Molecular Docking of Bacterial Protein Modulators and Pharmacotherapeutics of Carica papaya Leaves as a Promising Therapy for Sepsis: Synchronising In Silico and In Vitro Studies
Abstract
1. Introduction
2. Results
2.1. Molecular Docking In Silico Study
2.2. Percentage Yield
2.3. Phytochemical Screening of Extracts
2.4. Estimation of Total Flavonoids
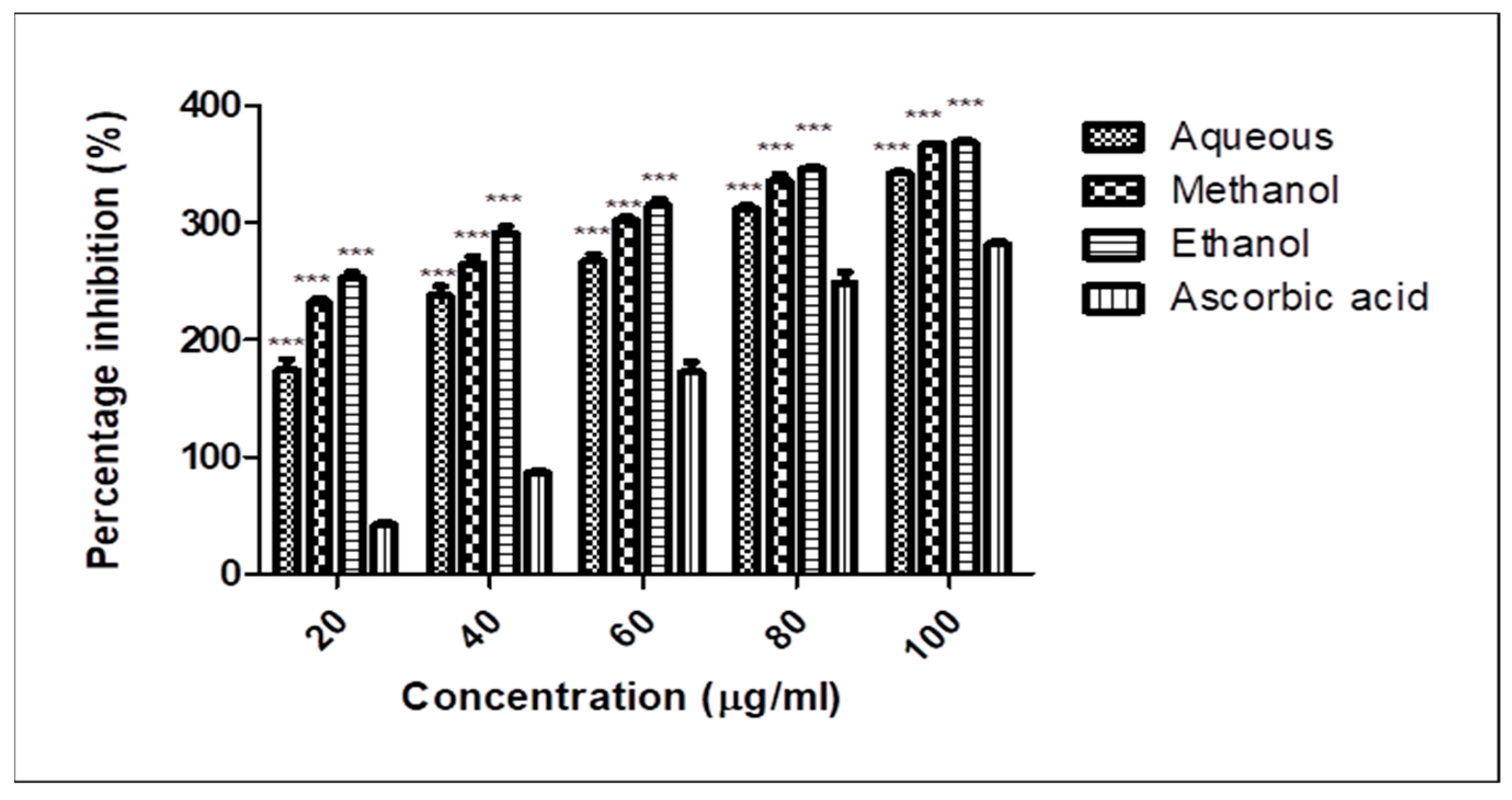
2.5. DPPH Radical Scavenging Activity
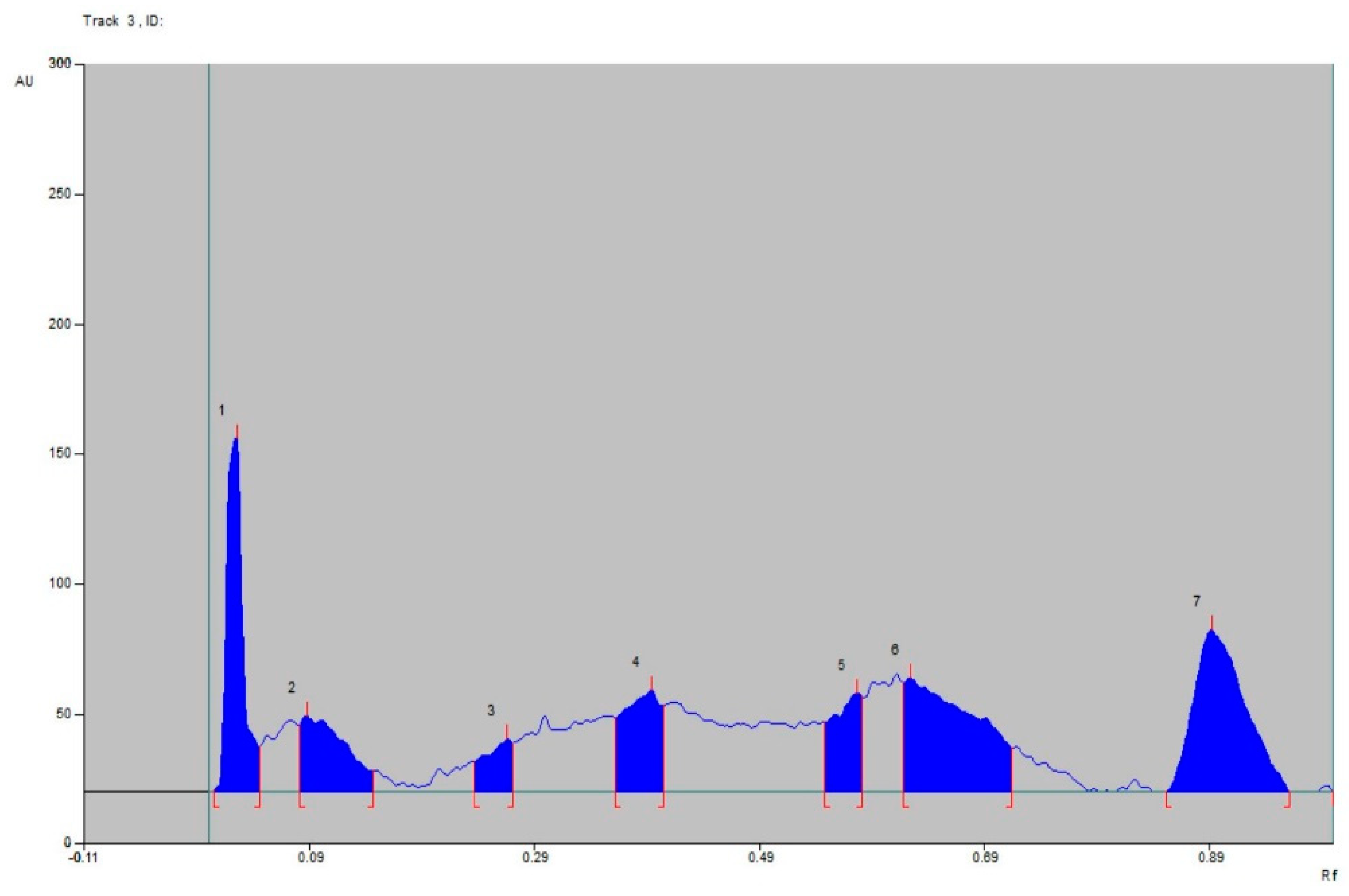
2.6. TLC Analysis
2.7. HPTLC Fingerprinting
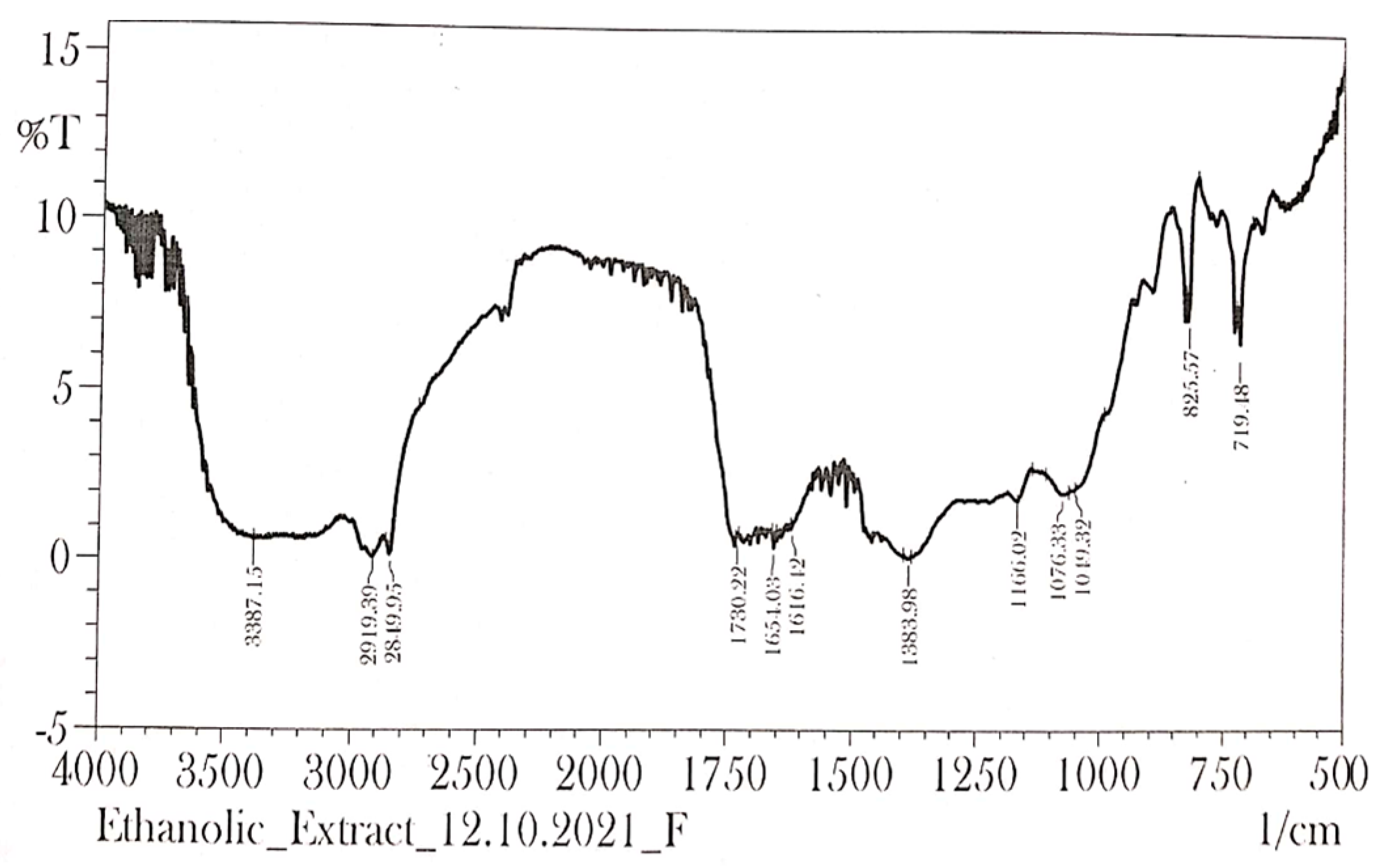
2.8. FTIR Spectrum
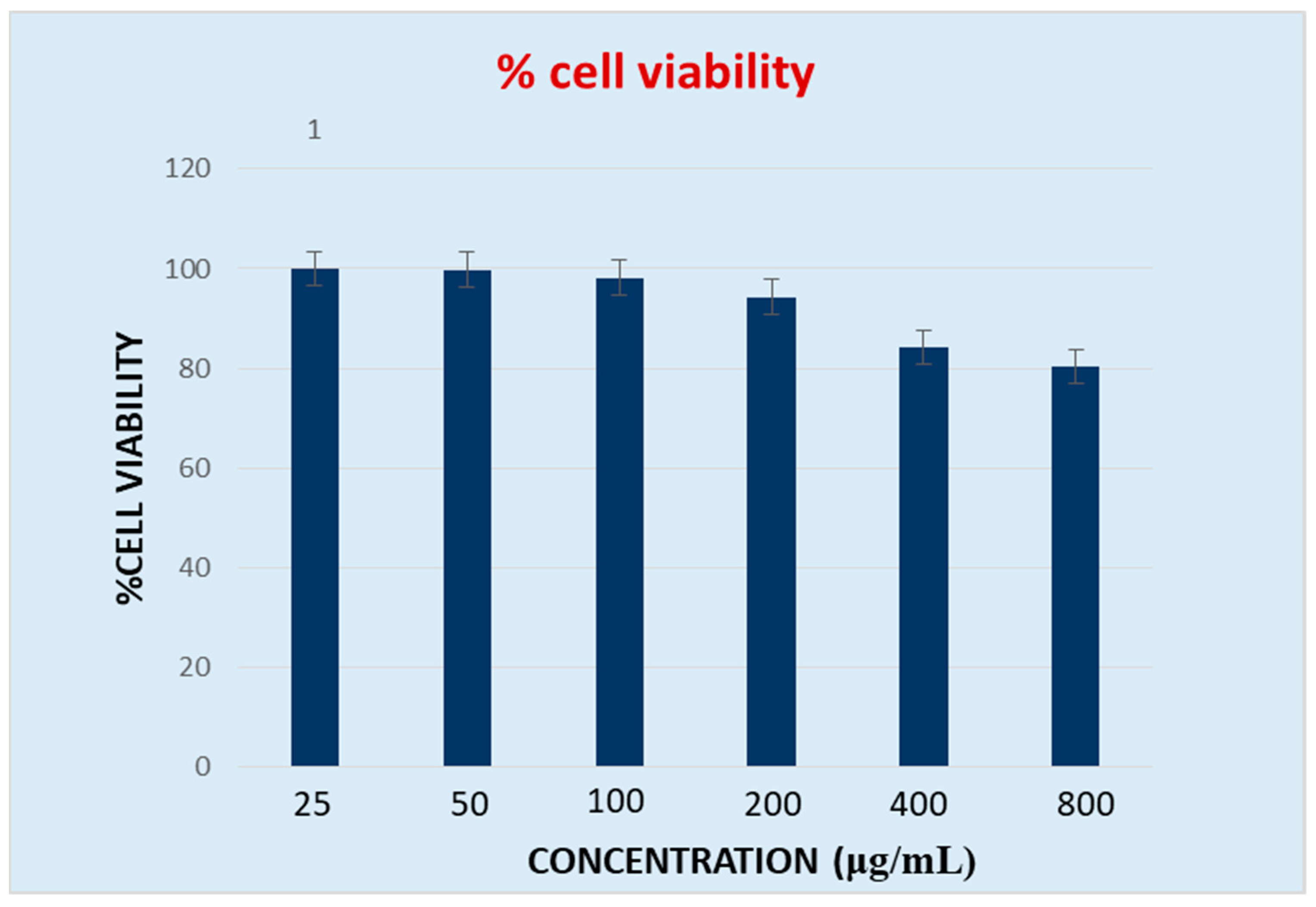
2.9. In Vitro % Cell Viability
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Authentication
4.2. In Silico Study
4.3. Preparation of Extracts
4.4. Preliminary Estimation of Phytoconstituents
4.5. Total Flavonoid Content
4.6. DPPH Radical Scavenging Activity
4.7. Thin Layer Chromatography Analysis
4.8. HPTLC Fingerprinting Profile of Ethanolic Extract of Carica Papaya Leaves
4.9. Fourier Transform Infrared Spectroscopy (FTIR)
4.10. Cytotoxicity Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rahmani, A.H.; Aldebasi, Y.H. Potential Role of Carica papaya and Their Active Constituents in the Prevention and Treatment of Diseases Implication of PTEN, Akt and Bcl2 Expressions and Its Co-Relation with Apoptotic Pathways in Oral Squamous Cell Carcinoma View Project Natural Product. Int. J. Pharm. Pharm. Sci. 2016, 8, 11–15. [Google Scholar]
- Priyadarshi, A.; Ram, B. A Review on Pharmacognosy, Phytochemistry and Pharmacological Activity of Carica papaya (Linn.) Leaf. Int. J. Pharm. Sci. Res. 2018, 9, 4071–4078. [Google Scholar]
- Santana, L.F.; Inada, A.C.; Santo, B.L.S.D.E.; Filiú, W.F.O.; Pott, A.; Alves, F.M.; Guimarães, R.D.C.A.; Freitas, K.D.C.; Hiane, P.A. Nutraceutical Potential of Carica papaya in Metabolic Syndrome. Nutrients 2019, 11, 1608. [Google Scholar] [CrossRef] [PubMed]
- Sagadevan, P.; Selvakumar, S.; Raghunath, M.; Megala, R.; Janarthanan, P.; Vinitha Ebziba, C.; Senthil Kumar, V. Medicinal Properties of Carica papaya Linn: Review. Madridge J. Nov. Drug Res. 2019, 3, 120–125. [Google Scholar] [CrossRef]
- Bhowmik, D. Traditional and Medicinal Uses of Carica papaya. J. Med. Plants Stud. Year 2013, 1, 7–15. [Google Scholar]
- Miean, K.H.; Mohamed, S. Flavonoid (Myricetin, Quercetin, Kaempferol, Luteolin, and Apigenin) Content of Edible Tropical Plants. J. Agric. Food Chem. 2001, 49, 3106–3112. [Google Scholar] [CrossRef] [PubMed]
- Khuzhaev, V.U.; Aripova, S.F.; Shakirov, R.S. Dynamics of the Accumulation of the Alkaloids of Arundo Donax. Chem. Nat. Compd. 1995, 30, 637–638. [Google Scholar] [CrossRef]
