Chemical Synthesis and Biological Evaluation of 3-Substituted Estrone/Estradiol Derivatives as 17β-Hydroxysteroid Dehydrogenase Type 1 Inhibitors Acting via a Reverse Orientation of the Natural Substrate Estrone
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemistry (General)
2.2. Chemistry (Chemical Synthesis)
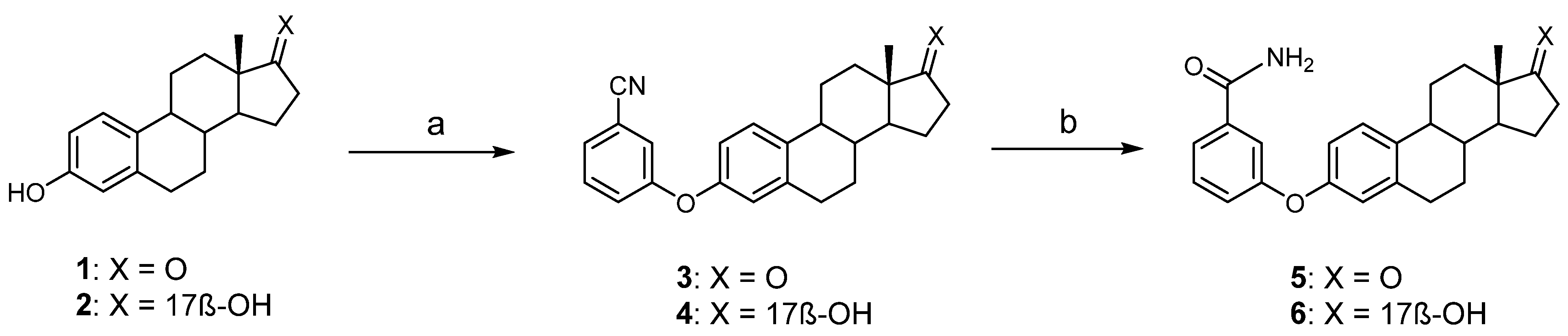
2.2.1. Synthesis of 3-(3-Cyanophenyloxy)-estra-1,3,5(10)-trien-17-one (3)
2.2.2. Synthesis of 3-(3-Cyanophenyloxy)-estra-1,3,5(10)-trien-17β-ol (4)
2.2.3. Synthesis of 3-(3-Carbamoylphenyloxy)-estra-1,3,5(10)-trien-17-one (5)
2.2.4. Synthesis of 3-(3-Carbamoylphenyloxy)-estra-1,3,5(10)-trien-17β-ol (6)
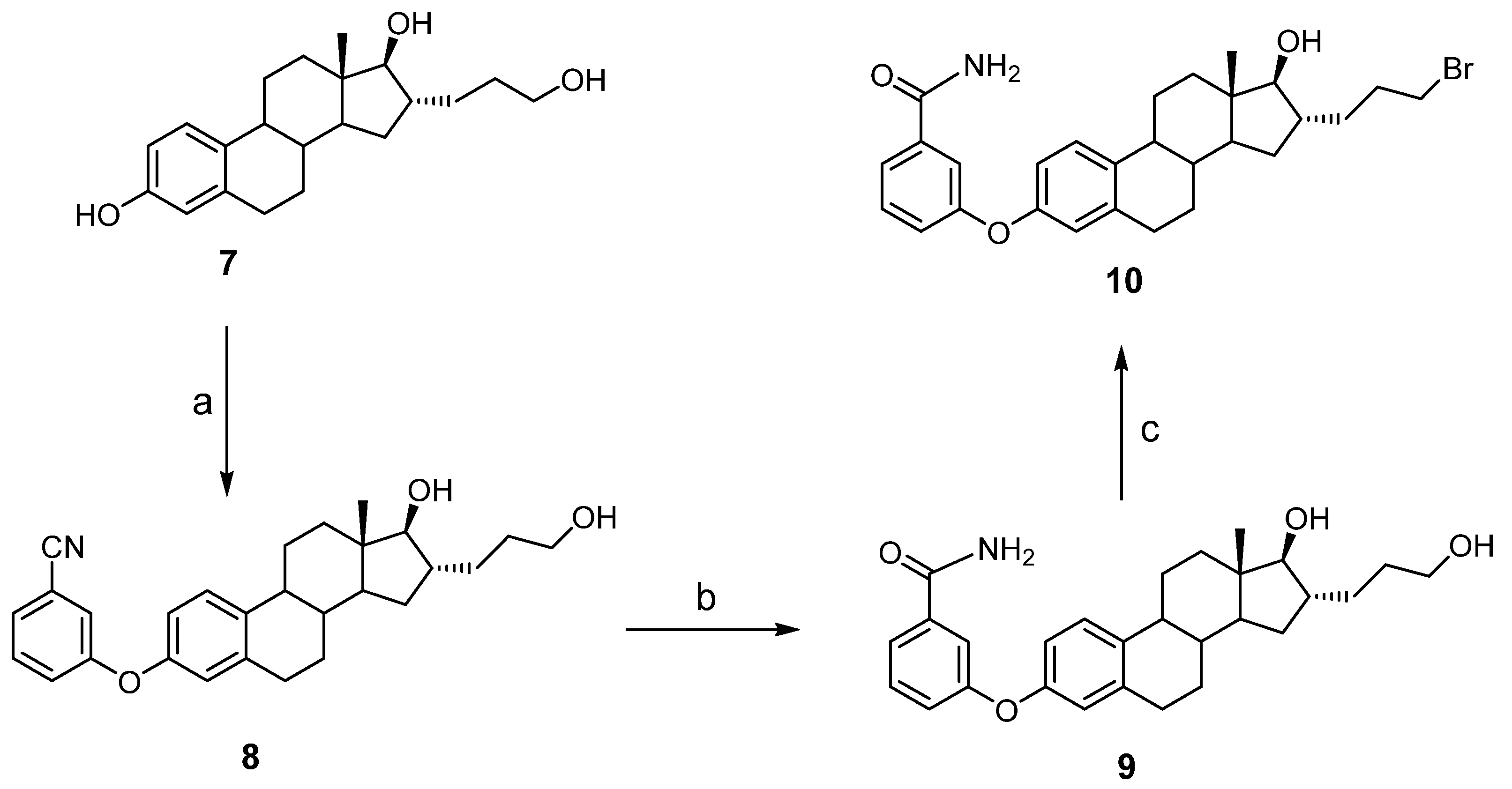
2.2.5. Synthesis of 3-(3-Cyanophenyloxy)-16α-(3-hydroxypropyl)-estra-1,3,5(10)-trien-17β-ol (8)
2.2.6. Synthesis of 3-(3-Carbamoylphenyloxy)-16α-(3-hydroxypropyl)-estra-1,3,5(10)-trien-17β-ol (9)
2.2.7. Synthesis of 3-(3-Carbamoylphenyloxy)-16α-(3-bromopropyl)-estra-1,3,5(10)-trien-17β-ol (10)
2.3. Biology
2.3.1. Cells for Enzymatic and Proliferative Assays
2.3.2. 17β-HSD1 Inhibition Assay
2.3.3. Cell Viability Assay
2.4. Molecular Modeling
3. Results and Discussion
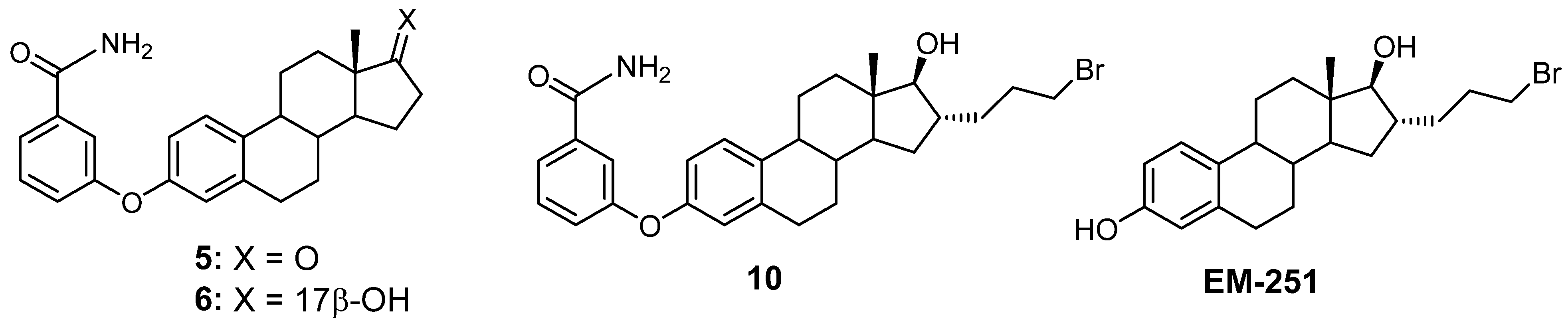
3.1. Chemical Synthesis
3.2. Inhibition of 17β-HSD1 Activity Transforming E1 into E2
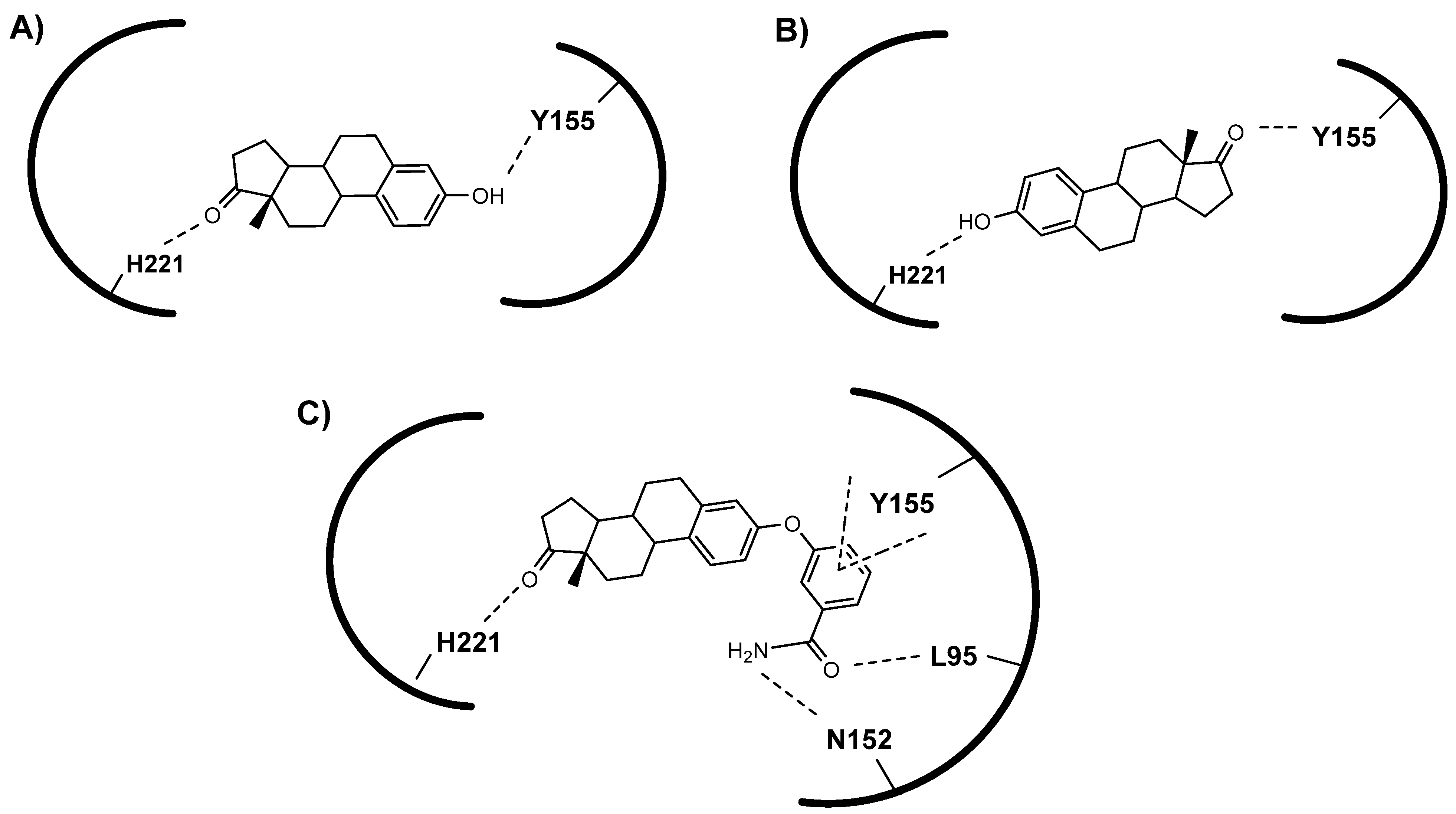
3.3. Molecular Docking
3.4. Antiproliferative Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Baulieu, E.E. Hormones, a complexe communication network. In Hormones, from Molecules to Disease; Baulieu, E.E., Kelly, P.A., Eds.; Hermann, publishers in arts and science: Paris, France, 1990; pp. 3–149. [Google Scholar]
- Thomas, D.B. Do hormones cause breast cancer? Cancer 1984, 53, 595–604. [Google Scholar] [CrossRef]
- Dickson, R.B.; Lippman, M.E. Estrogenic regulation of growth and polypeptide growth factor secretion in human breast carcinoma. Endocr. Rev. 1987, 8, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Delvoux, B.; D’Hooghe, T.; Kyama, C.; Koskimies, P.; Hermans, R.J.J.; Dunselman, G.A.; Romano, A. Inhibition of type 1 17β-hydroxysteroid dehydrogenase impairs the synthesis of 17β-estradiol in endometriosis lesions. J. Clin. Endocrinol. Metab. 