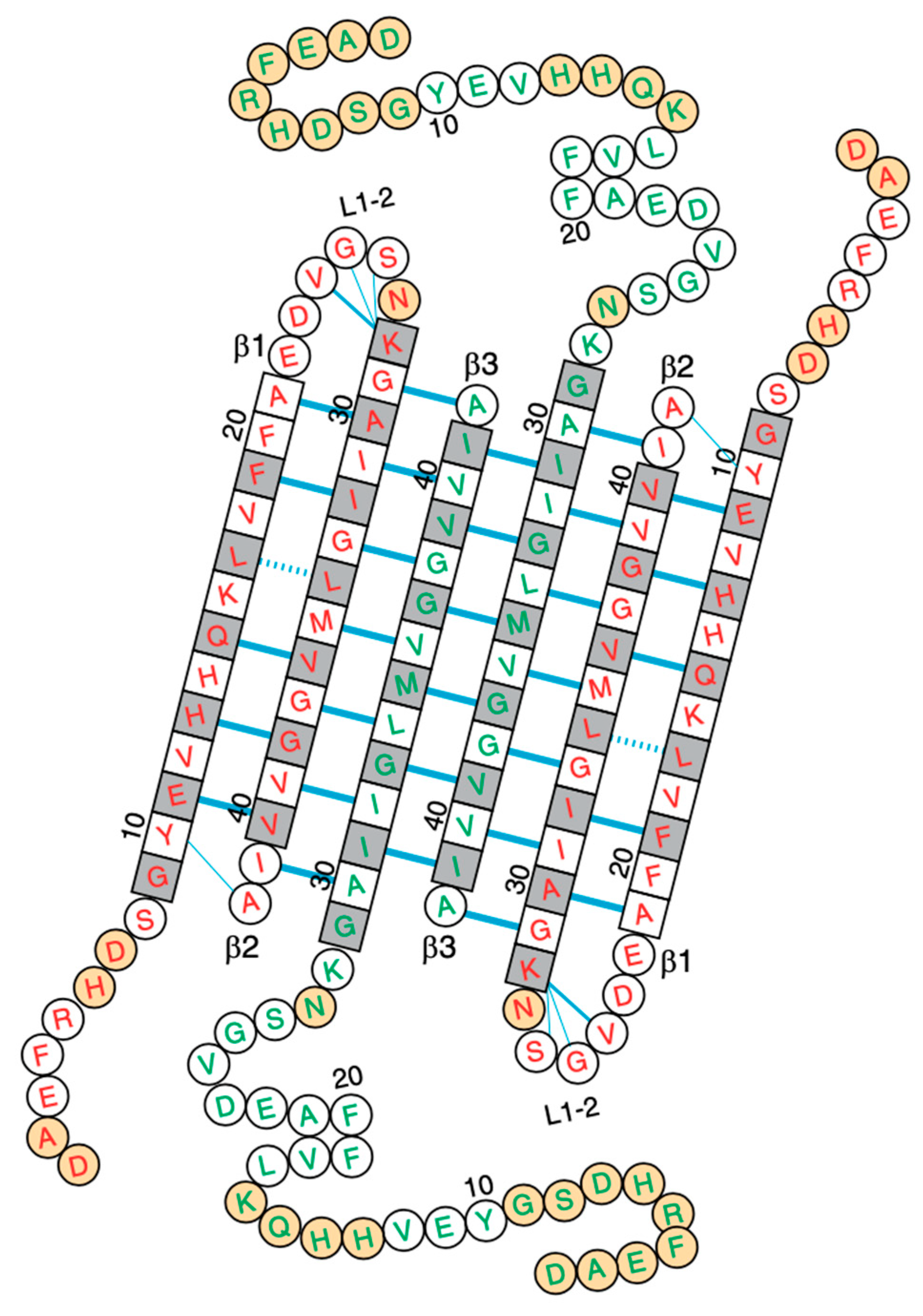
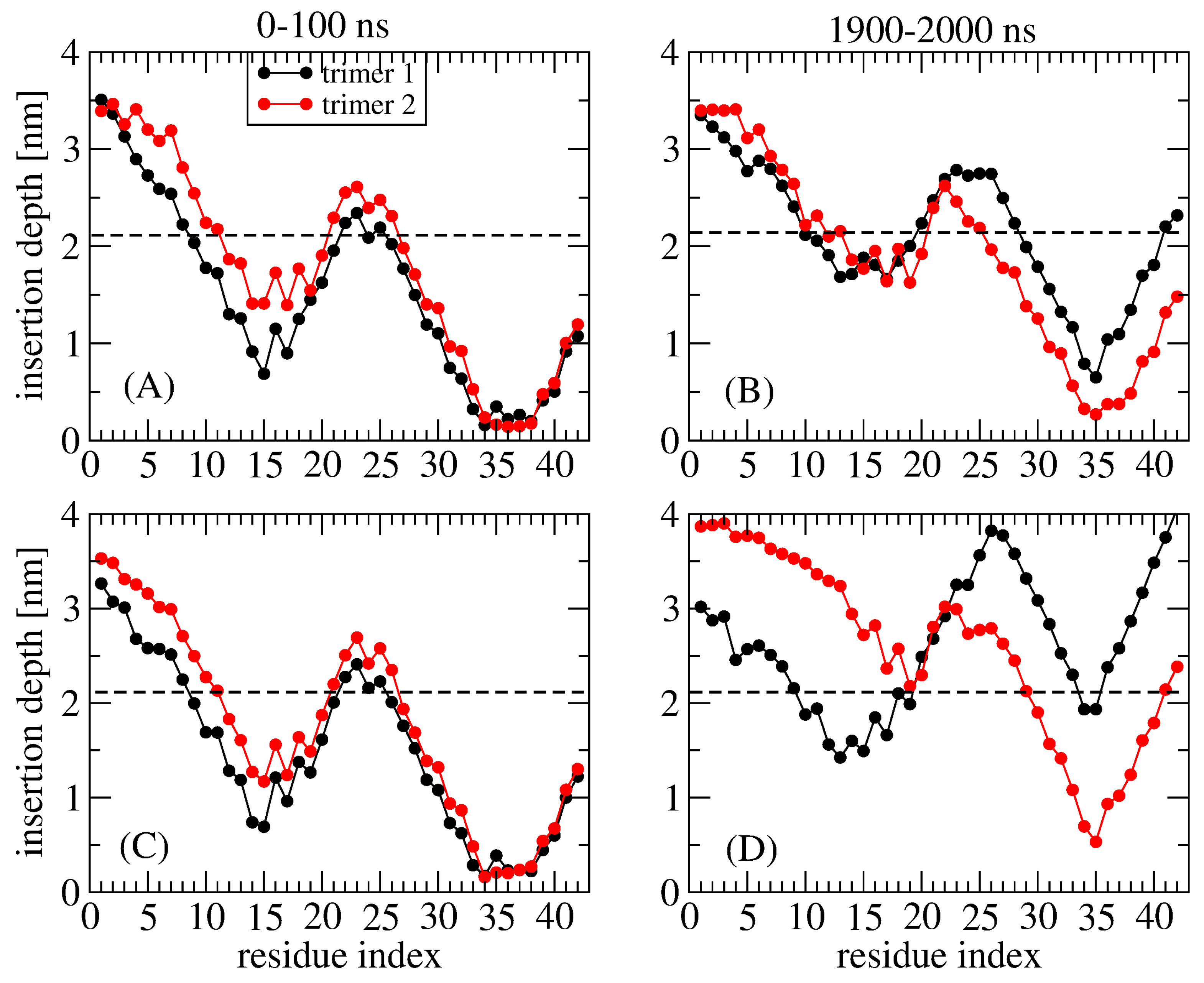
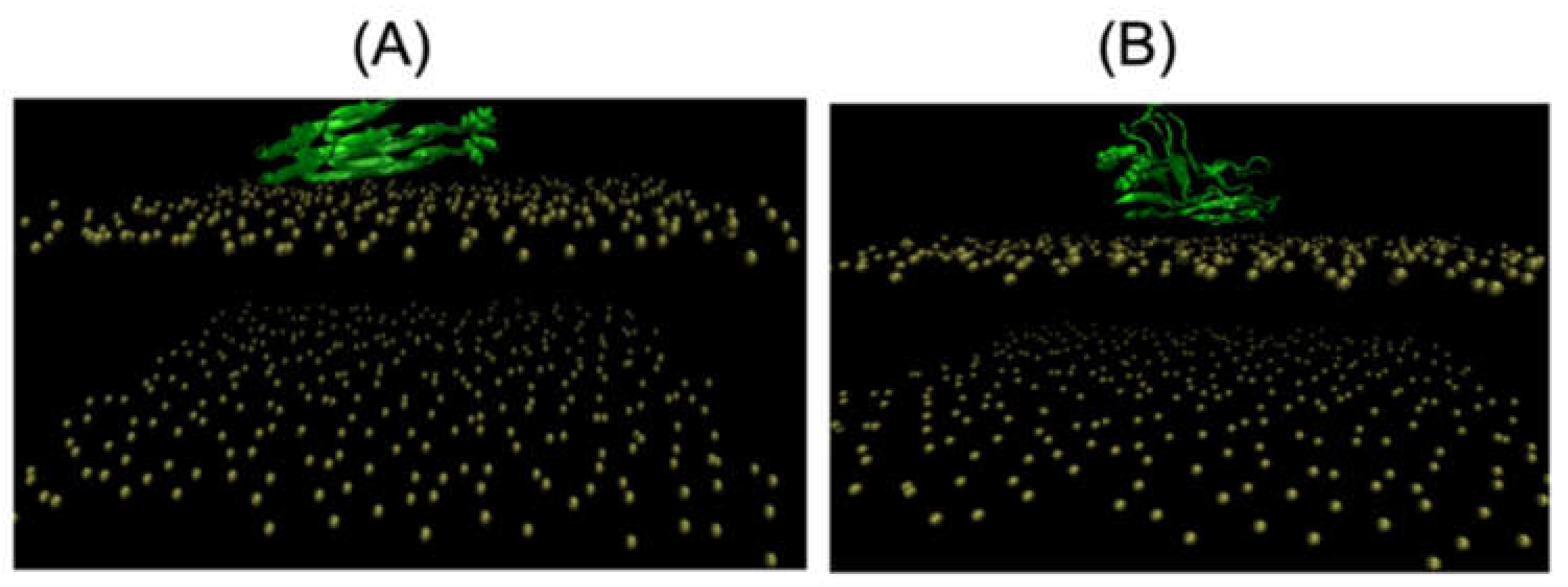
2. Adsorption and Insertion of Amyloid-β
3. Detergent Effect of Amyloid-β
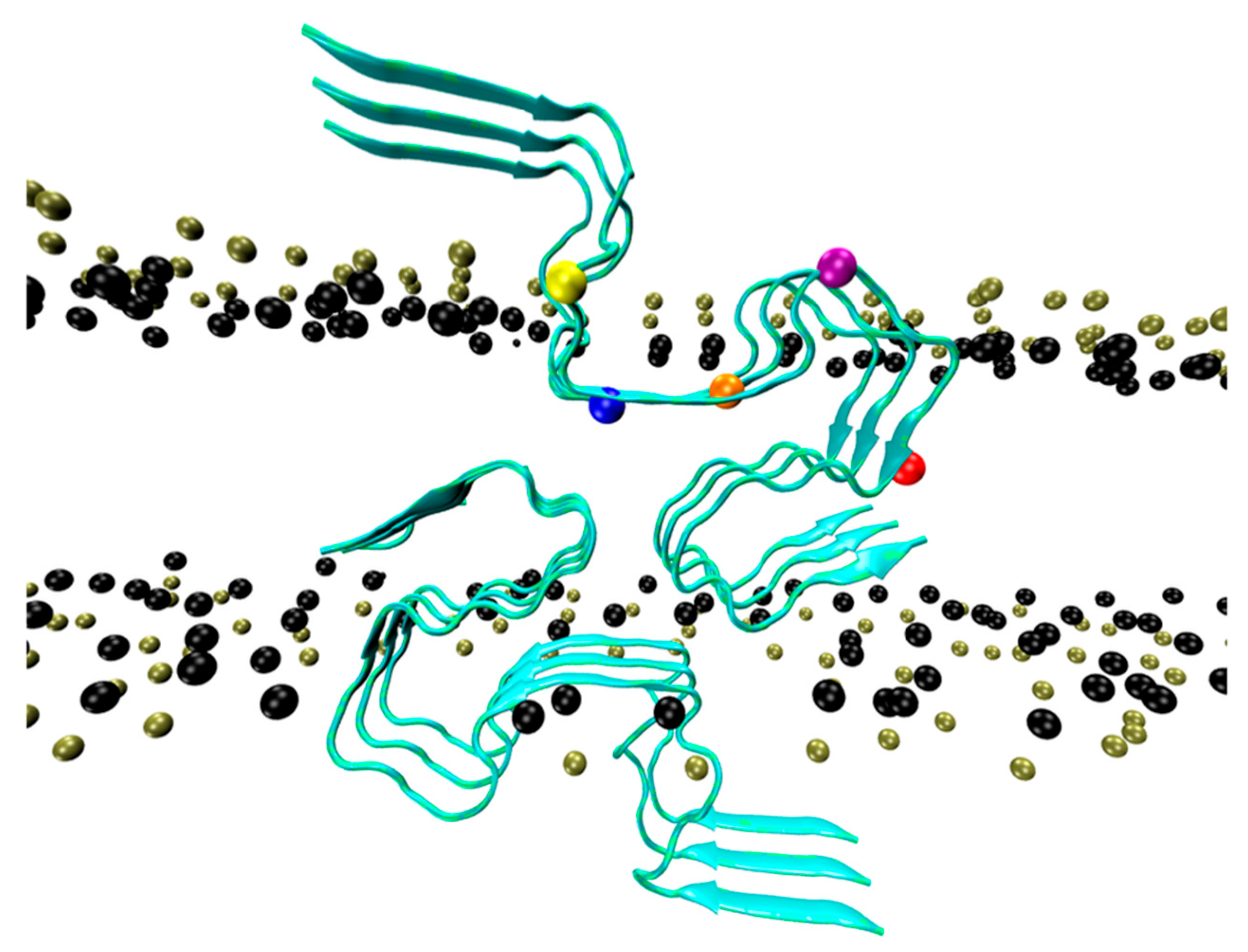
4. Amyloid-β Pore Formation
5. Tau–Membrane Interactions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nasica-Labouze, J.; Nguyen, P.H.; Sterpone, F.; Berthoumieu, O.; Buchete, N.V.; Coté, S.; De Simone, A.; Doig, A.J.; Faller, P.; Garcia, A.; et al. Amyloid β Protein and Alzheimer’s Disease: When computer simulations complement experimental studies. Chem. Rev. 2015, 115, 3518–3563. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.H.; Ramamoorthy, A.; Sahoo, B.R.; Zheng, J.; Faller, P.; Straub, J.E.; Dominguez, L.; Shea, J.E.; Dokholyan, N.V.; De Simone, A.; et al. Amyloid oligomers: A joint experimental/computational perspective on Alzheimer’s disease, Parkinson’s disease, type II diabetes, and amyotrophic lateral sclerosis. Chem. Rev. 2021, 121, 2545–2647. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhang, W.; Yang, Y.; Murzin, A.G.; Falcon, B.; Kotecha, A.; van Beers, M.; Tarutani, A.; Kametani, F.; Garringer, H.J.; et al. Structure-based classification of tauopathies. Nature 2021, 598, 359–363. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Yau, W.M.; Louis, J.M.; Tycko, R. Structures of brain-derived 42-residue amyloid-β fibril polymorphs with unusual molecular conformations and intermolecular interactions. Proc. Natl. Acad. Sci. USA 2023, 120, e2218831120. [Google Scholar] [CrossRef] [PubMed]
- Michaels, T.C.T.; Šarić, A.; Curk, S.; Bernfur, K.; Arosio, P.; Meisl, G.; Dear, A.J.; Cohen, S.I.A.; Dobson, C.M.; Vendruscolo, M.; et al. Dynamics of oligomer populations formed during the aggregation of Alzheimer’s Aβ42 peptide. Nat. Chem. 2020, 12, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J.; Hardy, J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 2016, 8, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Bigi, A.; Cascella, R.; Chito, F.; Cecchi, C. Amyloid fibrils act as a reservoir of soluble oligomers, the main culprits in protein deposition diseases. Bioessays 2022, 44, e2200086. [Google Scholar] [CrossRef]
- Tian, Y.; Viles, J.H. pH dependence of amyloid-β fibril assembly kinetics: Unravelling the microscopic molecular processes. Angew. Chem. Int. Ed. Engl. 2022, 61, e202210675. [Google Scholar] [CrossRef]
- Thacker, D.; Willas, A.; Dear, A.J.; Linse, S. Role of hydrophobicity at the N-terminal region of Aβ42 in secondary nucleation. ACS Chem. Neurosci. 2022, 13, 3477–3487. [Google Scholar] [CrossRef]
