The Hybrid Nano-Biointerface between Proteins/Peptides and Two-Dimensional Nanomaterials
Abstract
:1. Introduction
2. The Nano-Biointerface between 2D Nanomaterials and the Biological Medium
2.1. NP’s Properties Affecting the 2D Nano-Biointerface
2.1.1. Chemical Structure, Oxidation, and Wettability
2.1.2. Surface Chemical Defects and Roughness
2.1.3. NP Shape and Surface Curvature
2.1.4. Toxicity and Biofunctionalization
2.2. Advanced Research on the Hybrid Nano-Biointerface between Proteins/Peptides and 2D Nanomaterials
2.3. Basic Mechanisms and Peculiar Properties of the Nano-Biointerface: Driving Interaction Forces at the 2D Nano-Biointerface
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zhen, X.; Jia, L.; Tang, Q.; Zhao, Y.; Li, P.; Li, J.; Xie, X.; Wang, S. Hybrid biointerface engineering nanoplatform for dual-targeted tumor hypoxia relief and enhanced photodynamic therapy. J. Colloid Interface Sci. 2023, 647, 211–223. [Google Scholar] [CrossRef]
- Hao, Y.; Chen, Y.; He, X.; Han, R.; Yang, C.; Liu, T.; Yang, Y.; Liu, Q.; Qian, Z. RGD peptide modified platinum nanozyme Co-loaded glutathione-responsive prodrug nanoparticles for enhanced chemo-photodynamic bladder cancer therapy. Biomaterials 2023, 293, 121975. [Google Scholar] [CrossRef] [PubMed]
- Yanar, F.; Carugo, D.; Zhang, X. Hybrid Nanoplatforms Comprising Organic Nanocompartments Encapsulating Inorganic Nanoparticles for Enhanced Drug Delivery and Bioimaging Applications. Molecules 2023, 28, 5694. [Google Scholar] [CrossRef]
- Assali, A.; Razzazan, S.; Akhavan, O.; Mottaghitalab, F.; Adeli, M.; Atyabi, F. The bio-interface between functionalized Au NR@GO nanoplatforms with protein corona and their impact on delivery and release system. Colloids Surf. B Biointerfaces 2019, 173, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Rao, S.; Liu, R.; Wang, L.; Chen, W.; Tao, W.; Kong, N. 2D materials-based nanomedicine: From discovery to applications. Adv. Drug Deliv. Rev. 2022, 185, 114268. [Google Scholar] [CrossRef]
- Li, F.; Zhu, J.; Sun, P.; Zhang, M.; Li, Z.; Xu, D.; Gong, X.; Zou, X.; Geim, A.K.; Su, Y.; et al. Highly efficient and selective extraction of gold by reduced graphene oxide. Nat. Commun. 2022, 13, 4472. [Google Scholar] [CrossRef]
- Fatima, J.; Shah, A.N.; Tahir, M.B.; Mehmood, T.; Shah, A.A.; Tanveer, M.; Nazir, R.; Jan, B.L.; Alansi, S. Tunable 2D Nanomaterials; Their Key Roles and Mechanisms in Water Purification and Monitoring. Front. Environ. Sci. 2022, 10, 766743. [Google Scholar] [CrossRef]
- Guarnieri, D.; Sánchez-Moreno, P.; Del Rio Castillo, A.E.; Bonaccorso, F.; Gatto, F.; Bardi, G.; Martín, C.; Vázquez, E.; Catelani, T.; Sabella, S.; et al. Biotransformation and Biological Interaction of Graphene and Graphene Oxide during Simulated Oral Ingestion. Small 2018, 14, 1800227. [Google Scholar] [CrossRef]
- UN General Assembly. Transforming Our World: The 2030 Agenda for Sustainable Development, 21 October 2015, A/RES/70/1. Available online: https://www.refworld.org/docid/57b6e3e44.html (accessed on 30 June 2023).
- Liu, J.; Fu, S.; Yuan, B.; Li, Y.; Deng, Z. Toward a Universal “Adhesive Nanosheet” for the Assembly of Multiple Nanoparticles Based on a Protein-Induced Reduction/Decoration of Graphene Oxide. J. Am. Chem. Soc. 2010, 132, 7279–7281. [Google Scholar] [CrossRef]
- Mahmoudi, M.; Landry, M.P.; Moore, A.; Coreas, R. The protein corona from nanomedicine to environmental science. Nat. Rev. Mater. 2023, 8, 422–438. [Google Scholar] [CrossRef]
- Rampado, R.; Crotti, S.; Caliceti, P.; Pucciarelli, S.; Agostini, M. Recent Advances in Understanding the Protein Corona of Nanoparticles and in the Formulation of “Stealthy” Nanomaterials. Front. Bioeng. Biotechnol. 2020, 8, 166. [Google Scholar] [CrossRef]
- Cui, L.; Quagliarini, E.; Xiao, S.; Giulimondi, F.; Renzi, S.; Digiacomo, L.; Caracciolo, G.; Wang, J.; Amici, A.; Marchini, C.; et al. The protein corona reduces the anticancer effect of graphene oxide in HER-2-positive cancer cells. Nanoscale Adv. 2022, 4, 4009–4015. [Google Scholar] [CrossRef] [PubMed]
- Gebauer, J.S.; Malissek, M.; Simon, S.; Knauer, S.K.; Maskos, M.; Stauber, R.H.; Peukert, W.; Treuel, L. Impact of the Nanoparticle–Protein Corona on Colloidal Stability and Protein Structure. Langmuir 2012, 28, 9673–9679. [Google Scholar] [CrossRef]