2014, 99, 276–284. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). International Classification of Diseases (Fact Sheets: Cancer), 11th Revision (ICD-11) Geneva WHO, 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer (accessed on 14 November 2022).
- Zondervan, K.T.; Becker, C.M.; Missmer, S.A. Endometriosis. N. Engl. J. Med. 2020, 382, 1244–1256. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). International Classification of Diseases (Fact Sheets: Endometriosis), 11th Revision (ICD-11) Geneva WHO, 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/endometriosis (accessed on 14 November 2022).
- Payne, A.H.; Hales, D.B. Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones. Endocr. Rev. 2004, 25, 947–970. [Google Scholar] [CrossRef]
- Labrie, F. Intracrinology. Mol. Cell Endocrinol. 1991, 78, C113–C118. [Google Scholar] [CrossRef] [PubMed]
- Labrie, F.; Luu-The, V.; Lin, S.X.; Labrie, C.; Simard, J.; Breton, R.; Bélanger, A. The key role of 17β-hydroxysteroid dehydrogenases in sex steroid biology. Steroids 1997, 62, 148–158. [Google Scholar] [CrossRef]
- Penning, T.M. Molecular endocrinology of hydroxysteroid dehydrogenases. Endocr. Rev. 1997, 18, 281–305. [Google Scholar] [CrossRef]
- Vihko, P.; Herrala, A.; Harkonen, P.; Isomaa, V.; Kaija, H.; Kurkela, R.; Pulkka, A. Control of cell proliferation by steroids: The role of 17β-HSDs. Mol. Cell. Endocrinol. 2006, 248, 141–148. [Google Scholar] [CrossRef]
- Meier, M.; Moller, G.; Adamski, J. Perspectives in understanding the role of human 17β-hydroxysteroid dehydrogenases in health and disease. Steroid Enzym. Cancer Ann. N. Y. Acad. Sci. 2009, 1155, 15–24. [Google Scholar] [CrossRef]
- Poirier, D. Inhibitors of 17 beta-hydroxysteroid dehydrogenases. Curr. Med. Chem. 2003, 10, 453–477. [Google Scholar] [CrossRef]
- Brozic, P.; Lanisnik Rizner, T.; Gobec, S. Inhibitors of 17beta-hydroxysteroid dehydrogenase type 1. Curr. Med. Chem. 2008, 15, 137–150. [Google Scholar] [CrossRef]