- Nguyen, P.H.; Derreumaux, P. Insights into the mixture of Aβ24 and Aβ42 peptides from atomistic simulations. J. Phys. Chem. B 2022, 126, 10689–10696. [Google Scholar] [CrossRef]
- Popov, K.I.; Makepeace, K.A.T.; Petrotchenko, E.V.; Dokholyan, N.V.; Borchers, C.H. Insight into the structure of the "unstructured" tau protein. Structure 2019, 27, 1710–1715.e4. [Google Scholar] [CrossRef] [PubMed]
- Stelzl, L.S.; Pietrek, L.M.; Holla, A.; Oroz, J.; Sikora, M.; Köfinger, J.; Schuler, B.; Zweckstetter, M.; Hummer, G. Global structure of the intrinsically disordered protein tau emerges from its local structure. JACS Au 2022, 2, 673–686. [Google Scholar] [CrossRef]
- Chakraborty, D.; Straub, J.E.; Thirumalai, D. Differences in the free energies between the excited states of Aβ40 and Aβ42 monomers encode their aggregation propensities. Proc. Natl. Acad. Sci. USA 2020, 117, 19926–19937. [Google Scholar] [CrossRef] [PubMed]
- Santuz, H.; Nguyen, P.H.; Sterpone, F.; Derreumaux, P. Small Oligomers of Aβ42 Protein in the bulk solution with AlphaFold2. ACS Chem. Neurosci. 2022, 13, 711–713. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.H.; Sterpone, F.; Derreumaux, P. Self-assembly of amyloid-beta (Aβ) peptides from solution to near in vivo conditions. J. Phys. Chem. B 2022, 126, 10317–10326. [Google Scholar] [CrossRef] [PubMed]
- Jucker, M.; Walker, L.C. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 2013, 501, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Biasetti 2020; Al-Hilaly, Y.K.; Foster, B.E.; Biasetti, L.; Lutter, L.; Pollack, S.J.; Rickard, J.E.; Storey, J.M.D.; Harrington, C.R.; Xue, W.F.; et al. Tau (297-391) forms filaments that structurally mimic the core of paired helical filaments in Alzheimer’s disease brain. FEBS Lett. 2020, 594, 944–950. [Google Scholar] [CrossRef]
- Maina, M.B.; Al-Hilaly, Y.K.; Oakley, S.; Burra, G.; Khanom, T.; Biasetti, L.; Mengham, K.; Marshall, K.; Harrington, C.R.; Wischik, C.M.; et al. Dityrosine cross-links are present in Alzheimer’s disease-derived tau oligomers and paired helical filaments (PHF) which promote the stability of the PHF-core tau (297-391) in vitro. J. Mol. Biol. 2022, 434, 167785. [Google Scholar] [CrossRef]
- Banerjee, S.; Hashemi, M.; Zagorski, K.; Lyubchenko, Y.L. Interaction of Aβ42 with membranes triggers the self-assembly into oligomers. Int. J. Mol. Sci. 2020, 21, 1129. [Google Scholar] [CrossRef]