- Olafsdottir, E.S.; Bolt Jorgensen, L.; Jaroszewski, J.W. Cyanogenesis in Glucosinolate-Producing Plants: Carica papaya and Carica Quercifolia. Phytochemistry 2002, 60, 269–273. [Google Scholar] [CrossRef]
- Dharmarathna, S.L.C.A.; Wickramasinghe, S.; Waduge, R.N.; Rajapakse, R.P.V.J.; Kularatne, S.A.M. Does Carica papaya Leaf-Extract Increase the Platelet Count? An Experimental Study in a Murine Model. Asian Pac. J. Trop. Biomed. 2013, 3, 720–724. [Google Scholar] [CrossRef]
- Anjum, V.; Arora, P.; Ansari, S.H.; Najmi, A.K.; Ahmad, S. Antithrombocytopenic and Immunomodulatory Potential of Metabolically Characterized Aqueous Extract of Carica papaya Leaves. Pharm. Biol. 2017, 55, 2043–2056. [Google Scholar] [CrossRef]
- Kad, D.R.; Tambe, V.S. Phytochemical Screening and Evaluation of Platelet—Google Scholar. Adv. Plants Agric. Res. 2018, 8, 531–535. [Google Scholar]
- Pandey, S.; Cabot, P.J.; Shaw, P.N.; Hewavitharana, A.K. Anti-Inflammatory and Immunomodulatory Properties of Carica papaya. J. Immunotoxicol. 2016, 13, 590–602. [Google Scholar] [CrossRef] [PubMed]
- Renganathan, S.; Aroulmoji, V.; Shanmugam, G.; Devarajan, G.; Rao, K.V.; Rajendar, V.; Park, S.H. Silver Nanoparticle Synthesis from Carica papaya and Virtual Screening for Anti-Dengue Activity Using Molecular Docking. MRE 2019, 6, 035028. [Google Scholar] [CrossRef]
- Jiang, B.; Liang, P.; Deng, G.; Tu, Z.; Liu, M.; Xiao, X. Increased Stability of Bcl-2 in HSP70-Mediated Protection against Apoptosis Induced by Oxidative Stress. Cell Stress Chaperones 2010, 16, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Kuzmenko, A.; Wan, S.; Schaffer, L.; Weiss, A.; Fisher, J.H.; Kim, K.S.; McCormack, F.X. Surfactant Proteins A and D Inhibit the Growth of Gram-Negative Bacteria by Increasing Membrane Permeability. J. Clin. Invest. 2003, 111, 1589–1602. [Google Scholar] [CrossRef]
- Dickgießer, U.; Weiss, N.; Fritsche, D. Lactobacillus Gasseri as the Cause of Septic Urinary Infection. Infection 1984, 12, 14–16. [Google Scholar] [CrossRef]
- Canini, A.; Alesiani, D.; D’Arcangelo, G.; Tagliatesta, P. Gas Chromatography-Mass Spectrometry Analysis of Phenolic Compounds from Carica papaya, L. Leaf. J. Food Compos. Anal. 2007, 20, 584–590. [Google Scholar] [CrossRef]
- Comalada, M.; Camuesco, D.; Sierra, S.; Ballester, I.; Xaus, J.; Gálvez, J.; Zarzuelo, A. In Vivo Quercitrin Anti-Inflammatory Effect Involves Release of Quercetin, Which Inhibits Inflammation through down-Regulation of the NF-ΚB Pathway. Eur. J. Immunol. 2005, 35, 584–592. [Google Scholar] [CrossRef]