- Day, J.M.; Tutill, H.J.; Purohit, A. 17β-hydroxysteroid dehydrogenase inhibitors. Minerva Endocrinol. 2010, 35, 87–108. [Google Scholar] [PubMed]
- Poirier, D. 17β-Hydroxysteroid dehydrogenase inhibitors: A patent review. Expert Opin. Ther. Patents 2010, 20, 1123–1145. [Google Scholar] [CrossRef]
- Marchais-Oberwinkler, S.; Henn, C.; Möller, G.; Klein, T.; Negri, M.; Oster, A.; Spadaro, A.; Werth, R.; Wetzel, M.; Xu, K.; et al. 17β-Hydroxysteroid dehydrogenases (17β-HSDs) as therapeutic targets: Protein structures, functions, and recent progress in inhibitor development. J. Steroid Biochem. Mol. Biol. 2011, 125, 66–82. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Stephen, P.; Zhu, D.W.; Shi, R.; Lin, S.X. Crystal structures of human 17β-hydroxysteroid dehydrogenase type 1 complexed with estrone and NADP+ reveal the mechanism of substrate inhibition. FEBS J. 2019, 286, 2155–2166. [Google Scholar] [CrossRef] [PubMed]
- Negri, M.; Recenatini, M.; Hartmann, R.W. Insights in 17β-HSD1 enzyme kinetics and ligand binding by dynamic motion investigation. PLoS ONE 2010, 5, e12026. [Google Scholar] [CrossRef]
- Lespérance, M.; Roy, J.; Ngueta Djiemeny, A.; Maltais, R.; Poirier, D. Synthesis of 16β-derivatives of 3-(2-bromoethyl)-estra-1,3,5(10)-trien-17β-ol as inhibitors of 17β-HSD1 and/or steroid sulfatase for the treatment of estrogen-dependent diseases. Steroids 2021, 172, 108856. [Google Scholar] [CrossRef] [PubMed]
- Maltais, R.; Trottier, A.; Roy, J.; Ayan, D.; Bertrand, N.; Poirier, D. Pharmacokinetic profile of PBRM in rodents, a first selective covalent inhibitor of 17β-HSD1 for breast cancer and endometriosis treatments. J. Steroid Biochem. Mol. Biol. 2018, 178, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Maltais, R.; Ayan, D.; Trottier, A.; Barbeau, X.; Lagüe, P.; Bouchard, J.E.; Poirier, D. Discovery of a non-estrogenic irreversible inhibitor of 17β-hydroxysteroid dehydrogenase type 1 from 3-substituted-16β-(m-carbamoylbenzyl)-estradiol derivatives. J. Med. Chem. 2014, 57, 204–222. [Google Scholar] [CrossRef] [PubMed]