- Yan, Y.; Gause, K.T.; Kamphuis, M.M.J.; Ang, C.-S.; O’Brien-Simpson, N.M.; Lenzo, J.C.; Reynolds, E.C.; Nice, E.C.; Caruso, F. Differential Roles of the Protein Corona in the Cellular Uptake of Nanoporous Polymer Particles by Monocyte and Macrophage Cell Lines. ACS Nano 2013, 7, 10960–10970. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Han, P.; Xie, X.; Yin, X.; Duan, G.; Wen, L. Protein corona reduced graphene oxide cytotoxicity by inhibiting endocytosis. Colloid Interface Sci. Commun. 2021, 45, 100514. [Google Scholar] [CrossRef]
- Corbo, C.; Molinaro, R.; Parodi, A.; Toledano Furman, N.E.; Salvatore, F.; Tasciotti, E. The impact of nanoparticle protein corona on cytotoxicity, immunotoxicity and target drug delivery. Nanomedicine 2016, 11, 81–100. [Google Scholar] [CrossRef] [PubMed]
- Baimanov, D.; Wu, J.; Chu, R.; Cai, R.; Wang, B.; Cao, M.; Tao, Y.; Liu, J.; Guo, M.; Wang, J.; et al. Immunological Responses Induced by Blood Protein Coronas on Two-Dimensional MoS2 Nanosheets. ACS Nano 2020, 14, 5529–5542. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, X.; Huang, J.; Wang, J.; Wang, Z. Harnessing Protein Corona for Biomimetic Nanomedicine Design. Biomimetics 2022, 7, 126. [Google Scholar] [CrossRef]
- Li, H.; Fierens, K.; Zhang, Z.; Vanparijs, N.; Schuijs, M.J.; Van Steendam, K.; Feiner Gracia, N.; De Rycke, R.; De Beer, T.; De Beuckelaer, A.; et al. Spontaneous Protein Adsorption on Graphene Oxide Nanosheets Allowing Efficient Intracellular Vaccine Protein Delivery. ACS Appl. Mater. Interfaces 2016, 8, 1147–1155. [Google Scholar] [CrossRef]
- Verde, V.; Longo, A.; Cucci, L.M.; Sanfilippo, V.; Magrì, A.; Satriano, C.; Anfuso, C.D.; Lupo, G.; La Mendola, D. Anti-Angiogenic and Anti-Proliferative Graphene Oxide Nanosheets for Tumor Cell Therapy. Int. J. Mol. Sci. 2020, 21, 5571. [Google Scholar] [CrossRef]
- Mei, K.-C.; Ghazaryan, A.; Teoh, E.Z.; Summers, H.D.; Li, Y.; Ballesteros, B.; Piasecka, J.; Walters, A.; Hider, R.C.; Mailänder, V.; et al. Protein-Corona-by-Design in 2D: A Reliable Platform to Decode Bio-Nano Interactions for the Next-Generation Quality-by-Design Nanomedicines. Adv. Mater. 2018, 30, 1802732. [Google Scholar] [CrossRef]
- Di Santo, R.; Digiacomo, L.; Quagliarini, E.; Capriotti, A.L.; Laganà, A.; Zenezini Chiozzi, R.; Caputo, D.; Cascone, C.; Coppola, R.; Pozzi, D.; et al. Personalized Graphene Oxide-Protein Corona in the Human Plasma of Pancreatic Cancer Patients. Front. Bioeng. Biotechnol. 2020, 8, 491. [Google Scholar] [CrossRef]
- Pinals, R.L.; Chio, L.; Ledesma, F.; Landry, M.P. Engineering at the nano-bio interface: Harnessing the protein corona towards nanoparticle design and function. Analyst 2020, 145, 5090–5112. [Google Scholar] [CrossRef]
- Duan, Y.; Coreas, R.; Liu, Y.; Bitounis, D.; Zhang, Z.; Parviz, D.; Strano, M.; Demokritou, P.; Zhong, W. Prediction of protein corona on nanomaterials by machine learning using novel descriptors. NanoImpact 2020, 17, 100207. [Google Scholar] [CrossRef]
- Lundqvist, M.; Stigler, J.; Elia, G.; Lynch, I.; Cedervall, T.; Dawson, K.A. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. USA 2008, 105, 14265–14270. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Parekh, S.H. Linking graphene-based material physicochemical properties with molecular adsorption, structure and cell fate. Commun. Chem. 2020, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Sopotnik, M.; Leonardi, A.; Križaj, I.; Dušak, P.; Makovec, D.; Mesarič, T.; Ulrih, N.P.; Junkar, I.; Sepčić, K.; Drobne, D. Comparative study of serum protein binding to three different carbon-based nanomaterials. Carbon 2015, 95, 560–572. [Google Scholar] [CrossRef]
- Chong, Y.; Ge, C.; Yang, Z.; Garate, J.A.; Gu, Z.; Weber, J.K.; Liu, J.; Zhou, R. Reduced Cytotoxicity of Graphene Nanosheets Mediated by Blood-Protein Coating. ACS Nano 2015, 9, 5713–5724. [Google Scholar] [CrossRef]
- Shi, X.; Chang, H.; Chen, S.; Lai, C.; Khademhosseini, A.; Wu, H. Regulating Cellular Behavior on Few-Layer Reduced Graphene Oxide Films with Well-Controlled Reduction States. Adv. Funct. Mater. 2012, 22, 751–759. [Google Scholar] [CrossRef]
- Sengupta, B.; Gregory, W.E.; Zhu, J.; Dasetty, S.; Karakaya, M.; Brown, J.M.; Rao, A.M.; Barrows, J.K.; Sarupria, S.; Podila, R. Influence of carbon nanomaterial defects on the formation of protein corona. RSC Adv. 2015, 5, 82395–82402. [Google Scholar] [CrossRef]