- Wang, S.; Yao, J.; Zhou, B.; Yang, J.; Chaudry, M.T.; Wang, M.; Xiao, F.; Li, Y.; Yin, W. Bacteriostatic Effect of Quercetin as an Antibiotic Alternative In Vivo and Its Antibacterial Mechanism In Vitro. J. Food Prot. 2018, 81, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Avinash Shinde, A.; Hase, D. Isolation of Carpaine from Carica papaya Leaves by Using LCMS. J. Med. Plants Stud. 2020, 8, 1–5. [Google Scholar]
- Sherma, J. Basic TLC Techniques, Materials, and Apparatus; CRC Press: Boca Raton, FL, USA, 2003; pp. 25–85. [Google Scholar] [CrossRef]
- Ram, M.; Abdin, M.Z.; Khan, M.A.; Jha, P. HPTLC Fingerprint Analysis: A Quality Control for Authentication of Herbal Phytochemicals. In High-Performance Thin-Layer Chromatogr; Springer: Berlin/Heidelberg, Germany, 2011; pp. 105–116. [Google Scholar] [CrossRef]
- Yuliani, R.; Syahdeni, F. Cytotoxicity of Ethanolic Extract of Papaya Leaves (Carica papaya) and Its Fractions on T47D Cells. Pharma. J. Farm. Indones. 2020, 17, 17–23. [Google Scholar] [CrossRef]
- Joseph, B.; Sankarganesh, P.; Ichiyama, K.; Yamamoto, N. In Vitro Study on Cytotoxic Effect and Anti-DENV2 Activity of Carica papaya, L. Leaf. Front. Life Sci. 2015, 2, 18–22. [Google Scholar] [CrossRef]
- Mahajan, S.D.; Law, W.C.; Aalinkeel, R.; Reynolds, J.; Nair, B.B.; Yong, K.T.; Roy, I.; Prasad, P.N.; Schwartz, S.A. Nanoparticle-Mediated Targeted Delivery of Antiretrovirals to the Brain. Methods Enzymol. 2012, 509, 41–60. [Google Scholar] [CrossRef] [PubMed]
- Grela, E.; Kozłowska, J.; Grabowiecka, A. Current Methodology of MTT Assay in Bacteria—A Review. Acta Histochem. 2018, 120, 303–311. [Google Scholar] [CrossRef]
- Cecconi, M.; Evans, L.; Levy, M.; Rhodes, A. Sepsis and Septic Shock. Lancet 2018, 392, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Varon, J.; Arciniegas Rubio, A.; Amador-Munoz, D.; Corcoran, A.; DeCorte, J.A.; Isabelle, C.; Pinilla Vera, M.; Walker, K.; Brown, L.; Cernadas, M.; et al. Surfactant Protein D Influences Mortality During Abdominal Sepsis by Facilitating Escherichia Coli Colonization in the Gut. Crit. Care Explor. 2022, 4, e0699. [Google Scholar] [CrossRef]
- Usmani, J.; Khan, T.; Ahmad, R.; Sharma, M. Potential Role of Herbal Medicines as a Novel Approach in Sepsis Treatment. Biomed. Pharmacother. 2021, 144, 112337. [Google Scholar] [CrossRef]
- Zunjar, V.; Dash, R.P.; Jivrajani, M.; Trivedi, B.; Nivsarkar, M. Antithrombocytopenic Activity of Carpaine and Alkaloidal Extract of Carica papaya Linn. Leaves in Busulfan Induced Thrombocytopenic Wistar Rats. J. Ethnopharmacol. 2016, 181, 20–25. [Google Scholar] [CrossRef]
- Baskaran, C.; Bai, V.R.; Velu, S.; Kumaran, K. The Efficacy of Carica papaya Leaf Extract on Some Bacterial and a Fungal Strain by Well Diffusion Method. Asian Pacific J. Trop. Dis. 2012, 2, S658–S662. [Google Scholar] [CrossRef]