- Laplante, Y.; Cadot, Y.; Fournier, M.A.; Poirier, D. Estradiol and estrone C-16 derivatives as inhibitors of type 1 17β-hydroxysteroid dehydrogenase: Blocking of ER+ breast cancer cell proliferation induced by estrone. Bioorg. Med. Chem. 2008, 16, 1849–1860. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, M.R.; Poirier, D. Overview of a rational approach to design type 1 17beta-hydroxysteroid dehydrogenase inhibitors without estrogenic activity: Chemical synthesis and biological evaluation. J. Steroid Biochem. Mol. Biol. 1998, 66, 179–191. [Google Scholar] [CrossRef]
- Sam, K.M.; Boivin, R.P.; Tremblay, M.R.; Auger, S.; Poirier, D. C16 and C17 derivatives of estradiol as inhibitors of 17β-hydroxysteroid dehydrogenase type 1: Chemical synthesis and structure-activity relationships. Drug Design Discov. 1998, 15, 157–180. [Google Scholar] [PubMed]
- Tremblay, M.R.; Auger, S.; Poirier, D. Synthesis of 16-bromoalkyl)-estradiols having inhibitory effect on human placental estradiol 17β-hydroxysteroid dehydrogenase (17β-HSD type 1). Bioorg. Med. Chem. 1995, 3, 505–523. [Google Scholar] [CrossRef]
- Ayan, D.; Maltais, R.; Hospital, A.; Poirier, D. Chemical synthesis, cytotoxicity, selectivity and bioavailability of 5α-androstane-3α,17β-diol derivatives. Bioorg. Med. Chem. 2014, 22, 5847–5859. [Google Scholar] [CrossRef]
- Jegham, H.; Maltais, R.; Roy, J.; Doillon, C.; Poirier, D. Biological evaluation of a new family of aminosteroids that display a selective toxicity for various malignant cell lines. Anti-Cancer Drugs 2012, 23, 803–814. [Google Scholar] [CrossRef]
- Perreault, M.; Maltais, R.; Roy, J.; Dutour, R.; Poirier, D. Design of a mestranol 2-N-piperazino-substituted derivative showing potent and selective in vitro and in vivo activities in MCF-7 breast cancer models. ChemMedChem 2017, 12, 177–182. [Google Scholar] [CrossRef]
- Maltais, R.; Perreault, M.; Roy, J.; Poirier, D. Minor chemical modifications of the aminosteroid derivative RM-581 lead to major impact on its anticancer activity, metabolic stability and aqueous solubility. Eur. J. Med. Chem. 2020, 188, 111990. [Google Scholar] [CrossRef] [PubMed]
- RCSB Protein Data Bank (RCSB PDB). Available online: http://www.rcsb.org/ (accessed on 22 September 2022).