- Han, M.; Zhu, L.; Mo, J.; Wei, W.; Yuan, B.; Zhao, J.; Cao, C. Protein Corona and Immune Responses of Borophene: A Comparison of Nanosheet–Plasma Interface with Graphene and Phosphorene. ACS Appl. Bio Mater. 2020, 3, 4220–4229. [Google Scholar] [CrossRef] [PubMed]
- Gogotsi, Y.; Anasori, B. The Rise of MXenes. ACS Nano 2019, 13, 8491–8494. [Google Scholar] [CrossRef]
- Yu, L.; Lu, L.; Zhou, X.; Xu, L. Current Understanding of the Wettability of MXenes. Adv. Mater. Interfaces 2022, 10, 2201818. [Google Scholar] [CrossRef]
- Wu, X.; Tan, F.; Cheng, S.; Chang, Y.; Wang, X.; Chen, L. Investigation of interaction between MXene nanosheets and human plasma and protein corona composition. Nanoscale 2022, 14, 3777–3787. [Google Scholar] [CrossRef]
- Luan, B.; Zhou, R. Wettability and friction of water on a MoS2 nanosheet. Appl. Phys. Lett. 2016, 108, 131601. [Google Scholar] [CrossRef]
- Brito, A.M.M.; Belleti, E.; Menezes, L.R.; Lanfredi, A.J.C.; Nantes-Cardos, I.L. Proteins and Peptides at the Interfaces of Nanostructures. An. Acad. Bras. Ciências 2019, 91. [Google Scholar] [CrossRef] [PubMed]
- Guan, G.; Zhang, S.; Liu, S.; Cai, Y.; Low, M.; Teng, C.P.; Phang, I.Y.; Cheng, Y.; Duei, K.L.; Srinivasan, B.M.; et al. Protein Induces Layer-by-Layer Exfoliation of Transition Metal Dichalcogenides. J. Am. Chem. Soc. 2015, 137, 6152–6155. [Google Scholar] [CrossRef]
- Consiglio, G.; Di Pietro, P.; D’Urso, L.; Forte, G.; Grasso, G.; Sgarlata, C.; Cossement, D.; Snyders, R.; Satriano, C. Surface tailoring of polyacrylate-grafted graphene oxide for controlled interactions at the biointerface. J. Colloid Interface Sci. 2017, 506, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, S.; Montazeri, A.; Rafii-Tabar, H. Molecular dynamics study of the interfacial mechanical properties of the graphene–collagen biological nanocomposite. Comput. Mater. Sci. 2013, 69, 29–39. [Google Scholar] [CrossRef]
- Guo, Y.-n.; Lu, X.; Weng, J.; Leng, Y. Density Functional Theory Study of the Interaction of Arginine-Glycine-Aspartic Acid with Graphene, Defective Graphene, and Graphene Oxide. J. Phys. Chem. C 2013, 117, 5708–5717. [Google Scholar] [CrossRef]
- Qin, R.; Liu, Y.; Tao, F.; Li, C.; Cao, W.; Yang, P. Protein-Bound Freestanding 2D Metal Film for Stealth Information Transmission. Adv. Mater. 2018, 31, 1803377. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Yang, Z.; Chong, Y.; Ge, C.; Weber, J.K.; Bell, D.R.; Zhou, R. Surface Curvature Relation to Protein Adsorption for Carbon-based Nanomaterials. Sci. Rep. 2015, 5, 10886. [Google Scholar] [CrossRef]
- Yin, X.; Li, B.; Liu, S.; Gu, Z.; Zhou, B.; Yang, Z. Effect of the surface curvature on amyloid-β peptide adsorption for graphene. RSC Adv. 2019, 9, 10094–10099. [Google Scholar] [CrossRef] [PubMed]
- Manzi, B.M.; Werner, M.; Ivanova, E.P.; Crawford, R.J.; Baulin, V.A. Simulations of Protein Adsorption on Nanostructured Surfaces. Sci. Rep. 2019, 9, 4694. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Peng, C.; Lv, M.; Li, X.; Zhang, Y.; Chen, N.; Fan, C.; Huang, Q. Protein Corona-Mediated Mitigation of Cytotoxicity of Graphene Oxide. ACS Nano 2011, 5, 3693–3700. [Google Scholar] [CrossRef] [PubMed]
- Sitsanidis, E.D.; Schirmer, J.; Lampinen, A.; Mentel, K.K.; Hiltunen, V.-M.; Ruokolainen, V.; Johansson, A.; Myllyperkiö, P.; Nissinen, M.; Pettersson, M. Tuning protein adsorption on graphene surfaces via laser-induced oxidation. Nanoscale Adv. 2021, 3, 2065–2074. [Google Scholar] [CrossRef]
- Puglia, M.K.; Malhotra, M.; Kumar, C.V. Engineering functional inorganic nanobiomaterials: Controlling interactions between 2D-nanosheets and enzymes. Dalton Trans. 2020, 49, 3917–3933. [Google Scholar] [CrossRef]
- Pattammattel, A.; Puglia, M.; Chakraborty, S.; Deshapriya, I.K.; Dutta, P.K.; Kumar, C.V. Tuning the Activities and Structures of Enzymes Bound to Graphene Oxide with a Protein Glue. Langmuir 2013, 29, 15643–15654. [Google Scholar] [CrossRef]
- Coreas, R.; Castillo, C.; Li, Z.; Yan, D.; Gao, Z.; Chen, J.; Bitounis, D.; Parviz, D.; Strano, M.S.; Demokritou, P.; et al. Biological Impacts of Reduced Graphene Oxide Affected by Protein Corona Formation. Chem. Res. Toxicol. 2022, 35, 1244–1256. [Google Scholar] [CrossRef]