- Agada, R.; Usman, W.A.; Shehu, S.; Thagariki, D. In Vitro and in Vivo Inhibitory Effects of Carica papaya Seed on α-Amylase and α-Glucosidase Enzymes. Heliyon 2020, 6, e03618. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Halim, S.; Ibrahim, M.; Abdel Mohsen, M.; Abou-Setta, L.; Sleem, A.; El-Missiry, M. The Influence of the Extraction Method on Polyphenols, Flavonoids Composition and Anti-Hyperlipidemic Properties of Papaya Leaves (Carica papaya Linn.). Bull. Natl. Res. Cent. 2021, 45, 85. [Google Scholar] [CrossRef]
- Gulcin, İ. Antioxidants and Antioxidant Methods: An Updated Overview. Arch. Toxicol. 2020, 94, 651–715. [Google Scholar] [CrossRef] [PubMed]
- Alorkpa, E.J.; Boadi, N.O.; Badu, M.; Saah, S.A. Phytochemical Screening, Antimicrobial and Antioxidant Properties of Assorted Carica papaya Leaves in Ghana. J. Med. Plants Stud. 2016, 4, 193–198. [Google Scholar]
- Winikoff, S.E.; Zeh, H.J.; DeMarco, R.; Lotze, M.T. Cytolytic Assays. In Measuring Immunity: Basic Science and Clinical Practice; Elsevier: Amsterdam, The Netherlands, 2011; pp. 341–343. [Google Scholar]
- Zhang, P.; Leu, J.I.J.; Murphy, M.E.; George, D.L.; Marmorstein, R. Crystal Structure of the Stress-Inducible Human Heat Shock Protein 70 Substrate-Binding Domain in Complex with Peptide Substrate. PLoS ONE 2014, 9, e103518. [Google Scholar] [CrossRef] [PubMed]
- Shrive, A.K.; Tharia, H.A.; Strong, P.; Kishore, U.; Burns, I.; Rizkallah, P.J.; Reid, K.B.M.; Greenhough, T.J. High-Resolution Structural Insights into Ligand Binding and Immune Cell Recognition by Human Lung Surfactant Protein D. J. Mol. Biol. 2003, 331, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Raghunathan, K.; Harris, P.T.; Spurbeck, R.R.; Arvidson, C.G.; Arvidson, D.N. Crystal Structure of an Efficacious Gonococcal Adherence Inhibitor: An Enolase from Lactobacillus Gasseri. FEBS Lett. 2014, 588, 2212–2216. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Dhankhar, P.; Dalal, V.; Tomar, S.; Kumar, P. In-Silico Functional and Structural Annotation of Hypothetical Protein from Klebsiella Pneumonia: A Potential Drug Target. J. Mol. Graph. Model. 2022, 116, 108262. [Google Scholar] [CrossRef]
- Singh, V.; Dhankhar, P.; Dalal, V.; Tomar, S.; Golemi-Kotra, D.; Kumar, P. Drug-Repurposing Approach To Combat Staphylococcus Aureus: Biomolecular and Binding Interaction Study. ACS Omega 2022, 7, 38448–38458. [Google Scholar] [CrossRef]
- Akbar, S.; Das, S.; Iqubal, A.; Ahmed, B. Synthesis, Biological Evaluation and Molecular Dynamics Studies of Oxadiazine Derivatives as Potential Anti-Hepatotoxic Agents. J. Biomol. Struct. Dyn. 2021, 40, 9974–9991. [Google Scholar] [CrossRef]
- DUD—A Directory of Useful Decoys. Available online: http://dud.docking.org/ (accessed on 1 December 2022).