- Li, F.; Wang, Q.; Ding, Z.; Tao, G. Microwave-assisted synthesis of diaryl ethers without catalyst. Org. Lett. 2003, 5, 2169–2171. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lin, J.-H.; Xiao, J.-C. Halogenation through deoxygenation of alcohols and aldehydes. Org. Lett. 2018, 20, 3061–3064. [Google Scholar] [CrossRef]
- Maltais, R.; Poirier, D. Development of a gram-scale synthesis of PBRM, an irreversible inhibitor of 17β-hydroxysteroid dehydrogenase type 1. Org. Process Res. Dev. 2019, 23, 2323–2335. [Google Scholar] [CrossRef]
- Laplante, Y.; Rancourt, C.; Poirier, D. Relative involvement of three 17β-hydroxysteroid dehydrogenases (types 1, 7 and 12) in the formation of estradiol in various breast cancer cell lines using selective inhibitors. Mol. Cell. Endocrinol. 2009, 301, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Poirier, D.; Roy, J.; Maltais, R. A targeted-covalent inhibitor of 17β-HSD1 blocks two estrogen-biosynthesis pathways: In vitro (metabolism) and in vivo (xenograft) studies in T-47D breast cancer models. Cancers 2021, 13, 1841. [Google Scholar] [CrossRef] [PubMed]
- Poirier, D.; Nyachieo, A.; Romano, A.; Roy, J.; Maltais, R.; Chai, D.; Delvous, B.; Tomassetti, C.; Vanhie, A. An irreversible inhibitor of 17β-hydroxysteroid dehydrogenase type 1 inhibits estradiol synthesis in human endometriosis lesions an induces regression of the non-human primate endometriosis. J. Steroid Biochem. Mol. Biol. 2022, 222, 106136. [Google Scholar] [CrossRef]