- Riela, L.; Cucci, L.M.; Hansson, Ö.; Marzo, T.; La Mendola, D.; Satriano, C. A Graphene Oxide-Angiogenin Theranostic Nanoplatform for the Therapeutic Targeting of Angiogenic Processes: The Effect of Copper-Supplemented Medium. Inorganics 2022, 10, 188. [Google Scholar] [CrossRef]
- Naletova, I.; Cucci, L.M.; D’Angeli, F.; Anfuso, C.D.; Magrì, A.; La Mendola, D.; Lupo, G.; Satriano, C. A Tunable Nanoplatform of Nanogold Functionalised with Angiogenin Peptides for Anti-Angiogenic Therapy of Brain Tumours. Cancers 2019, 11, 1322. [Google Scholar] [CrossRef]
- Cucci, L.M.; Satriano, C.; Marzo, T.; La Mendola, D. Angiogenin and Copper Crossing in Wound Healing. Int. J. Mol. Sci. 2021, 22, 10704. [Google Scholar] [CrossRef] [PubMed]
- Magrì, A.; Munzone, A.; Peana, M.; Medici, S.; Zoroddu, M.; Hansson, O.; Satriano, C.; Rizzarelli, E.; La Mendola, D. Coordination Environment of Cu(II) Ions Bound to N-Terminal Peptide Fragments of Angiogenin Protein. Int. J. Mol. Sci. 2016, 17, 1240. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, G.; Sanfilippo, V.; Chiaverini, L.; Satriano, C.; Marzo, T.; Merlino, A.; La Mendola, D. Cisplatin binding to angiogenin protein: New molecular pathways and targets for the drug’s anticancer activity. Dalton Trans. 2023, 52, 9058–9067. [Google Scholar] [CrossRef]
- Marzo, T.; Ferraro, G.; Cucci, L.M.; Pratesi, A.; Hansson, Ö.; Satriano, C.; Merlino, A.; La Mendola, D. Oxaliplatin inhibits angiogenin proliferative and cell migration effects in prostate cancer cells. J. Inorg. Biochem. 2022, 226, 111657. [Google Scholar] [CrossRef]
- Xu, W.; Li, Y.; Wang, H.; Du, Q.; Li, M.; Sun, Y.; Cui, M.; Li, L. Study on the Adsorption Performance of Casein/Graphene Oxide Aerogel for Methylene Blue. ACS Omega 2021, 6, 29243–29253. [Google Scholar] [CrossRef]
- Mei, K.C.; Costa, P.M.; Kreuzer, M.; Al-Jamal, K.T. Interpreting 2D Materials Bio-Nano Interactions: Influence of Aggregation Status, Protein Corona, Cell Culture Media, and Cell Types. Adv. Mater. Interfaces 2021, 8, 2100251. [Google Scholar] [CrossRef]
- Liu, J.-W.; Zhang, Q.; Chen, X.-W.; Wang, J.-H. Surface Assembly of Graphene Oxide Nanosheets on SiO2 Particles for the Selective Isolation of Hemoglobin. Chem. A Eur. J. 2011, 17, 4864–4870. [Google Scholar] [CrossRef]
- Dobson, C.M. Protein folding and misfolding. Nature 2003, 426, 884–890. [Google Scholar] [CrossRef]
- Bashiri, G.; Padilla, M.S.; Swingle, K.L.; Shepherd, S.J.; Mitchell, M.J.; Wang, K. Nanoparticle protein corona: From structure and function to therapeutic targeting. Lab A Chip 2023, 23, 1432–1466. [Google Scholar] [CrossRef]
- Stefani, M. Protein Folding and Misfolding on Surfaces. Int. J. Mol. Sci. 2008, 9, 2515–2542. [Google Scholar] [CrossRef] [PubMed]
- Wells, C.; Brennan, S.; Keon, M.; Ooi, L. The role of amyloid oligomers in neurodegenerative pathologies. Int. J. Biol. Macromol. 2021, 181, 582–604. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, C.; Guo, S.; Zhang, J. Interactions of graphene and graphene oxide with proteins and peptides. Nanotechnol. Rev. 2013, 2, 27–45. [Google Scholar] [CrossRef]
- Joshi, S.; Sharma, P.; Siddiqui, R.; Kaushal, K.; Sharma, S.; Verma, G.; Saini, A. A review on peptide functionalized graphene derivatives as nanotools for biosensing. Microchim. Acta 2019, 187, 27. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, K.; Xing, R.; Yan, X. Peptide self-assembly: Thermodynamics and kinetics. Chem. Soc. Rev. 2016, 45, 5589–5604. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, T.; Wang, Y. Mechanistic understanding and binding analysis of two-dimensional MoS2 nanosheets with human serum albumin by the biochemical and biophysical approach. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 211, 18–25. [Google Scholar] [CrossRef]
- Katoch, J.; Kim, S.N.; Kuang, Z.; Farmer, B.L.; Naik, R.R.; Tatulian, S.A.; Ishigami, M. Structure of a Peptide Adsorbed on Graphene and Graphite. Nano Lett. 2012, 12, 2342–2346. [Google Scholar] [CrossRef]
- Reches, M.; Gazit, E. Casting Metal Nanowires Within Discrete Self-Assembled Peptide Nanotubes. Science 2003, 300, 625–627. [Google Scholar] [CrossRef]
- Ménard-Moyon, C.; Venkatesh, V.; Krishna, K.V.; Bonachera, F.; Verma, S.; Bianco, A. Self-Assembly of Tyrosine into Controlled Supramolecular Nanostructures. Chem. A Eur. J. 2015, 21, 11681–11686. [Google Scholar] [CrossRef]