- Habchi, J.; Chia, S.; Galvagnion, C.; Michaels, T.C.T.; Bellaiche, M.M.J.; Ruggeri, F.S.; Sanguanini, M.; Idini, I.; Kumita, J.R.; Sparr, E.; et al. Cholesterol catalyses Aβ42 aggregation through a heterogeneous nucleation pathway in the presence of lipid membranes. Nat. Chem. 2018, 10, 673–683. [Google Scholar] [CrossRef]
- Petersen, R.B.; Peng, A. Probing the interactions between amyloidogenic proteins and bio-membranes. Biophys. Chem. 2023, 296, 106984. [Google Scholar]
- Viles, J.H. Imaging amyloid-β membrane interactions; ion-channel pores and lipid-bilayer permeability in Alzheimer’s disease. Angew. Chem. Int. Ed. Engl. 2023, 62, e202215785. [Google Scholar] [CrossRef] [PubMed]
- Ciudad, S.; Puig, E.; Botzanowski, T.; Meigooni, M.; Arango, A.S.; Do, J.; Mayzel, M.; Bayoumi, M.; Chaignepain, S.; Maglia, G.; et al. Aβ(1-42) tetramer and octamer structures reveal edge conductivity pores as a mechanism for membrane damage. Nat. Commun. 2020, 11, 3014. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Liang, R.; Kumar, A.; Szwedziak, P.; Viles, J.H. 3D-visualization of amyloid-β oligomer interactions with lipid membranes by cryo-electron tomography. Chem. Sci. 2021, 12, 6896–6907. [Google Scholar] [CrossRef] [PubMed]
- Flagmeier, P.; De, S.; Michaels, T.C.T.; Yang, X.; Dear, A.J.; Emanuelsson, C.; Vendruscolo, M.; Linse, S.; Klenerman, D.; Knowles, T.P.J.; et al. Direct measurement of lipid membrane disruption connects kinetics and toxicity of Aβ42 aggregation. Nat. Struct. Mol. Biol. 2020, 27, 886–891. [Google Scholar] [CrossRef] [PubMed]
- Saha, J.; Bose, P.; Dhakal, S.; Ghosh, P.; Rangachari, V. Ganglioside-enriched phospholipid vesicles induce cooperative Aβ oligomerization and membrane disruption. Biochemistry 2022, 61, 2206–2220. [Google Scholar] [CrossRef] [PubMed]
- Rana, M.; Sharma, A.K. Cu and Zn interactions with Aβ peptides: Consequence of coordination on aggregation and formation of neurotoxic soluble Aβ oligomers. Metallomics 2019, 11, 64–84. [Google Scholar] [CrossRef] [PubMed]
- Budvytyte, R.; Valincius, G. The interactions of amyloid β aggregates with phospholipid membranes and the implications for neurodegeneration. Biochem. Soc. Trans. 2023, 51, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Liu, W.; Zhao, W.; Chang, Z.; Yang, J. The co-effect of copper and lipid vesicles on Aβ aggregation. Biochim. Biophys. Acta Biomembr. 2023, 1865, 184082. [Google Scholar] [CrossRef]
- Zhu, M.; Zeng, L.; Li, Z.; Wang, C.; Wu, L.; Jiang, X. Revealing the nanoarchitectonics of amyloid β-aggregation on two-dimensional biomimetic membranes by surface-enhanced infrared absorption spectroscopy. ChemistryOpen 2023, 12, e202200253. [Google Scholar] [CrossRef]