- Beranic, N.; Lanisnik Rizner, T. Effects of progestins on local estradiol biosynthesis and action in the Z-12 endometriotic epithelial cell line. J. Steroid Biochem. Mol. Biol. 2012, 132, 303–310. [Google Scholar] [CrossRef]
- Banu, S.K.; Lee, J.; Starzinski-Powitz, A.; Arosh, J.A. Gene expression profiles and functional characterization of human immortalized endometriotic epithelial and stromal cells. Fertil. Steril. 2008, 90, 972–987. [Google Scholar] [CrossRef]
- Zeitvogel, A.; Baumann, R.; Starzinski-Powitz, A. Identification of an invasive, N-cadherin-expressing epithelial cell type in endometriosis using a new cell culture model. Am. J. Pathol. 2001, 159, 1839–1852. [Google Scholar] [CrossRef]
- Azzi, A.; Rhese, P.H.; Zhu, D.W.; Campbell, R.L.; Labrie, F.; Lin, S.X. Estrogenic 17-beta Hydroxysteroid Dehydrogenase Complexed 17-beta-estradiol. Available online: https://www.rcsb.org/structure/1iol (accessed on 22 September 2022). [CrossRef]
- Azzi, A.; Rhese, P.H.; Zhu, D.W.; Campbell, R.L.; Labrie, F.; Lin, S.X. Crystal structure of human estrogenic 17β-hydroxysteroid dehydrogenase complexed with 17β-estradiol. Nat. Struct. Biol. 1996, 3, 665–668. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Maltais, R.; Poirier, D.; Lin, S.X. Combined biophysical chemistry reveals a new covalent inhibitor with a low-reactivity alkyl halide. J. Phys. Chem. Lett. 2018, 9, 5275–5280. [Google Scholar] [CrossRef]