- Nguyen, V.; Zhu, R.; Jenkins, K.; Yang, R. Self-assembly of diphenylalanine peptide with controlled polarization for power generation. Nat. Commun. 2016, 7, 13566. [Google Scholar] [CrossRef]
- Vasilev, S.; Zelenovskiy, P.; Vasileva, D.; Nuraeva, A.; Shur, V.Y.; Kholkin, A.L. Piezoelectric properties of diphenylalanine microtubes prepared from the solution. J. Phys. Chem. Solids 2016, 93, 68–72. [Google Scholar] [CrossRef]
- Seabra, A.B.; Durán, N. Biological applications of peptides nanotubes: An overview. Peptides 2013, 39, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Ryan, K.; Neumayer, S.M.; Maraka, H.V.R.; Buchete, N.-V.; Kholkin, A.L.; Rice, J.H.; Rodriguez, B.J. Thermal and aqueous stability improvement of graphene oxide enhanced diphenylalanine nanocomposites. Sci. Technol. Adv. Mater. 2017, 18, 172–179. [Google Scholar] [CrossRef]
- Trapani, G.; Caruso, V.C.L.; Cucci, L.M.; Attanasio, F.; Tabbì, G.; Forte, G.; La Mendola, D.; Satriano, C. Graphene Oxide Nanosheets Tailored With Aromatic Dipeptide Nanoassemblies for a Tuneable Interaction with Cell Membranes. Front. Bioeng. Biotechnol. 2020, 8, 427. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Feng, D.; Shi, W.; Li, X.; Ma, H. A graphene oxide-peptide fluorescence sensor for proteolytically active prostate-specific antigen. Mol. BioSyst. 2012, 8, 1441–1445. [Google Scholar] [CrossRef]
- Yang, F.; Yan, Z.; Zhao, J.; Miao, S.; Wang, D.; Yang, P. Rapid capture of trace precious metals by amyloid-like protein membrane with high adsorption capacity and selectivity. J. Mater. Chem. A 2020, 8, 3438–3449. [Google Scholar] [CrossRef]
- Ou, L.; Luo, Y.; Wei, G. Atomic-Level Study of Adsorption, Conformational Change, and Dimerization of an α-Helical Peptide at Graphene Surface. J. Phys. Chem. B 2011, 115, 9813–9822. [Google Scholar] [CrossRef]
- Chen, S.H.; Bell, D.R.; Luan, B. Understanding interactions between biomolecules and two-dimensional nanomaterials using in silico microscopes. Adv. Drug Deliv. Rev. 2022, 186, 114336. [Google Scholar] [CrossRef]
- Gu, Z.; Song, W.; Chen, S.H.; Li, B.; Li, W.; Zhou, R. Defect-assisted protein HP35 denaturation on graphene. Nanoscale 2019, 11, 19362–19369. [Google Scholar] [CrossRef]
- Guo, J.; Yao, X.; Ning, L.; Wang, Q.; Liu, H. The adsorption mechanism and induced conformational changes of three typical proteins with different secondary structural features on graphene. RSC Adv. 2014, 4, 9953–9962. [Google Scholar] [CrossRef]
- Chiticaru, E.A.; Pilan, L.; Damian, C.-M.; Vasile, E.; Burns, J.S.; Ioniţă, M. Influence of Graphene Oxide Concentration when Fabricating an Electrochemical Biosensor for DNA Detection. Biosensors 2019, 9, 113. [Google Scholar] [CrossRef]
- Kim, S.N.; Kuang, Z.; Slocik, J.M.; Jones, S.E.; Cui, Y.; Farmer, B.L.; McAlpine, M.C.; Naik, R.R. Preferential Binding of Peptides to Graphene Edges and Planes. J. Am. Chem. Soc. 2011, 133, 14480–14483. [Google Scholar] [CrossRef]
- Rajesh, C.; Majumder, C.; Mizuseki, H.; Kawazoe, Y. A theoretical study on the interaction of aromatic amino acids with graphene and single walled carbon nanotube. J. Chem. Phys. 2009, 130, 124911. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Yin, B.-C.; Wang, X.-F.; Ye, B.-C. Interaction of peptides with graphene oxide and its application for real-time monitoring of protease activity. Chem. Commun. 2011, 47, 2399–2401. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Wei, S.; Jasensky, J.; Xiao, M.; Wang, Q.; Brooks, C.L.; Chen, Z. Molecular Interactions between Graphene and Biological Molecules. J. Am. Chem. Soc. 2017, 139, 1928–1936. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, V.C.; Jachak, A.; Hurt, R.H.; Kane, A.B. Biological Interactions of Graphene-Family Nanomaterials: An Interdisciplinary Review. Chem. Res. Toxicol. 2011, 25, 15–34. [Google Scholar] [CrossRef]
- Liu, B.; Salgado, S.; Maheshwari, V.; Liu, J. DNA adsorbed on graphene and graphene oxide: Fundamental interactions, desorption and applications. Curr. Opin. Colloid Interface Sci. 2016, 26, 41–49. [Google Scholar] [CrossRef]
- Schultz, J.V.; Tonel, M.Z.; Martins, M.O.; Fagan, S.B. Graphene oxide and flavonoids as potential inhibitors of the spike protein of SARS-CoV-2 variants and interaction between ligands: A parallel study of molecular docking and DFT. Struct. Chem. 2023, 34, 1857–1867. [Google Scholar] [CrossRef]