- Kenyaga, J.M.; Cheng, Q.; Qiang, W. Early stage β-amyloid-membrane interactions modulate lipid dynamics and influence structural interfaces and fibrillation. J. Biol. Chem. 2022, 298, 102491. [Google Scholar] [CrossRef] [PubMed]
- Feuillie, C.; Lambert, E.; Ewald, M.; Azouz, M.; Henry, S.; Marsaudon, S.; Cullin, C.; Lecomte, S.; Molinari, M. High speed AFM and nanoinfrared spectroscopy investigation of Aβ1-42 peptide variants and their interaction with POPC/SM/Chol/GM1 model membranes. Front. Mol. Biosci. 2020, 7, 571696. [Google Scholar] [CrossRef] [PubMed]
- Ida, A.; Abe, M.; Nochi, M.; Soga, C.; Unoura, K.; Nabika, H. Promoted aggregation of Aβ on lipid bilayers in an open flowing system. J. Phys. Chem. Lett. 2021, 12, 4453–4460. [Google Scholar] [CrossRef] [PubMed]
- Baumann, K.N.; Šneiderienė, G.; Sanguanini, M.; Schneider, M.; Rimon, O.; González Díaz, A.; Greer, H.; Thacker, D.; Linse, S.; Knowles, T.P.J.; et al. A kinetic map of the influence of biomimetic lipid model membranes on Aβ42 aggregation. ACS Chem. Neurosci. 2023, 14, 323–329. [Google Scholar] [CrossRef]
- Azouz, M.; Feuillie, C.; Lafleur, M.; Molinari, M.; Lecomte, S. Interaction of tau construct K18 with model lipid membranes. Nanoscale Adv. 2021, 3, 4244–4253. [Google Scholar] [CrossRef]
- Sallaberry, C.A.; Voss, B.J.; Majewski, J.; Biernat, J.; Mandelkow, E.; Chi, E.Y.; Vander Zanden, C.M. Tau and membranes: Interactions that promote folding and condensation. Front. Cell Dev. Biol. 2021, 9, 725241. [Google Scholar] [CrossRef]
- Lasagna-Reeves, C.A.; Sengupta, U.; Castillo-Carranza, D.; Gerson, J.E.; Guerrero-Munoz, M.; Troncoso, J.C.; Jackson, G.R.; Kayed, R. The formation of tau pore-like structures is prevalent and cell specific: Possible implications for the disease phenotypes. Acta Neuropathol. Commun. 2014, 2, 56. [Google Scholar] [CrossRef]
- Ait-Bouziad, N.; Lv, G.; Mahul-Mellier, A.L.; Xiao, S.; Zorludemir, G.; Eliezer, D.; Walz, T.; Lashuel, H.A. Discovery and characterization of stable and toxic Tau/phospholipid oligomeric complexes. Nat. Commun. 2017, 8, 1678. [Google Scholar] [CrossRef]
- Yao, Q.Q.; Wen, J.; Perrett, S.; Wu, S. Distinct lipid membrane-mediated pathways of Tau assembly revealed by single-molecule analysis. Nanoscale 2022, 14, 4604–4613. [Google Scholar] [CrossRef]
- Fatafta, H.; Kav, B.; Bundschuh, B.F.; Loschwitz, J.; Strodel, B. Disorder-to-order transition of the amyloid-β peptide upon lipid binding. Biophys. Chem. 2022, 280, 106700. [Google Scholar] [CrossRef]