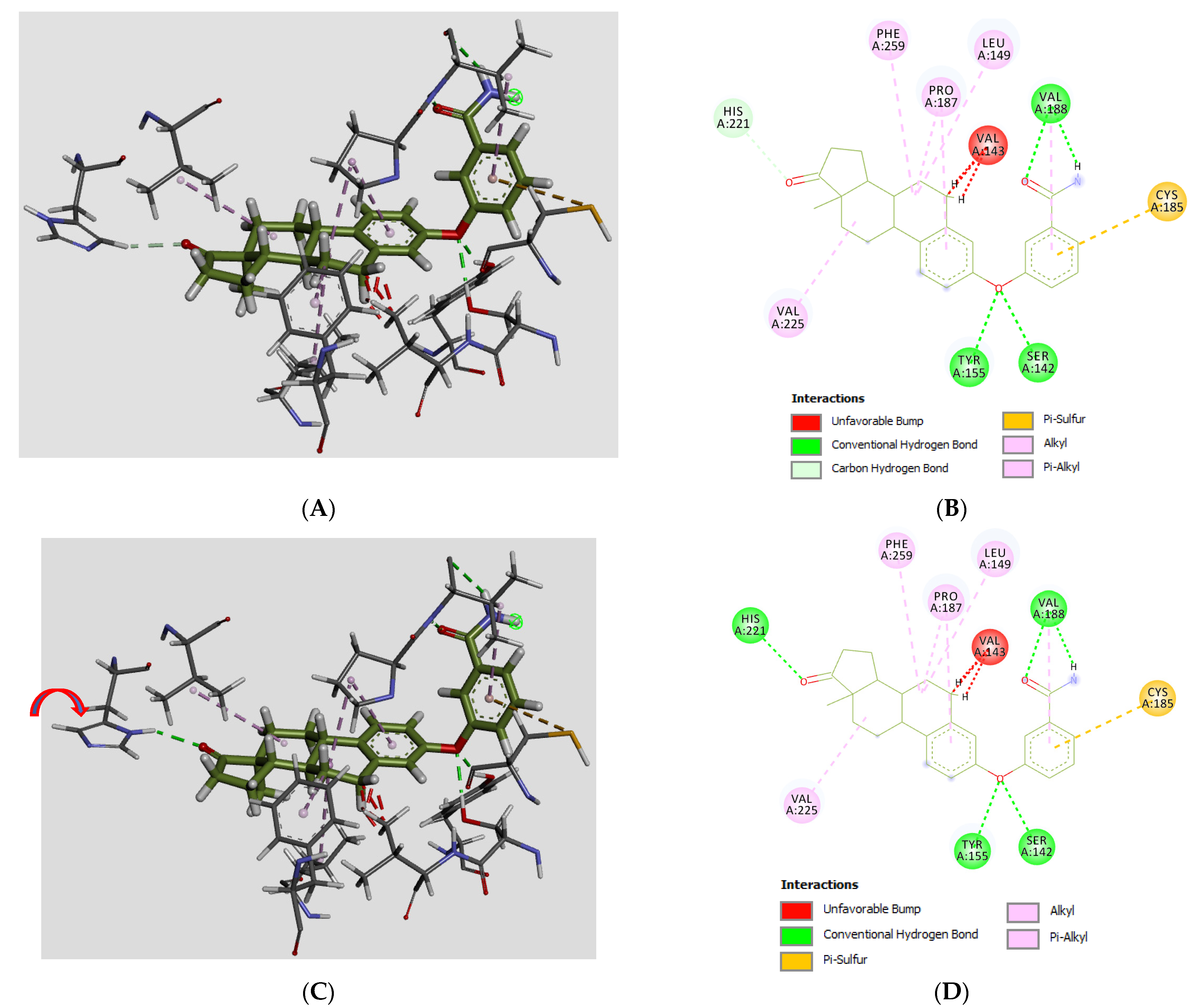

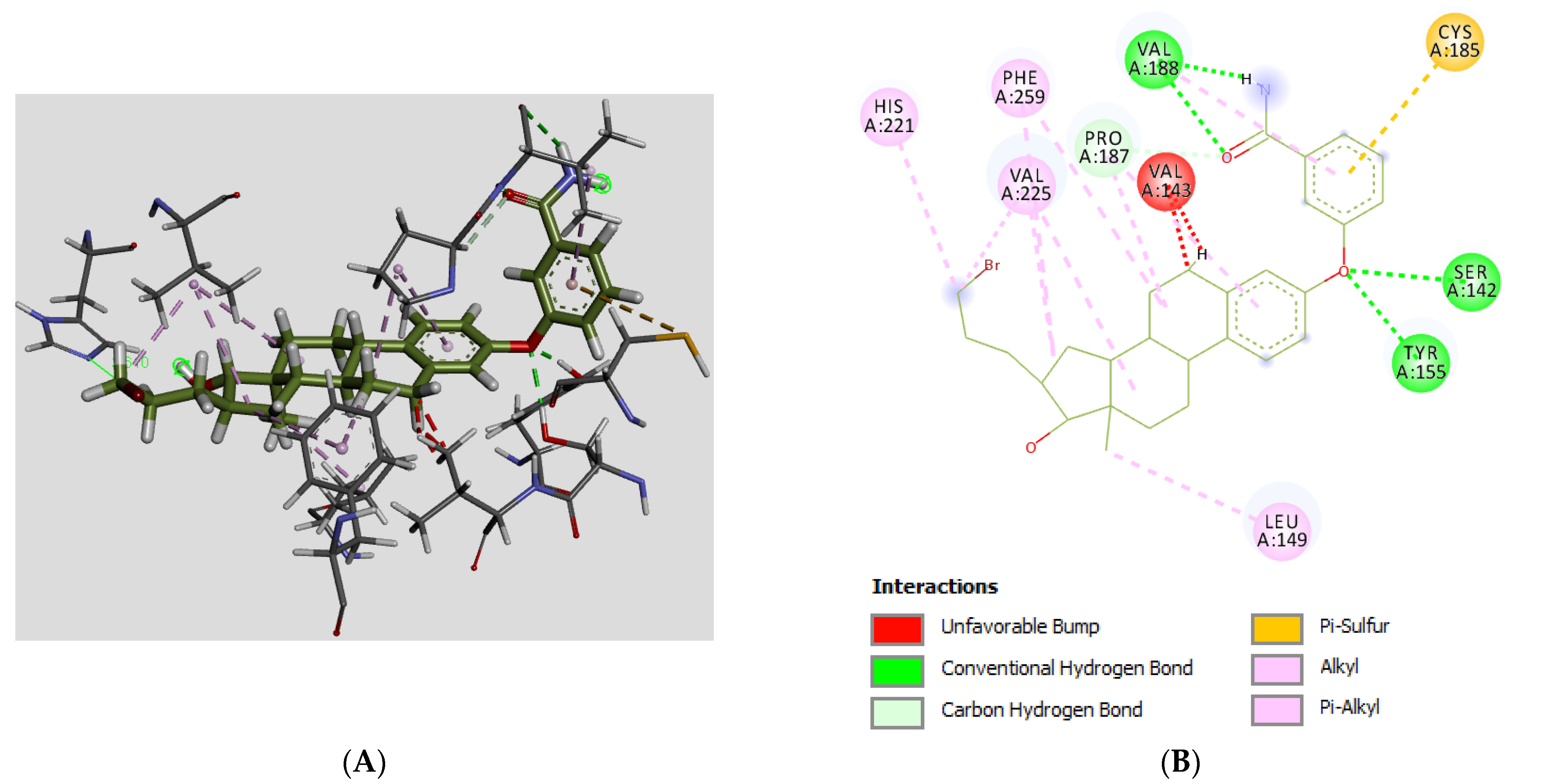
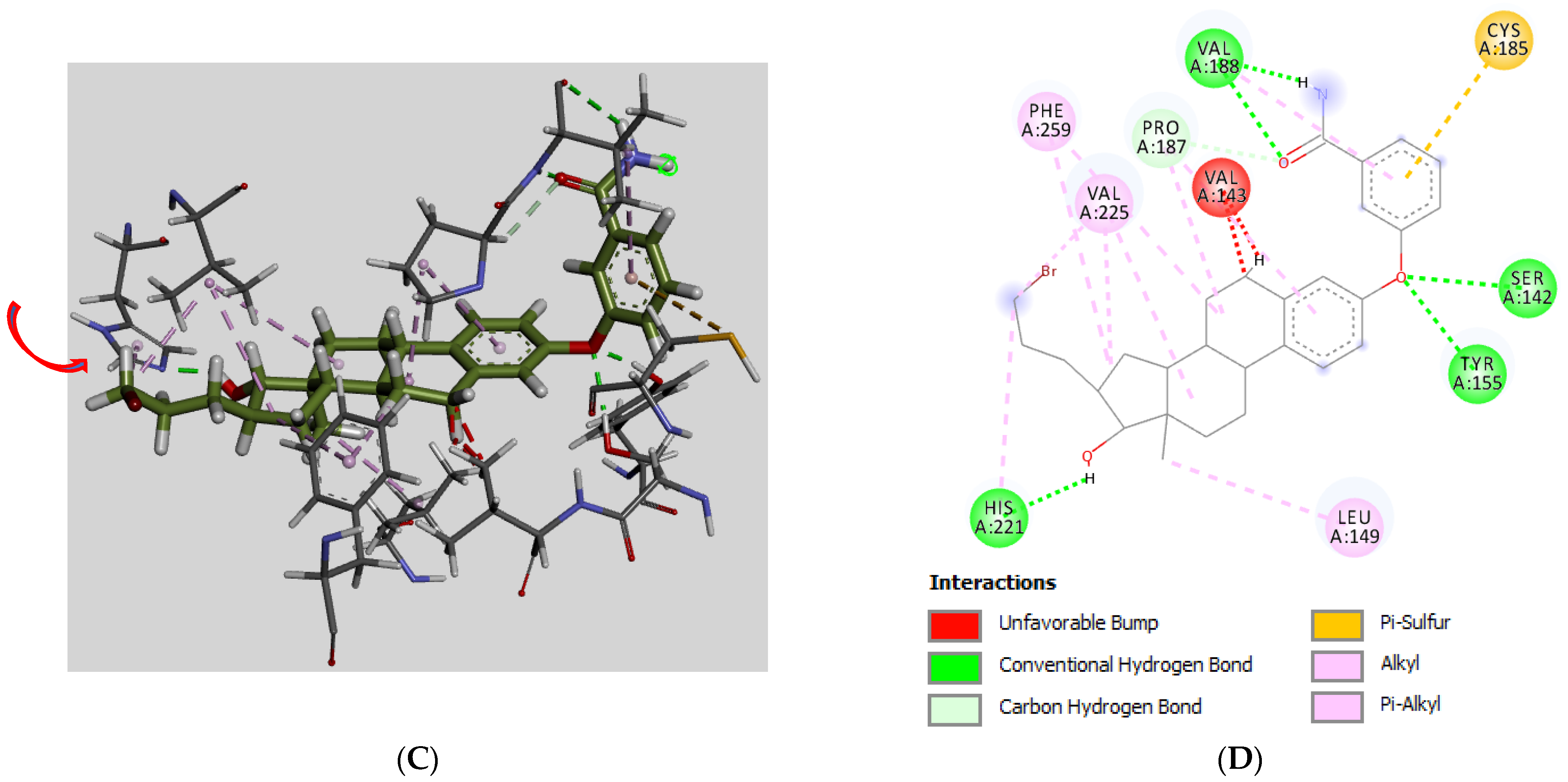
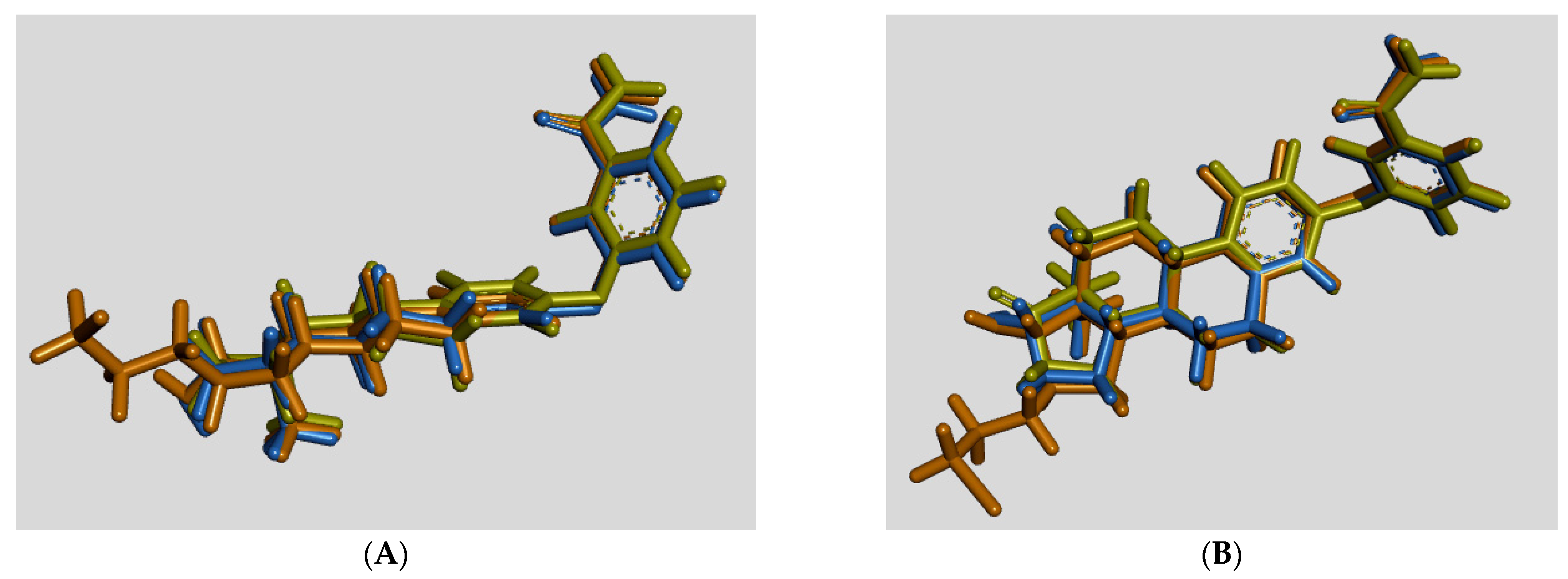
| # | C3/C17/C16-Groups | Inhibition (% at 0.1 μM) | Inhibition (% at 1 μM) | Inhibition (% at 10 μM) | Inhibition (IC50 in μM) |
|---|---|---|---|---|---|
| 5 (ether) | 3-R′ / 17-C=O / H | 29.1 ± 6.2 | 70.7 ± 14.1 | 98.9 ± 2.8 | 0.31 |
| 6 (ether) | 3-R′ / 17β-OH / H | 40.0 ± 6.5 | 48.6 ± 15.7 | 59.6 ± 1.5 | 1.2 |
| 10 (ether) | 3-R′ / 17β-OH / 16α-(CH2)3Br | 28.2 ± 5.4 | 37.0 ± 14.5 | 82.1 ± 5.7 | 1.3 |
| E1 (phenol) | OH / C=O / H | 44.9 ± 14.7 | 60.9 ± 8.3 | 78.1 ± 2.2 | 0.21 |
| EM-251 (phenol) | OH / 17β-OH / 16α-(CH2)3Br | 43.8 ± 0.2 | 36.9 ± 9.3 | 86.6 ± 8.0 | 0.58 |
| PBRM (bromoethyl) | 3-(2′-(CH2)2Br) / 17β-OH / 16β-R″ | 43.8 ± 3.4 | 74.8 ± 3.2 | 91.0 ± 3.8 | 0.15 |
| # | C3/C17/C16-Groups | T-47D Cells (Breast Cancer) (IC50 in μM) | Z-12 Cells (Endometriosis) (IC50 in μM) |
|---|---|---|---|
| 5 | 3-R / 17-C=O / H | 49 | >30 |
| 6 | 3-R / 17β-OH / H | 37 | >30 |
| 10 | 3-R / 17β-OH / 16α-(CH2)3Br | 14 | 20 |