- Shuhel, M.A.; Easwari, T.S.; Sen, S. Stability Study and Haematological Profile of Aqueous Leaves Extract of Carica papaya. Der Pharm. Lett. 2016, 8, 182–187. [Google Scholar]
- Evans, W.C. Trease and Evans’ Pharmacognosy, 16th ed.; Elsevier: Amsterdam, The Netherlands, 2009; Volume 16, pp. 1–603. [Google Scholar]
- Boham, B.A.; Kocipai-Abyazan, A.C. Flavonoids and Condensed Tannins from Seed of Carica papaya. Pac. Sci. 1994, 8, 458–463. [Google Scholar]
- Lee, S.E.; Hwang, H.J.; Ha, J.S.; Jeong, H.S.; Kim, J.H. Screening of Medicinal Plant Extracts for Antioxidant Activity. Life Sci. 2003, 73, 167–179. [Google Scholar] [CrossRef]
- Mishra, K.; Ojha, H.; Chaudhury, N.K. Estimation of Antiradical Properties of Antioxidants Using DPPH Å Assay: A Critical Review and Results. Food Chem. 2011, 130, 1036–1046. [Google Scholar] [CrossRef]
- De Britto, J.A.; Roshan Sebastian, S.; Sujin, M.R. Phytochemical Analysis of Eight Medicinal Plants of Lamiaceae. J. Res. Plant Sci. 2011, 1, 001–006. [Google Scholar]
- Hussain, S.Z.; Razvi, N.; Ali, S.I.; Hasan, S.M.F. Development of Quality Standard and Phytochemical Analysis of Carica papaya Linn Leaves. Pak J. Pharm Sci. 2018, 31, 2169–2177. [Google Scholar]
- Hernández-Martínez, M.; Gallardo-Velázquez, T.; Osorio-Revilla, G.; Almaraz-Abarca, N.; Castañeda-Pérez, E. Application of MIR-FTIR Spectroscopy and Chemometrics to the Rapid Prediction of Fish Fillet Quality. CyTA J. Food 2014, 12, 369–377. [Google Scholar] [CrossRef]
- Fanelli, S.; Zimmermann, A.; Totóli, E.G.; Salgado, H.R.N. FTIR Spectrophotometry as a Green Tool for Quantitative Analysis of Drugs: Practical Application to Amoxicillin. J. Chem. 2018, 2018, 3920810. [Google Scholar] [CrossRef]