- Ahyayauch, H.; García-Arribas, A.B.; Masserini, M.E.; Pantano, S.; Goñi, F.M.; Alonso, A. β-Amyloid (1-42) peptide adsorbs but does not insert into ganglioside-containing phospholipid membranes in the liquid-disordered state: Modelling and experimental studies. Int. J. Biol. Macromol. 2020, 164, 2651–2658. [Google Scholar] [CrossRef] [PubMed]
- Ahyayauch, H.; Masserini, M.; Goñi, F.M.; Alonso, A. The interaction of Aβ42 peptide in monomer, oligomer or fibril forms with sphingomyelin/cholesterol/ganglioside bilayers. Int. J. Biol. Macromol. 2021, 168, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Fatafta, H.; Poojari, C.; Sayyed-Ahmad, A.; Strodel, B.; Owen, M.C. Role of oxidized Gly25, Gly29, and Gly33 residues on the interactions of Aβ1-42 with lipid membranes. ACS Chem. Neurosci. 2020, 11, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Boopathi, S.; Garduño-Juárez, R. Calcium inhibits penetration of Alzheimer’s Aβ1 -42monomers into the membrane. Proteins 2022, 90, 2124–2143. [Google Scholar] [CrossRef] [PubMed]
- Drabik, D.; Chodaczek, G.; Kraszewski, S. Effect of amyloid-β monomers on lipid membrane mechanical parameters-potential implications for mechanically driven neurodegeneration in Alzheimer’s disease. Int. J. Mol. Sci. 2020, 22, 18. [Google Scholar] [CrossRef] [PubMed]
- Tempra, C.; La Rosa, C.; Lolicato, F. The role of alpha-helix on the structure-targeting drug design of amyloidogenic proteins. Chem. Phys. Lipids. 2021, 236, 105061. [Google Scholar] [CrossRef] [PubMed]
- Cholko, T.; Barnum, J.; Chang, C.A. Amyloid-β (Aβ42) peptide aggregation rate and mechanism on surfaces with widely varied properties: Insights from Brownian dynamics simulations. J. Phys. Chem. B 2020, 124, 5549–5558. [Google Scholar] [CrossRef] [PubMed]
- Fatafta, H.; Khaled, M.; Owen, M.C.; Sayyed-Ahmad, A.; Strodel, B. Amyloid-β peptide dimers undergo a random coil to β-sheet transition in the aqueous phase but not at the neuronal membrane. Proc. Natl. Acad. Sci. USA 2021, 118, e2106210118. [Google Scholar] [CrossRef]
- Kargar, F.; Emadi, S.; Fazli, H. Dimerization of Aβ40 inside dipalmitoylphosphatidyl-choline bilayer and its effect on bilayer integrity: Atomistic simulation at three temperatures. Proteins 2020, 88, 1540–1552. [Google Scholar] [CrossRef]
- Wang, B.; Guo, C. Concentration-dependent effects of cholesterol on the dimerization of Amyloid-β peptides in lipid bilayers. ACS Chem. Neurosci. 2022, 13, 2709–2718. [Google Scholar] [CrossRef]