| RM-581 | -- | 0.61 | 1.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ngueta, A.D.; Roy, J.; Maltais, R.; Poirier, D. Chemical Synthesis and Biological Evaluation of 3-Substituted Estrone/Estradiol Derivatives as 17β-Hydroxysteroid Dehydrogenase Type 1 Inhibitors Acting via a Reverse Orientation of the Natural Substrate Estrone. Molecules 2023, 28, 632. https://doi.org/10.3390/molecules28020632
Ngueta AD, Roy J, Maltais R, Poirier D. Chemical Synthesis and Biological Evaluation of 3-Substituted Estrone/Estradiol Derivatives as 17β-Hydroxysteroid Dehydrogenase Type 1 Inhibitors Acting via a Reverse Orientation of the Natural Substrate Estrone. Molecules. 2023; 28(2):632. https://doi.org/10.3390/molecules28020632
Chicago/Turabian StyleNgueta, Adrien Djiemeny, Jenny Roy, René Maltais, and Donald Poirier. 2023. "Chemical Synthesis and Biological Evaluation of 3-Substituted Estrone/Estradiol Derivatives as 17β-Hydroxysteroid Dehydrogenase Type 1 Inhibitors Acting via a Reverse Orientation of the Natural Substrate Estrone" Molecules 28, no. 2: 632. https://doi.org/10.3390/molecules28020632
APA StyleNgueta, A. D., Roy, J., Maltais, R., & Poirier, D. (2023). Chemical Synthesis and Biological Evaluation of 3-Substituted Estrone/Estradiol Derivatives as 17β-Hydroxysteroid Dehydrogenase Type 1 Inhibitors Acting via a Reverse Orientation of the Natural Substrate Estrone. Molecules, 28(2), 632. https://doi.org/10.3390/molecules28020632