- Khedri, M.; Maleki, R.; Dahri, M.; Sadeghi, M.M.; Rezvantalab, S.; Santos, H.A.; Shahbazi, M.-A. Engineering of 2D nanomaterials to trap and kill SARS-CoV-2: A new insight from multi-microsecond atomistic simulations. Drug Deliv. Transl. Res. 2021, 12, 1408–1422. [Google Scholar] [CrossRef]
- Unal, M.A.; Bayrakdar, F.; Nazir, H.; Besbinar, O.; Gurcan, C.; Lozano, N.; Arellano, L.M.; Yalcin, S.; Panatli, O.; Celik, D.; et al. Graphene Oxide Nanosheets Interact and Interfere with SARS-CoV-2 Surface Proteins and Cell Receptors to Inhibit Infectivity. Small 2021, 17, 2101483. [Google Scholar] [CrossRef]
- Heskins, M.; Guillet, J.E. Solution Properties of Poly(N-isopropylacrylamide). J. Macromol. Sci. Part A Chem. 1968, 2, 1441–1455. [Google Scholar] [CrossRef]
- Wang, X.; Qiu, X.; Wu, C. Comparison of the Coil-to-Globule and the Globule-to-Coil Transitions of a Single Poly(N-isopropylacrylamide) Homopolymer Chain in Water. Macromolecules 1998, 31, 2972–2976. [Google Scholar] [CrossRef]
- Consiglio, G.; Forte, G. Molecular dynamics study of coil-to-globule transition in a thermo-responsive oligomer bound to various surfaces: Hydrophilic surfaces stabilize the coil form. Phys. Chem. Chem. Phys. 2018, 20, 29754–29763. [Google Scholar] [CrossRef] [PubMed]
- Putri, A.D.; Murti, B.T.; Kanchi, S.; Sabela, M.I.; Bisetty, K.; Tiwari, A.; Inamuddin; Asiri, A.M. Computational studies on the molecular insights of aptamer induced poly(N-isopropylacrylamide)-graft-graphene oxide for on/off- switchable whole-cell cancer diagnostics. Sci. Rep. 2019, 9, 7873. [Google Scholar] [CrossRef] [PubMed]
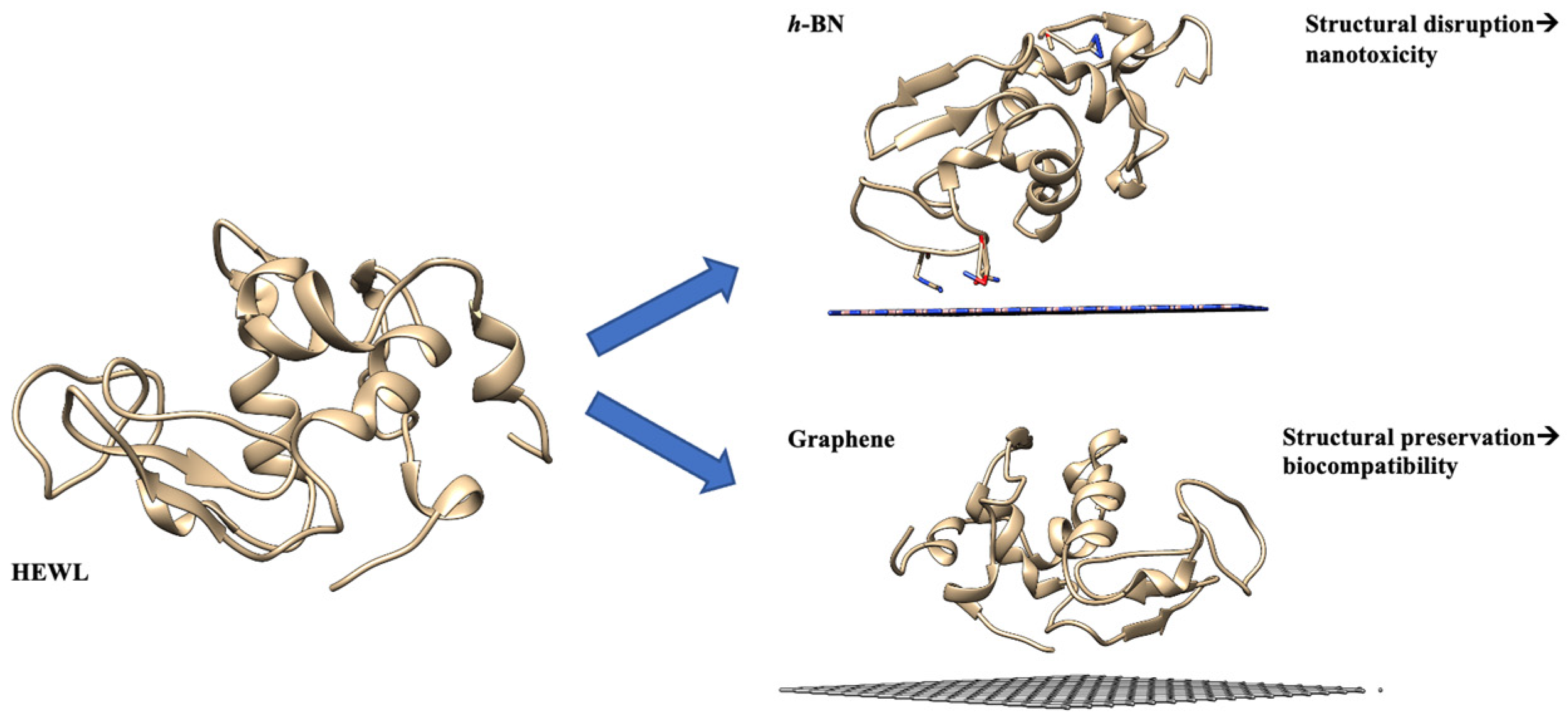
- Paul, S.; Mukhopadhyay, T.K.; Paul, S. In Silico Investigation on the Selective Nanotoxicity of Two-Dimensional Materials to Hen Egg White Lysozyme Protein. ACS Appl. Nano Mater. 2023, 6, 6504–6517. [Google Scholar] [CrossRef]
- Brljak, N.; Jin, R.; Walsh, T.R.; Knecht, M.R. Selective manipulation of peptide orientation on hexagonal boron nitride nanosheets. Nanoscale 2021, 13, 5670–5678. [Google Scholar] [CrossRef]
- Ebrahim-Habibi, M.-B.; Ghobeh, M.; Mahyari, F.A.; Rafii-Tabar, H.; Sasanpour, P. An investigation into non-covalent functionalization of a single-walled carbon nanotube and a graphene sheet with protein G:A combined experimental and molecular dynamics study. Sci. Rep. 2019, 9, 1273. [Google Scholar] [CrossRef]
- Luan, B.; Huynh, T.; Zhao, L.; Zhou, R. Potential Toxicity of Graphene to Cell Functions via Disrupting Protein–Protein Interactions. ACS Nano 2014, 9, 663–669. [Google Scholar] [CrossRef]
- Pandini, G.; Satriano, C.; Pietropaolo, A.; Gianì, F.; Travaglia, A.; La Mendola, D.; Nicoletti, V.G.; Rizzarelli, E. The Inorganic Side of NGF: Copper(II) and Zinc(II) Affect the NGF Mimicking Signaling of the N-Terminus Peptides Encompassing the Recognition Domain of TrkA Receptor. Front. Neurosci. 2016, 10, 569. [Google Scholar] [CrossRef]
- Travaglia, A.; Pietropaolo, A.; Di Martino, R.; Nicoletti, V.G.; La Mendola, D.; Calissano, P.; Rizzarelli, E. A Small Linear Peptide Encompassing the NGF N-Terminus Partly Mimics the Biological Activities of the Entire Neurotrophin in PC12 Cells. ACS Chem. Neurosci. 2015, 6, 1379–1392. [Google Scholar] [CrossRef]
- Naletova, I.; Grasso, G.I.; Satriano, C.; Travaglia, A.; La Mendola, D.; Arena, G.; Rizzarelli, E. Copper complexes of synthetic peptides mimicking neurotrophin-3 enhance neurite outgrowth and CREB phosphorylation. Metallomics 2019, 11, 1567–1578. [Google Scholar] [CrossRef] [PubMed]
- Redigolo, L.; Sanfilippo, V.; La Mendola, D.; Forte, G.; Satriano, C. Bioinspired Nanoplatforms Based on Graphene Oxide and Neurotrophin-Mimicking Peptides. Membranes 2023, 13, 489. [Google Scholar] [CrossRef]
- Song, J.; Li, M.; Liang, H.; Lou, H. Adsorption patterns of aromatic amino acids on monolayer MoS2 and Au-modified MoS2 surfaces: A first-principles study. Comput. Theor. Chem. 2017, 1118, 115–122. [Google Scholar] [CrossRef]
- Pham, L.N.; Walsh, T.R. Predicting biomolecule adsorption on MoS2 nanosheets with high structural fidelity. Chem. Sci. 2022, 13, 5186–5195. [Google Scholar] [CrossRef]
- Gu, Z.; Yang, Z.; Kang, S.-g.; Yang, J.R.; Luo, J.; Zhou, R. Robust Denaturation of Villin Headpiece by MoS2 Nanosheet: Potential Molecular Origin of the Nanotoxicity. Sci. Rep. 2016, 6, 28252. [Google Scholar] [CrossRef]