- Jayasinghe, C.D.; Gunasekera, D.S.; De Silva, N.; Jayawardena, K.K.M.; Udagama, P.V. Mature Leaf Concentrate of Sri Lankan Wild Type Carica papaya Linn. Modulates Nonfunctional and Functional Immune Responses of Rats. BMC Complement. Altern. Med. 2017, 17, 230. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.K.; Singh, S. Phytochemical Analysis and in Vitro Cytostatic Potential of Ethnopharmacological Important Medicinal Plants. Toxicol. Reports 2020, 7, 443–452. [Google Scholar] [CrossRef] [PubMed]
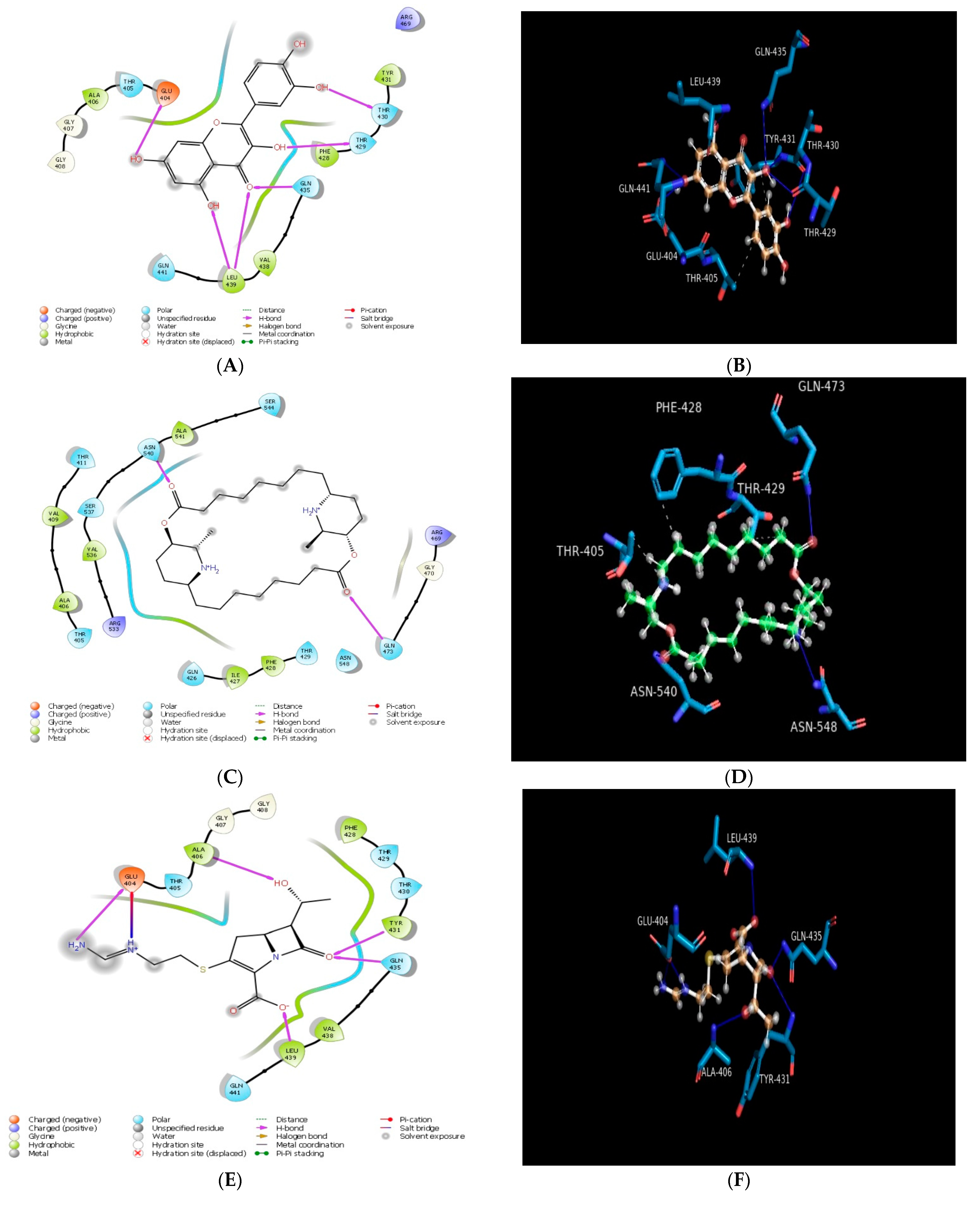
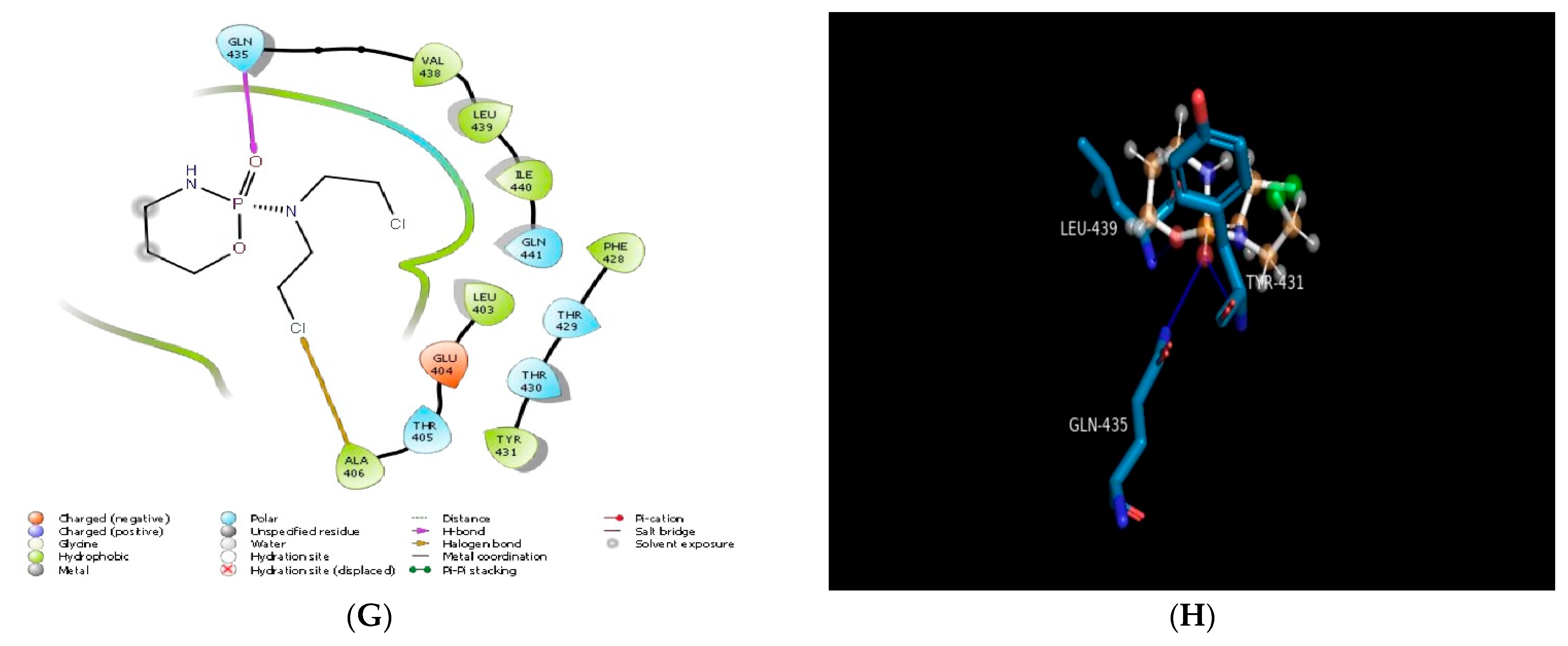
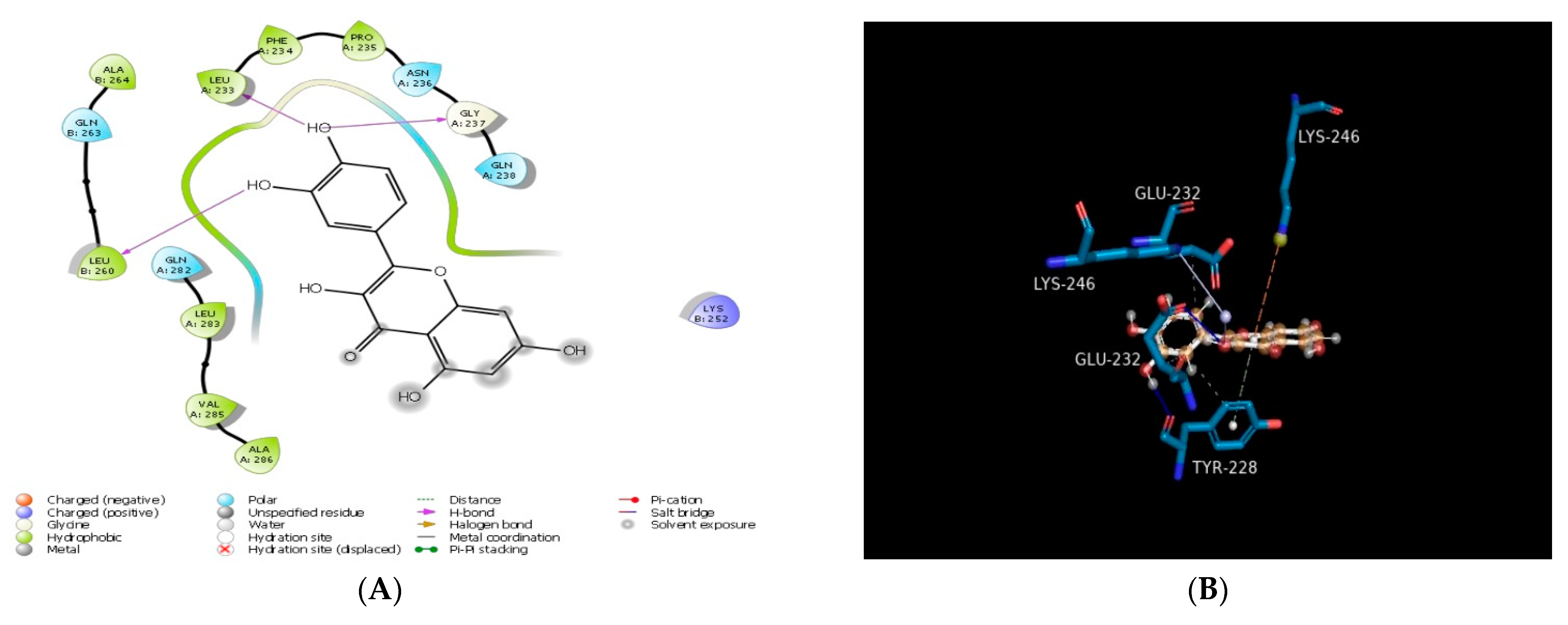
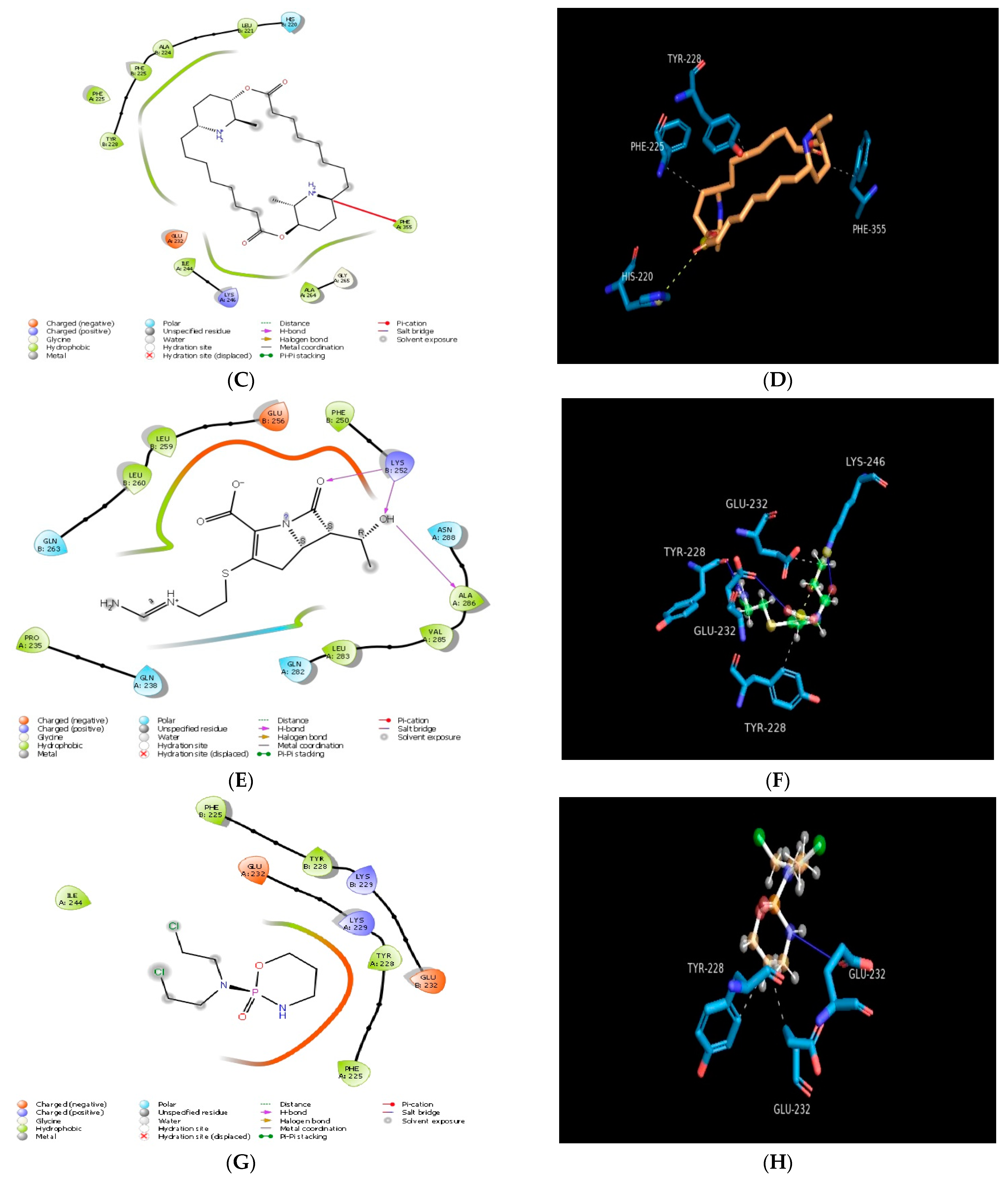
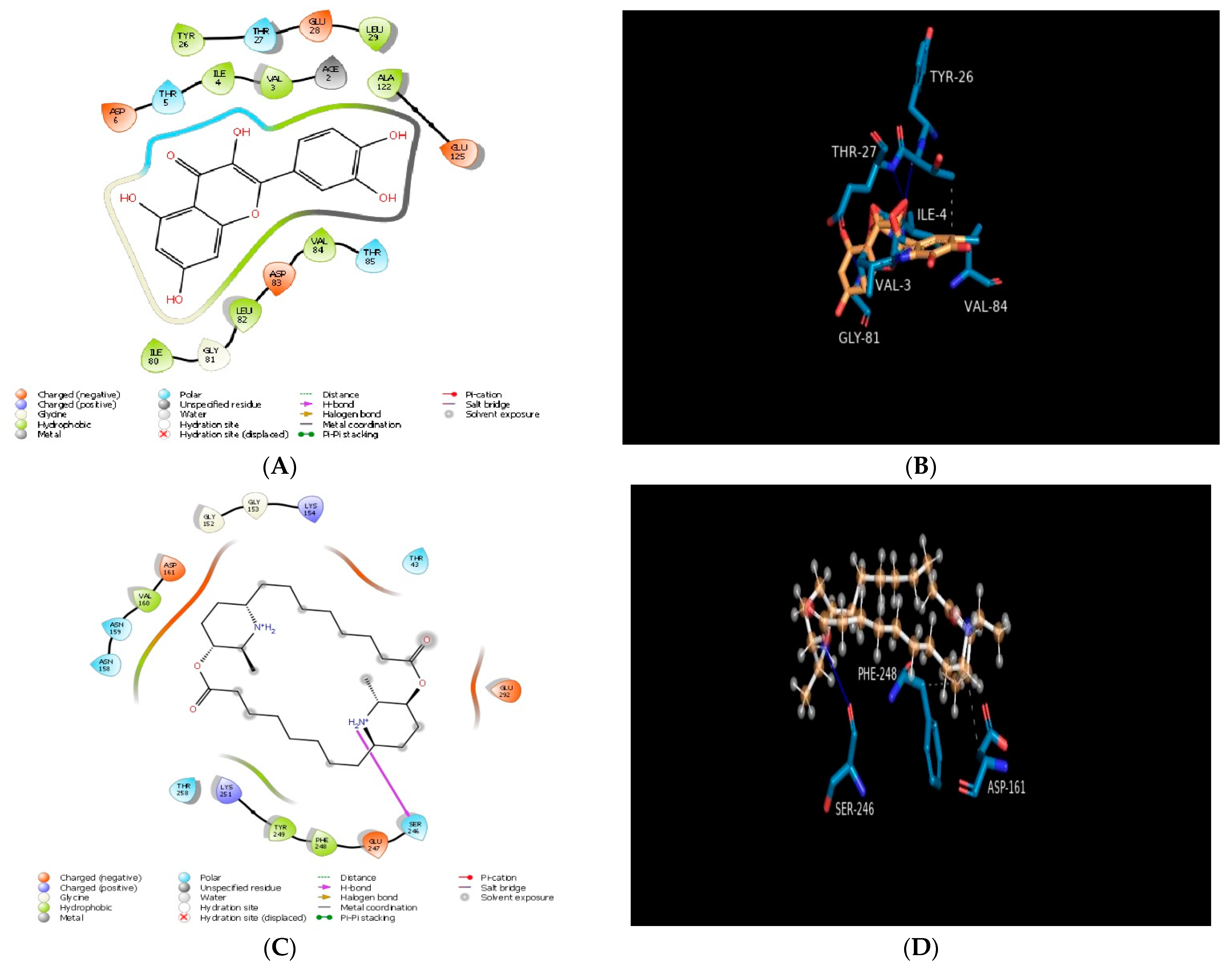

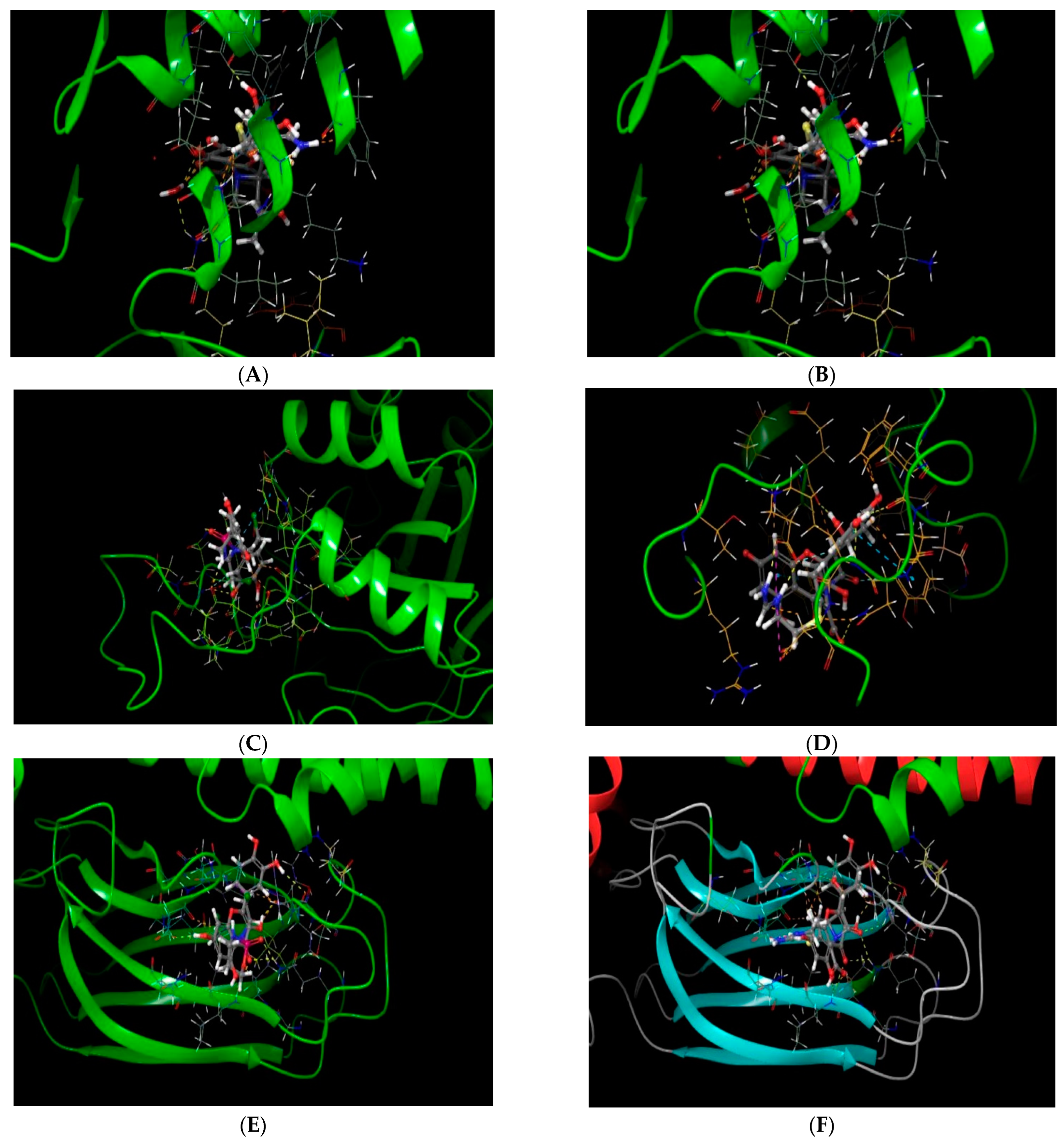
| PDB ID | Compound | Docking Score | No. of Hydrogen Bond | Hydrogen Bond-Forming Residue | Another Interacting Residue | PRIME-MMGBSA Binding Free Energy (Kcal/mol) |
|---|---|---|---|---|---|---|
| 1PW9 | Carpaine | −2.71 | n.f* | n.f* | TYR228, PHE225, ALA224, LEU221, HIS220, GLU232, ILE244, LYS246, ALA246, GLY265, PHE355 | −24.65 |
| Quercetin | −4.48 | 02 | TYR228 | GLU232, LYS229, PHE225, TYR228, VAL231 | −40.54 | |
| Imipenem | −4.20 | 03 | TYR228, LYS246 | LEU233, GLU232, LYS229, PHE225, TYR228, VAL231, ILE244 | −51.31 | |
| Cyclophosphamide | −4.35 | n.f* | n.f* | ILE244, GLU232, PHE225, TYR228, LYS229 | −11.03 | |
| 4PO2 | Carpaine | −3.44 | 02 | GLN473, ASN540 | THR405, ALA406, VAL409, THR411, ARG533, VAL536, SER537, ASN540, ALA541, SER544, GLN426, ILE427, PHE428, THR429, ASN548, GLY470, ARG469 | −41.41 |