- Agrawal, N.; Skelton, A.A.; Parisini, E. A coarse-grained molecular dynamics investigation on spontaneous binding of Aβ1-40 fibrils with cholesterol-mixed DPPC bilayers. Comput. Struct. Biotechnol. J. 2023, 21, 2688–2695. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.Y.; Cao, Y.; Rouzbehani, M.; Cheng, K.H. Coarse-grained MD simulations reveal beta-amyloid fibrils of various sizes bind to interfacial liquid-ordered and liquid-disordered regions in phase separated lipid rafts with diverse membrane-bound conformational states. Biophys. Chem. 2020, 260, 106355. [Google Scholar] [CrossRef] [PubMed]
- Dias, C.L.; Jalali, S.; Yang, Y.; Cruz, L. Role of cholesterol on Binding of amyloid Fibrils to lipid bilayers. J. Phys. Chem. B 2020, 124, 3036–3042. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.L.; Linh, H.Q.; Krupa, P.; La Penna, G.; Li, M.S. Amyloid β dodecamer disrupts the neuronal membrane more strongly than the mature fibril: Understanding the role of oligomers in neurotoxicity. J. Phys. Chem. B 2022, 126, 3659–3672. [Google Scholar] [CrossRef] [PubMed]
- Panchal, M.; Loeper, J.; Cossec, J.C.; Perruchini, C.; Lazar, A.; Pompon, D.; Duyckaerts, C. Enrichment of cholesterol in microdissected Alzheimer’s disease senile plaques as assessed by mass spectrometry. J. Lipid Res. 2010, 51, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Cutler, R.G.; Kelly, J.; Storie, K.; Pedersen, W.A.; Tammara, A.; Hatanpaa, K.; Troncoso, J.C.; Mattson, M.P. Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2004, 101, 2070–2075. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.; Nguyen, P.H.; Ngo, S.T.; Derreumaux, P. Effect of cholesterol molecules on Aβ1-42 wild-type and mutants trimers. Molecules 2022, 27, 1395. [Google Scholar] [CrossRef] [PubMed]
- Ngo, S.T.; Nguyen, P.H.; Derreumaux, P. Cholesterol molecules alter the energy landscape of small Aβ1-42 Oligomers. J. Phys. Chem. B 2021, 125, 2299–2307. [Google Scholar] [CrossRef]
- Hashemi, M.; Banerjee, S.; Lyubchenko, Y.L. Free cholesterol accelerates Aβ self-assembly on membranes at physiological concentration. Int. J. Mol. Sci. 2022, 23, 2803. [Google Scholar] [CrossRef]
- Chakravorty, A.; McCalpin, S.D.; Sahoo, B.R.; Ramamoorthy, A.; Brooks, C.L. 3rd. Free gangliosides can alter amyloid-β aggregation. J. Phys. Chem. Lett. 2022, 13, 9303–9308. [Google Scholar] [CrossRef]
- Khatua, P.; Jana, A.K.; Hansmann, U.H.E. Effect of lauric acid on the stability of Aβ42 oligomers. ACS Omega 2021, 6, 5795–5804. [Google Scholar] [CrossRef] [PubMed]