- Xiao, M.; Wei, S.; Li, Y.; Jasensky, J.; Chen, J.; Brooks, C.L.; Chen, Z. Molecular interactions between single layered MoS2 and biological molecules. Chem. Sci. 2018, 9, 1769–1773. [Google Scholar] [CrossRef]
- Sun, L.; Li, P.; Seki, T.; Tsuchiya, S.; Yatsu, K.; Narimatsu, T.; Sarikaya, M.; Hayamizu, Y. Chiral Recognition of Self-Assembled Peptides on MoS2 via Lattice Matching. Langmuir 2021, 37, 8696–8704. [Google Scholar] [CrossRef]
- Braz, B.A.; Hospinal-Santiani, M.; Martins, G.; Beirão, B.C.B.; Bergamini, M.F.; Marcolino-Junior, L.H.; Soccol, C.R.; Thomaz-Soccol, V. Disposable electrochemical platform based on solid-binding peptides and carbon nanomaterials: An alternative device for leishmaniasis detection. Microchim. Acta 2023, 190, 321. [Google Scholar] [CrossRef]
- Ligorio, C.; Zhou, M.; Wychowaniec, J.K.; Zhu, X.; Bartlam, C.; Miller, A.F.; Vijayaraghavan, A.; Hoyland, J.A.; Saiani, A. Graphene oxide containing self-assembling peptide hybrid hydrogels as a potential 3D injectable cell delivery platform for intervertebral disc repair applications. Acta Biomater. 2019, 92, 92–103. [Google Scholar] [CrossRef]
- Li, R.; Gao, R.; Zhao, Y.; Zhang, F.; Wang, X.; Li, B.; Wang, L.; Ma, L.; Du, J. pH-responsive graphene oxide loaded with targeted peptide and anticancer drug for OSCC therapy. Front. Oncol. 2022, 12, 930920. [Google Scholar] [CrossRef]
- Guilbaud-Chéreau, C.; Dinesh, B.; Wagner, L.; Chaloin, O.; Ménard-Moyon, C.; Bianco, A. Aromatic Dipeptide Homologue-Based Hydrogels for Photocontrolled Drug Release. Nanomaterials 2022, 12, 1643. [Google Scholar] [CrossRef]
- Wu, T.; Jiang, J.; Wang, H.; He, Y.; Zhao, X.; Xue, Y.; Xu, H. Design, synthesis, and in vivo evaluation of GO-SWL-Ahx-K-SWL. Bioorganic Med. Chem. Lett. 2022, 70, 128802. [Google Scholar] [CrossRef] [PubMed]
- Meng, C.; Jiang, W.; Huang, Z.; Liu, T.; Feng, J. Fabrication of a Highly Conductive Silk Knitted Composite Scaffold by Two-Step Electrostatic Self-Assembly for Potential Peripheral Nerve Regeneration. ACS Appl. Mater. Interfaces 2020, 12, 12317–12327. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.K.; Chen, P.; Lee, F.L.; Moochhala, S.; Liedberg, B. Peptide-Assembled Graphene Oxide as a Fluorescent Turn-On Sensor for Lipopolysaccharide (Endotoxin) Detection. Anal. Chem. 2015, 87, 9408–9412. [Google Scholar] [CrossRef]
- Wychowaniec, J.K.; Iliut, M.; Zhou, M.; Moffat, J.; Elsawy, M.A.; Pinheiro, W.A.; Hoyland, J.A.; Miller, A.F.; Vijayaraghavan, A.; Saiani, A. Designing Peptide/Graphene Hybrid Hydrogels through Fine-Tuning of Molecular Interactions. Biomacromolecules 2018, 19, 2731–2741. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Zhang, B.; Liu, S.; Tan, C.; Tan, Y.; Jiang, Y. A New Strategy Involving the Use of Peptides and Graphene Oxide for Fluorescence Turn-on Detection of Proteins. Sensors 2018, 18, 385. [Google Scholar] [CrossRef]
- Ren, T.; Wang, Y.; Yu, Q.; Li, M. Synthesis of antimicrobial peptide-grafted graphene oxide nanosheets with high antimicrobial efficacy. Mater. Lett. 2019, 235, 42–45. [Google Scholar] [CrossRef]
- Pramanik, A.; Gates, K.; Gao, Y.; Zhang, Q.; Han, F.X.; Begum, S.; Rightsell, C.; Sardar, D.; Ray, P.C. Composites Composed of Polydopamine Nanoparticles, Graphene Oxide, and ε-Poly-l-lysine for Removal of Waterborne Contaminants and Eradication of Superbugs. ACS Appl. Nano Mater. 2019, 2, 3339–3347. [Google Scholar] [CrossRef]
- Meng, F.; Sun, H.; Huang, Y.; Tang, Y.; Chen, Q.; Miao, P. Peptide cleavage-based electrochemical biosensor coupling graphene oxide and silver nanoparticles. Anal. Chim. Acta 2019, 1047, 45–51. [Google Scholar] [CrossRef]
- Song, B.; Zhao, H.; Yang, H.; Wang, S. Efficacy of graphene oxide-loaded cationic antimicrobial peptide AWRK6 on the neutralization of endotoxin activity and in the treatment of sepsis. Aging 2021, 13, 19867–19877. [Google Scholar] [CrossRef] [PubMed]
- Puah, P.Y.; Moh, P.Y.; Sipaut, C.S.; Lee, P.C.; How, S.E. Peptide Conjugate on Multilayer Graphene Oxide Film for the Osteogenic Differentiation of Human Wharton’s Jelly-Derived Mesenchymal Stem Cells. Polymers 2021, 13, 3290. [Google Scholar] [CrossRef] [PubMed]