| Quercetin | −6.04 | 05 | GLU404, LEU439, GLN435, THR429, THR430 | GLY408, GLY407, ALA406, THR405, GLU404, TYR431, PHE428, VAL438, GLN441 | −38.38 | |
| Imipenem | −6.64 | 05 | GLU404, ALA406, TYR431, GLN435, LEU439 | THR405, GLY407, GLY408, PHE428, THR429, THR430, VAL438, GLN441 | −45.18 | |
| Cyclophosphamide | −4.97 | 01 | GLN435 | VAL438, LEU439, ILE440, GLN441, LEU403, GLU404, THR405, ALA406, PHE428, THR429, THR430, TYR431 | −39.28 | |
| 4MKS | Carpaine | −4.36 | 01 | SER246 | ASN158, ASN159, VAL160, ASP161, GLY152, GLY153, LYS154, THR43, GLU292, GLU247, PHE248, TYR249, LYS251, THR258 | −32.87 |
| Quercetin | −5.86 | n.f* | n.f* | ILE4, VAL3, ALA122, TYR26, LEU29, ILE80, GLY81, LEU82, VAL84, ASP6, THR5, ACE2, GLU28, THR27, GLU125, THR85, ASP83 | −38.71 | |
| Imipenem | −5.34 | 03 | GLU268, GLU269, ASN250 | ARG263, THR266, TRP270, ASP291, LEU290, PRO289, ALA245, PHE248, TYR249, LYS251 | −36.96 | |
| Cyclophosphamide | −4.12 | 01 | GLU269 | GLU268, TRP270, LEU290, PRO289, ALA245, PHE248, TYR249, ASN250, LYS251, ASP252 | −20.46 |
| Phytoconstituents | Extracts of Carica papaya Leaves | ||
|---|---|---|---|
| Aqueous | Methanol | Ethanol | |
| Alkaloid | + | ++ | +++ |
| Flavonoids | ++ | + | +++ |
| Phenolic compound | − | + | − |
| Terpenoids | − | − | +++ |
| Saponins | − | + | ++ |
| Glycosides | + | − | + |
| Concentration (μg/mL) | Types of Extracts | |||
|---|---|---|---|---|
| Aqueous (%) | Methanol (%) | Ethanol (%) | Ascorbic Acid (%) | |
| 20 | 51.26 ± 1.47 | 70.43 ± 3.47 | 77.86 ± 3.08 | 7.23 ± 1.67 |
| 40 | 65.90 ± 1.41 | 75.10 ± 0.53 | 83.73 ± 5.28 | 15.40 ± 2.24 |
| 60 | 69.30 ± 1.26 | 80.66 ± 2.46 | 85.23 ± 3.93 | 37.46 ± 7.18 |
| 80 | 77.33 ± 1.31 | 85.23 ± 4.60 | 88.76 ± 1.35 | 56.16 ± 7.98 |