- Andrews, B.; Ruggiero, T.; Urbanc, B. How do salt and lipids affect conformational dynamics of Aβ42 monomers in water? Phys. Chem. Chem. Phys. 2023, 25, 2566–2583. [Google Scholar] [CrossRef] [PubMed]
- Österlund, N.; Moons, R.; Ilag, L.L.; Sobott, F.; Gräslund, A. Native ion mobility-mass spectrometry reveals the formation of β-barrel shaped amyloid-β hexamers in a membrane-mimicking environment. J. Am. Chem. Soc. 2019, 141, 10440–10450. [Google Scholar] [CrossRef] [PubMed]
- Quist, A.; Doudevski, I.; Lin, H.; Azimova, R.; Ng, D.; Frangione, B.; Kagan, B.; Ghiso, J.; Lal, R. Amyloid ion channels: A common structural link for protein-misfolding disease. Proc. Natl. Acad. Sci. USA 2005, 102, 10427–10432. [Google Scholar] [CrossRef] [PubMed]
- Ngo, S.T.; Nguyen, P.H.; Derreumaux, P. Impact of A2T and D23N mutations on tetrameric Aβ42 barrel within a dipalmitoylphosphatidylcholine lipid bilayer membrane by replica exchange molecular dynamics. J. Phys. Chem. B 2020, 124, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Durell, S.R.; Kayed, R.; Guy, H.R. The amyloid concentric β-barrel hypothesis: Models of amyloid beta 42 oligomers and annular protofibrils. Proteins 2022, 90, 1190–1209. [Google Scholar] [CrossRef] [PubMed]
- Sepehri, A.; Lazaridis, T. Putative structures of membrane-embedded amyloid β oligomers. ACS Chem. Neurosci. 2023, 14, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Matthes, D.; de Groot, B.L. Molecular dynamics simulations reveal the importance of amyloid-beta oligomer β-sheet edge conformations in membrane permeabilization. J. Biol. Chem. 2023, 299, 103034. [Google Scholar] [CrossRef]
- Rando, C.; Grasso, G.; Sarkar, D.; Sciacca, M.F.M.; Cucci, L.M.; Cosentino, A.; Forte, G.; Pannuzzo, M.; Satriano, C.; Bhunia, A.; et al. GxxxG motif stabilize ion-channel like pores through Cα-H···O interaction in Aβ (1-40). Int. J. Mol. Sci. 2023, 24, 2192. [Google Scholar] [CrossRef]
- Nguyen, P.H.; Derreumaux, P. An S-shaped Aβ42 cross-β hexamer embedded into a lipid bilayer reveals membrane disruption and permeability. ACS Chem. Neurosci. 2023, 14, 936–946. [Google Scholar] [CrossRef]
- Press-Sandler, O.; Miller, Y. Molecular insights into the primary nucleation of polymorphic amyloid β dimers in DOPC lipid bilayer membrane. Protein Sci. 2022, 31, e4283. [Google Scholar] [CrossRef]
- MacAinsh, M.; Zhou, H.X. Partial mimicry of the microtubule binding of tau by its membrane binding. Protein Sci. 2023, 32, e4581. [Google Scholar] [CrossRef] [PubMed]
- Georgieva, E.R.; Xiao, S.; Borbat, P.P.; Freed, J.H.; Eliezer, D. Tau binds to lipid membrane surfaces via short amphipathic helices located in its microtubule-binding repeats. Biophys. J. 2014, 107, 1441–1452. [Google Scholar] [CrossRef] [PubMed]