- Braz, B.A.; Hospinal-Santiani, M.; Martins, G.; Pinto, C.S.; Zarbin, A.J.G.; Beirão, B.C.B.; Thomaz-Soccol, V.; Bergamini, M.F.; Marcolino-Junior, L.H.; Soccol, C.R. Graphene-Binding Peptide in Fusion with SARS-CoV-2 Antigen for Electrochemical Immunosensor Construction. Biosensors 2022, 12, 885. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, H.; Nakamura, Y.; Tezuka, S.; Seki, T.; Yatsu, K.; Narimatsu, T.; Nakata, Y.; Hayamizu, Y. Self-assembled GA-Repeated Peptides as a Biomolecular Scaffold for Biosensing with MoS2 Electrochemical Transistors. ACS Appl. Mater. Interfaces 2023, 15, 14058–14066. [Google Scholar] [CrossRef] [PubMed]
- Begum, S.; Pramanik, A.; Gates, K.; Gao, Y.; Ray, P.C. Antimicrobial Peptide-Conjugated MoS2-Based Nanoplatform for Multimodal Synergistic Inactivation of Superbugs. ACS Appl. Bio Mater. 2018, 2, 769–776. [Google Scholar] [CrossRef] [PubMed]



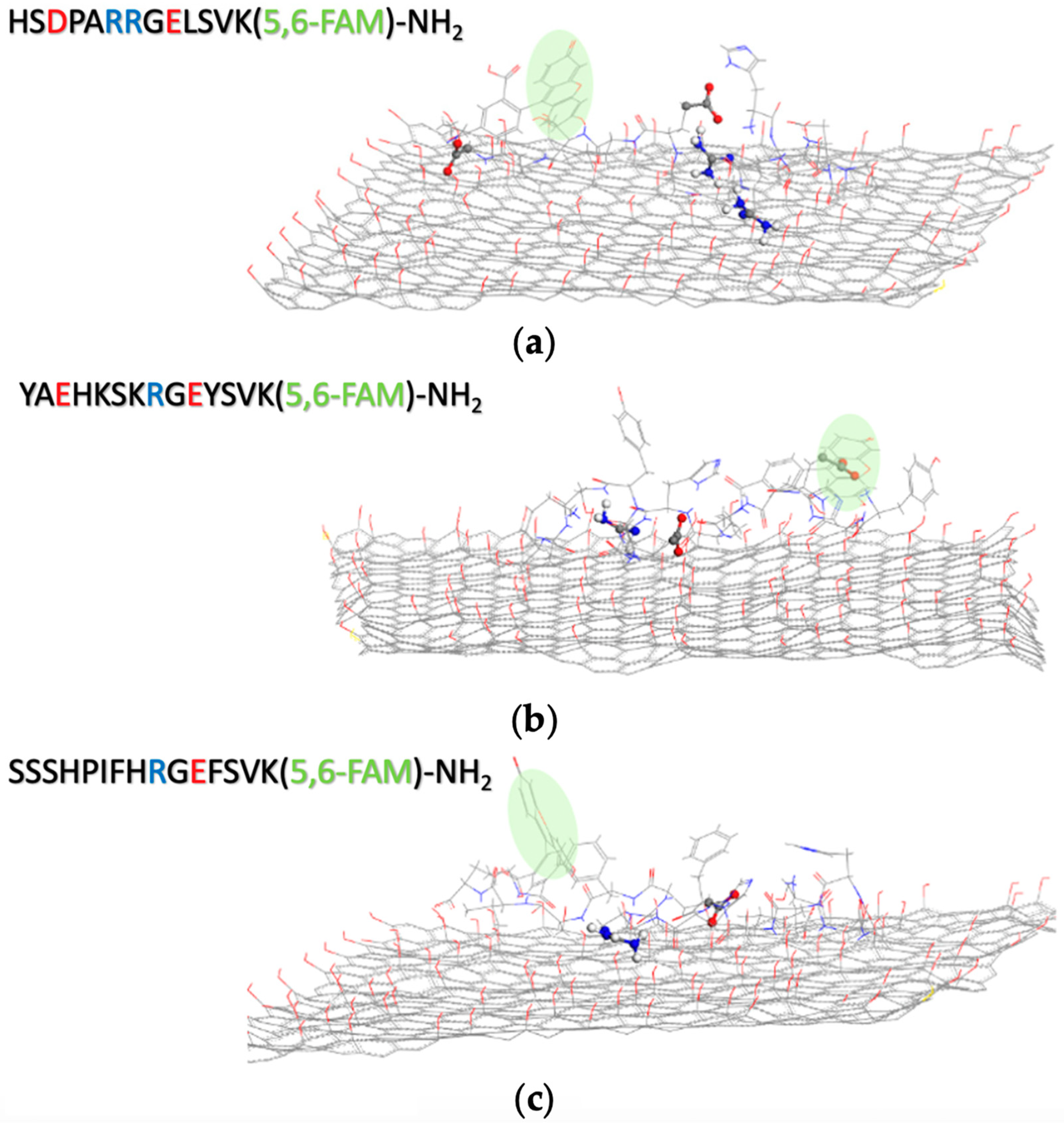
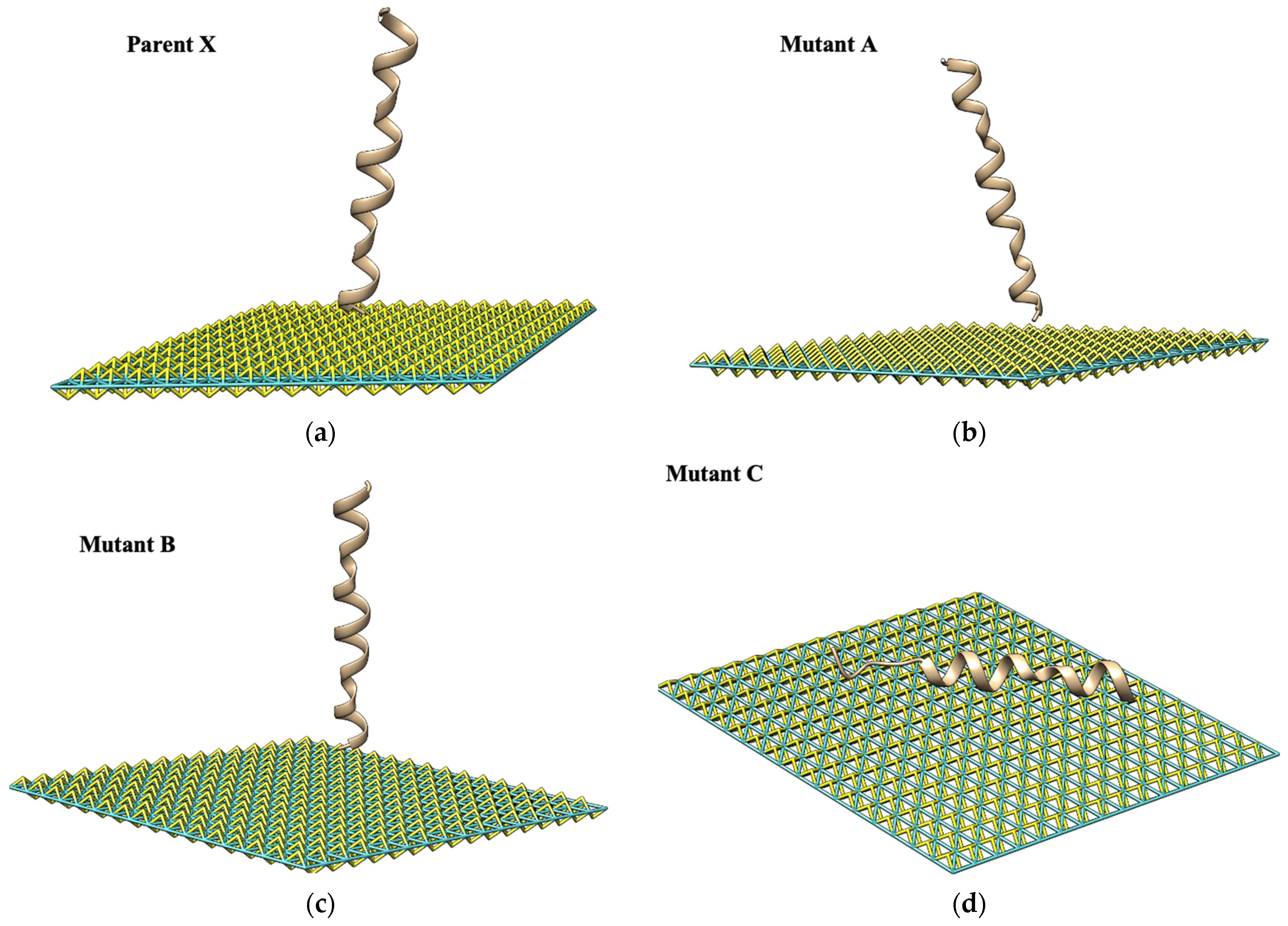
| Nanomaterial | Peptide Core Sequence and/or Name | Lead Interaction | 2D Platform Application | Ref. |
|---|---|---|---|---|
| GO, GQD 1 | 395-KKG; 395-G | covalent | Immunosensing for leishmaniasis detection | [109] |
| GO | FEFKFEFK | electrostatic | 3D injectable cell delivery | [110] |
| GO | GRPR 2 antagonists bombesin (BBN) peptides: GGGRNFQWAVGHL (MATBBN); FQWAVGHL (ATBBN) | electrostatic; π stacking | pH-sensitive drug release | [111] |
| GO, CNTs 3 | Boc-FF; Boc-YY | π stacking | Photocontrolled drug release | [112] |
| GO | YSAYPDSVPMMS (YSA); SWLAYPGAVSYR (SWL) | covalent | Targeted therapeutic drug for non-small cell lung cancer | [113] |
| graphene | Silk amino acids | H-bonding; electrostatic | Composite scaffold for peripheral nerve regeneration | [114] |
| GO | KKNYSSSISSIHC (dye-labeled LPS-binding peptide) 4 | electrostatic | Fluorescent turn-on sensor for endotoxin detection | [115] |
| GO, rGO | VEVKVEVK (V8); FEFKFEFK (F8); FEFEFKFE (FE) | π stacking | Hybrid peptide hydrogels for biomedical applications | [116] |
| GO | RRRRNLWAAQRYGRELRRMSDKFVD (R4); RRRRRRNLWAAQRYGRELRRMSDKFVD (R6); RRRRRRRRNLWAAQRYGRELRRMSDKFVD (R8) | electrostatic; π stacking | Fluorescence ‘turn-on’ protein sensing | [117] |
| GO | FLGVVFKLASKVFPAVFGKV (D28) | covalent grafting | AMP 5-based nanomaterial against Candida albicans and Escherichia coli | [118] |
| GO | ε-poly-L-lysine | electrostatic | Removal of waterborne contaminants; superbug eradication | [119] |
| GO | CGGHSSKLQFWYFWY | electrostatic | Electrochemical biosensor for the sensitive analysis of PSA 6 | [120] |
| GO | SWVGKHGKKFGLKKHKKH (AWRK6) | electrostatic; π stacking | AMP-based nanomaterial for the neutralization of endotoxin activity in the treatment of sepsis | [121] |
| GO | YIGSRWYQNMIRIKVAV (PepS1); QHREDGSYIGSRIKVAV (PepS2); WQPPRARIYIGSRIKVAV (PepS3); DGEARGDSPKRSR (PepS4) | electrostatic; π stacking | Multifunctional osteogenic differentiation of WJ-MSCs 7 in bone tissue regeneration | [122] |
| rGO | EPLQLKM- S protein (SARS-CoV-2-antibody binding site) | electrostatic; π stacking | Electrochemical immunosensor for the SARS-CoV-2 virus | [123] |
| MoS2 | EYGAGAGAGAGAYE (EY5); RYGAGAGAGAGAYR (RY5); QYGAGAGAGAGAYQ (QY5); YGAGAGAGAGAY (Bio-Y5Y) | electrostatic | Biomolecular scaffold for biosensing | [124] |
| MoS2 | GIGAVLKVLTTGLPALISWIKRKRQQ (melittin) | covalent grafting | An AMP 5-based theranostic nanoplatform for PTT 8 and PDT 9 processes in superbug inactivation | [125] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Forte, G.; La Mendola, D.; Satriano, C. The Hybrid Nano-Biointerface between Proteins/Peptides and Two-Dimensional Nanomaterials. Molecules 2023, 28, 7064. https://doi.org/10.3390/molecules28207064
Forte G, La Mendola D, Satriano C. The Hybrid Nano-Biointerface between Proteins/Peptides and Two-Dimensional Nanomaterials. Molecules. 2023; 28(20):7064. https://doi.org/10.3390/molecules28207064
Chicago/Turabian StyleForte, Giuseppe, Diego La Mendola, and Cristina Satriano. 2023. "The Hybrid Nano-Biointerface between Proteins/Peptides and Two-Dimensional Nanomaterials" Molecules 28, no. 20: 7064. https://doi.org/10.3390/molecules28207064
APA StyleForte, G., La Mendola, D., & Satriano, C. (2023). The Hybrid Nano-Biointerface between Proteins/Peptides and Two-Dimensional Nanomaterials. Molecules, 28(20), 7064. https://doi.org/10.3390/molecules28207064