| 100 | 80.66 ± 0.09 | 89.00 ± 1.20 | 89.63 ± 1.73 | 60.76 ± 2.28 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Usmani, J.; Kausar, H.; Akbar, S.; Sartaj, A.; Mir, S.R.; Hassan, M.J.; Sharma, M.; Ahmad, R.; Rashid, S.; Ansari, M.N. Molecular Docking of Bacterial Protein Modulators and Pharmacotherapeutics of Carica papaya Leaves as a Promising Therapy for Sepsis: Synchronising In Silico and In Vitro Studies. Molecules 2023, 28, 574. https://doi.org/10.3390/molecules28020574
Usmani J, Kausar H, Akbar S, Sartaj A, Mir SR, Hassan MJ, Sharma M, Ahmad R, Rashid S, Ansari MN. Molecular Docking of Bacterial Protein Modulators and Pharmacotherapeutics of Carica papaya Leaves as a Promising Therapy for Sepsis: Synchronising In Silico and In Vitro Studies. Molecules. 2023; 28(2):574. https://doi.org/10.3390/molecules28020574
Chicago/Turabian StyleUsmani, Juveria, Hina Kausar, Saleem Akbar, Ali Sartaj, Showkat R. Mir, Mohammed Jaseem Hassan, Manju Sharma, Razi Ahmad, Summaya Rashid, and Mohd Nazam Ansari. 2023. "Molecular Docking of Bacterial Protein Modulators and Pharmacotherapeutics of Carica papaya Leaves as a Promising Therapy for Sepsis: Synchronising In Silico and In Vitro Studies" Molecules 28, no. 2: 574. https://doi.org/10.3390/molecules28020574
APA StyleUsmani, J., Kausar, H., Akbar, S., Sartaj, A., Mir, S. R., Hassan, M. J., Sharma, M., Ahmad, R., Rashid, S., & Ansari, M. N. (2023). Molecular Docking of Bacterial Protein Modulators and Pharmacotherapeutics of Carica papaya Leaves as a Promising Therapy for Sepsis: Synchronising In Silico and In Vitro Studies. Molecules, 28(2), 574. https://doi.org/10.3390/molecules28020574