- Brotzakis, Z.F.; Lindstedt, P.R.; Taylor, R.J.; Rinauro, D.J.; Gallagher, N.C.T.; Bernardes, G.J.L.; Vendruscolo, M. A structural ensemble of a tau-microtubule complex reveals regulatory tau phosphorylation and acetylation mechanisms. ACS Cent. Sci. 2021, 7, 1986–1995. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.H.; Derreumaux, P. Molecular dynamics simulations of the tau R3-R4 domain monomer in the bulk Solution and at the surface of a lipid bilayer model. J. Phys. Chem. B 2022, 126, 3431–3438. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Commins, C.; DeVos, S.L.; Nobuhara, C.K.; Wegmann, S.; Roe, A.D.; Costantino, I.; Fan, Z.; Nicholls, S.B.; Sherman, A.E.; et al. Seed-competent high-molecular-weight tau species accumulates in the cerebrospinal fluid of Alzheimer’s disease mouse model and human patients. Ann. Neurol. 2016, 80, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.H.; Derreumaux, P. Molecular dynamics simulations of the tau amyloid fibril core dimer at the surface of a lipid bilayer model: I. In Alzheimer’s disease. J. Phys. Chem. B 2022, 126, 4849–4856. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.H.; Graf, A.; Lewis, A.; Pham, T.; Acharya, A. Exploring membrane binding targets of disordered human tau aggregates on lipid rafts using multiscale molecular dynamics simulations. Membranes 2022, 12, 1098. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, U.D.; Paul, A.; Bhargava, B.L. The effect of lipid composition on the dynamics of tau fibrils. Proteins 2022, 90, 2103–2115. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, P.H.; Derreumaux, P. Recent Computational Advances Regarding Amyloid-β and Tau Membrane Interactions in Alzheimer’s Disease. Molecules 2023, 28, 7080. https://doi.org/10.3390/molecules28207080
Nguyen PH, Derreumaux P. Recent Computational Advances Regarding Amyloid-β and Tau Membrane Interactions in Alzheimer’s Disease. Molecules. 2023; 28(20):7080. https://doi.org/10.3390/molecules28207080
Chicago/Turabian StyleNguyen, Phuong H., and Philippe Derreumaux. 2023. "Recent Computational Advances Regarding Amyloid-β and Tau Membrane Interactions in Alzheimer’s Disease" Molecules 28, no. 20: 7080. https://doi.org/10.3390/molecules28207080
APA StyleNguyen, P. H., & Derreumaux, P. (2023). Recent Computational Advances Regarding Amyloid-β and Tau Membrane Interactions in Alzheimer’s Disease. Molecules, 28(20), 7080. https://doi.org/10.3390/molecules28207080




